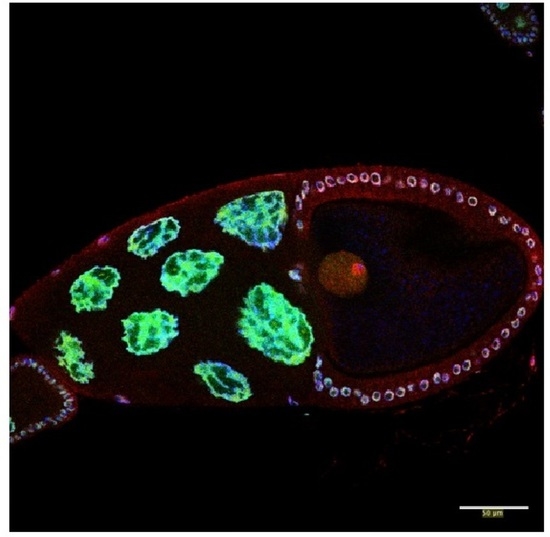
Roles for the RNA-Binding Protein Caper in Reproductive Output in Drosophila melanogaster
Abstract
1. Introduction
2. Materials and Methods
2.1. Fly Lines
2.2. Immunoblotting
2.3. Immunofluorescence
2.4. Fecundity Assay
2.5. Determining Fertilization and Embryonic Lethality
2.6. Larval and Pupal Lethality Assay
2.7. Mating Assay
2.8. Ovary Size Analysis
2.9. Statistical Analyses
3. Results
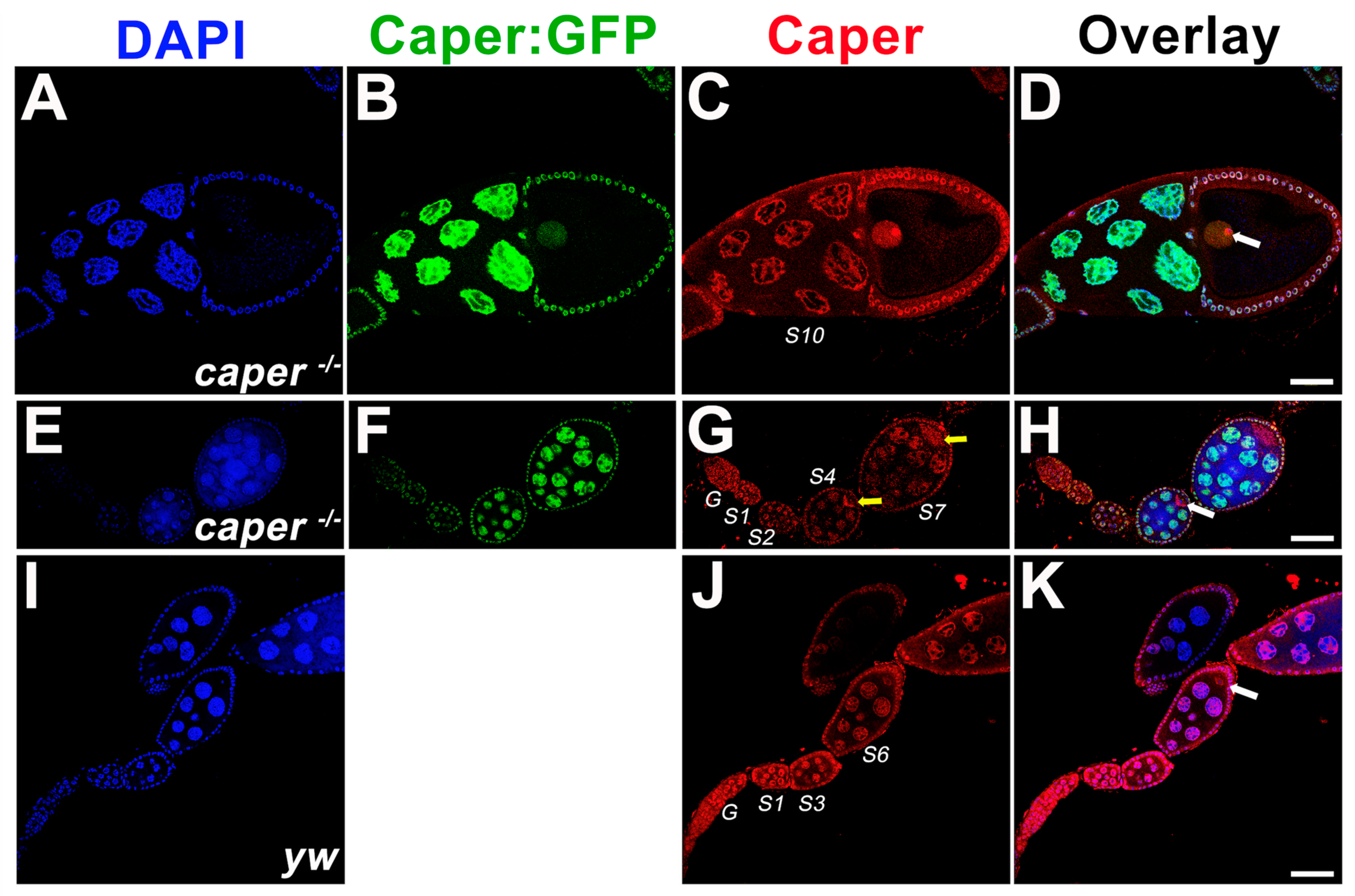
3.1. Caper Is Expressed during All Stages of Oogenesis
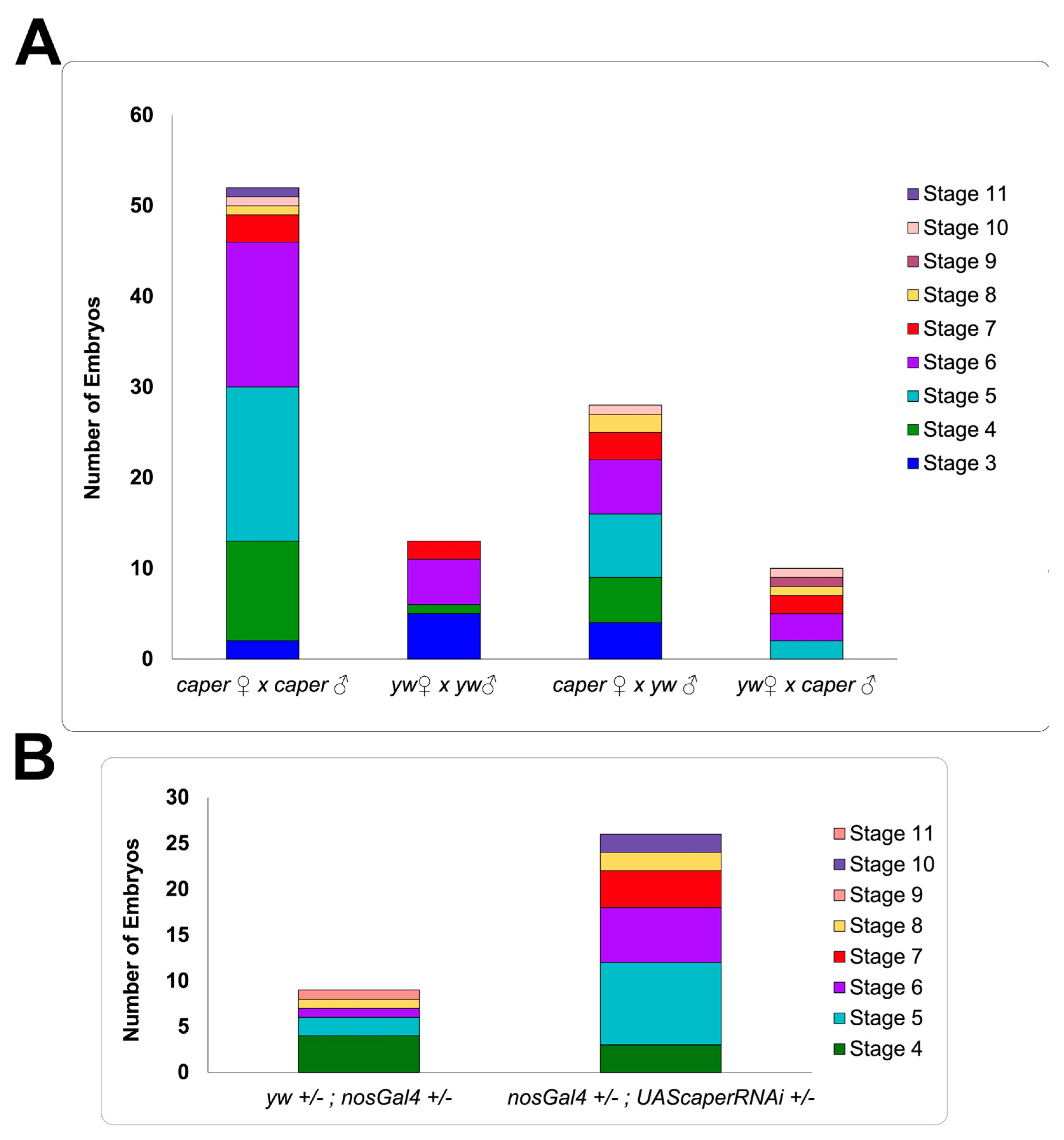
3.2. Caper Dysfunction Results in Lowered Reproductive Output for Females
3.3. Caper Dysfunction Results in Partial Embryonic and Larval Lethality
3.4. Caper Dysfunction Results in a Developmental Delay in Oogenesis
3.5. Reproductive Output of Caper−/−, but Not Caper Knockdown, Females May Be Impacted by Reduced Matings
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ameku, T.; Niwa, R. Mating-induced increase in germline stem cells via the neuroendocrine system in female Drosophila. PloS Genet. 2016, 12, e1006123. [Google Scholar] [CrossRef] [PubMed]
- Antonacci, S.; Forand, D.; Wolf, M.; Tyus, C.; Barney, J.; Kellogg, L.; Simon, M.A.; Kerr, G.; Wells, K.L.; Younes, S.; et al. Conserved RNA-binding proteins required for dendrite morphogenesis in Caenorhabditis elegans sensory neurons. G3: Genes Genomes Genet. 2015, 5, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Becalska, A.N.; Gavis, E.R. Lighting up mRNA localization in Drosophila oogenesis. Development 2009, 136, 2493–2503. [Google Scholar] [CrossRef] [PubMed]
- Brechbiel, J.L.; Gavis, E.R. Spatial regulation of nanos is required for its function in dendrite morphogenesis. Curr. Biol. 2008, 18, 745–750. [Google Scholar] [CrossRef]
- Brooks, M.E.; Kristensen, K.; van Benthem, K.J.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Maechler, M.; Bolker, B.M. glmmTMB Balances Speed and Flexibility Among Packages for Zero-inflated Generalized Linear Mixed Modeling. R J. 2017, 9, 378–400. [Google Scholar] [CrossRef]
- Buszczak, M.; Paterno, S.; Lighthouse, D.; Bachman, J.; Planck, J.; Owen, S.; Skora, A.D.; Nystul, T.G.; Ohlstein, B.; Allen, A.; et al. The carnegie protein trap library: A versatile tool for Drosophila developmental studies. Genetics 2007, 175, 1505–1531. [Google Scholar] [CrossRef]
- Campos-Ortega, J.A.; Hartenstein, V. The Embryonic Development of Drosophila melanogaster, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1985. [Google Scholar]
- Castagnetti, S.; Hentze, M.W.; Ephrussi, A.; Gebauer, F. Control of oskar mRNA translation by Bruno in a novel cell-free system from Drosophila ovaries. Development 2000, 127, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Ceron, J.; Rual, J.F.; Chandra, A.; Dupuy, D.; Vidal, M.; van den Heuvel, S. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev. Biol. 2007, 7, 30. [Google Scholar] [CrossRef]
- Christou-Kent, M.; Dhellemmes, M.; Lambert, E.; Ray, P.F.; Arnoult, C. Diversity of RNA-binding proteins modulating post-transcriptional regulation of protein expression in the maturing mammalian oocyte. Cells 2020, 9, 662. [Google Scholar] [CrossRef]
- Dowhan, D.H.; Hong, E.P.; Auboeuf, D.; Dennis, A.P.; Wilson, M.M.; Berget, S.M.; O’Malley, B.W. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERα and CAPERβ. Mol. Cell 2005, 17, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Driever, W.; Nüsslein-Volhard, C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 1988, 54, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Filardo, P.; Ephrussi, A. Bruno regulates gurken during Drosophila oogenesis. Mech. Dev. 2003, 120, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Gavis, E.R.; Lehmann, R. Localization of nanos RNA controls embryonic polarity. Cell 1992, 71, 301–313. [Google Scholar] [CrossRef]
- Gennarino, V.A.; Palmer, E.E.; McDonell, L.M.; Wang, L.; Adamski, C.J.; Koire, A.; See, L.; Chen, C.-A.; Schaaf, C.P.; Rosenfeld, J.A.; et al. A mild PUM1 mutation is associated with adult-onset ataxia, whereas haploinsufficiency causes developmental delay and seizures. Cell 2018, 172, 924–936. [Google Scholar] [CrossRef]
- González-Reyes, A.; Elliott, H.; St Johnston, D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature 1995, 375, 654–658. [Google Scholar] [CrossRef]
- Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package version 0.1.5. 2017. Available online: http://cran.nexr.com/web/packages/DHARMa/vignettes/DHARMa.html (accessed on 20 May 2022).
- Herndon, L.A.; Wolfner, M.F. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc. Natl. Acad. Sci. USA 1995, 92, 10114–10118. [Google Scholar] [CrossRef]
- Kadrmas, J.L.; Smith, M.A.; Pronovost, S.M.; Beckerle, M.C. Characterization of RACK1 function in Drosophila development. Dev. Dyn. 2007, 236, 2207–2215. [Google Scholar] [CrossRef]
- Kanke, M.; Jambor, H.; Reich, J.; Marches, B.; Gstir, R.; Ryu, Y.H.; Ephrussi, A.; Macdonald, P.M. oskar RNA plays multiple noncoding roles to support oogenesis and maintain integrity of the germline/soma distinction. Rna 2015, 21, 1096–1109. [Google Scholar] [CrossRef]
- Kato, Y.; Nakamura, A. Roles of cytoplasmic RNP granules in intracellular RNA localization and translational control in the drosophila oocyte. Dev. Growth Differ. 2012, 54, 19–31. [Google Scholar] [CrossRef]
- Kugler, J.M.; Lasko, P. Localization, anchoring and translational control of oskar, gurken, bicoid and nanos mRNA during Drosophila oogenesis. Fly 2009, 3, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Kwasnieski, J.C.; Orr-Weaver, T.L.; Bartel, D.P. Early genome activation in Drosophila is extensive with an initial tendency for aborted transcripts and retained introns. Genome Res. 2019, 29, 1188–1197. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R.; Nüsslein-Volhard, C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in drosophila. Cell 1986, 47, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Lenth, R.V. Tukey’s test: Estimated Marginal Means, aka Least-Squares Means. R Package version 1.6.0. 2021. Available online: https://CRAN.R-project.org/package=Tukey’stest (accessed on 20 May 2022).
- Lin, W.H.; Giachello, C.N.; Baines, R.A. Seizure control through genetic and pharmacological manipulation of Pumilio in Drosophila: A key component of neuronal homeostasis. Dis. Model. Mech. 2017, 10, 141–150. [Google Scholar] [PubMed]
- Massey, J.H.; Chung, D.; Siwanowicz, I.; Stern, D.L.; Wittkopp, P.J. The yellow gene influences Drosophila male mating success through sex comb melanization. eLife 2019, 8, e49388. [Google Scholar] [CrossRef]
- McDermott, S.M.; Meignin, C.; Rappsilber, J.; Davis, I. Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biol. Open 2012, 1, 488–497. [Google Scholar] [CrossRef]
- McGirr, J.A.; Johnson, L.M.; Kelly, W.; Markow, T.A.; Bono, J.M. Reproductive isolation among drosophila arizonae from geographically isolated regions of north america. Evol. Biol. 2017, 44, 82–90. [Google Scholar]
- Misra, M.; Edmund, H.; Ennis, D.; Schlueter, M.A.; Marot, J.E.; Tambasco, J.; Barlow, I.; Sigurbjornsdottir, S.; Mathew, R.; Vallés, A.M.; et al. A genome-wide screen for dendritically localized RNAs identifies genes required for dendrite morphogenesis. G3 Genes Genomes Genet. 2016, 6, 2397–2405. [Google Scholar] [CrossRef]
- Neelamraju, Y.; Hashemikhabir, S.; Janga, S.C. The human RBPome: From genes and proteins to human disease. J. Proteom. 2015, 127, 61–70. [Google Scholar] [CrossRef]
- Neuman-Silberberg, F.S.; Schupbach, T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development 1994, 120, 2457–2463. [Google Scholar] [CrossRef]
- Ni, J.-Q.; Zhou, R.; Czech, B.; Liu, L.-P.; Holderbaum, L.; Yang-Zhou, D.; Shim, H.-S.; Tao, R.; Handler, D.; Karpowicz, P.; et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 2011, 8, 405–407. [Google Scholar] [CrossRef] [PubMed]
- Olesnicky, E.C.; Bhogal, B.; Gavis, E.R. Combinatorial use of translational co-factors for cell type-specific regulation during neuronal morphogenesis in Drosophila. Dev. Biol. 2012, 365, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Olesnicky, E.C.; Bono, J.M.; Bell, L.; Schachtner, L.T.; Lybecker, M.C. The RNA-binding protein caper is required for sensory neuron development in Drosophila melanogaster. Dev. Dyn. 2017, 246, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Olesnicky, E.C.; Killian, D.J.; Garcia, E.; Morton, M.C.; Rathjen, A.R.; Sola, I.E.; Gavis, E.R. Extensive use of RNA-binding proteins in Drosophila sensory neuron dendrite morphogenesis. G3 Genes Genomes Genet. 2014, 4, 297–306. [Google Scholar]
- Olesnicky, E.C.; Killian, D.J. The cytoplasmic polyadenylation element binding protein (CPEB), Orb, is important for dendrite development and neuron fate specification in Drosophila melanogaster. Gene 2020, 738, 144473. [Google Scholar] [CrossRef]
- Olesnicky, E.C.; Wright, E.G. Drosophila as a model for assessing the function of RNA-binding proteins during neurogenesis and neurological disease. J. Dev. Biol. 2018, 6, 21. [Google Scholar] [CrossRef]
- Papasaikas, P.; Rao, A.; Huggins, P.; Valcarcel, J.; Lopez, A.J. Reconstruction of composite regulator-target splicing networks from high-throughput transcriptome data. BMC Genom. 2015, 16, S7. [Google Scholar] [CrossRef]
- Qin, X.; Ahn, S.; Speed, T.P.; Rubin, G.M. Global analyses of mRNA translational control during early drosophila embryogenesis. Genome Biol. 2007, 8, R63. [Google Scholar] [CrossRef]
- Rayburn, L.Y.; Gooding, H.C.; Choksi, S.P.; Maloney, D.; Kidd III, A.R.; Siekhaus, D.E.; Bender, M. Amontillado, the Drosophila homolog of the prohormone processing protease PC2, is required during embryogenesis and early larval development. Genetics 2003, 163, 227–237. [Google Scholar] [CrossRef]
- Ruhmann, H.; Wensing, K.U.; Neuhalfen, N.; Specker, J.; Fricke, C. Early reproductive success in Drosophila males is dependent on maturity of the accessory gland. Behav. Ecol. 2016, 27, 1859–1868. [Google Scholar]
- Samuelson, A.V.; Klimczak, R.R.; Thompson, D.B.; Carr, C.E.; Ruvkun, G. Identification of caenorhabditis elegans genes regulating longevity using enhanced RNAi-sensitive strains. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 489–497. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanford, J.R.; Bruzik, J.P. Regulation of SR protein localization during development. Proc. Natl. Acad. Sci. USA 2001, 98, 10184–10189. [Google Scholar] [CrossRef]
- Smith, C.M.; Hayamizu, T.F.; Finger, J.H.; Bello, S.M.; McCright, I.J.; Xu, J.; Baldarelli, R.M.; Beal, J.S.; Campbell, J.W.; Corbani, L.E.; et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019, 47, D774–D779. [Google Scholar] [CrossRef] [PubMed]
- Spendier, K.; Olesnicky, E.C.; Forand, D.; Wolf, M.; Killian, D.J. CPB-3 and CGH-1 localize to motile particles within dendrites in C. elegans PVD sensory neurons. BMC Res. Notes 2021, 14, 311. [Google Scholar] [CrossRef]
- Sturtevant, A.H. Experiments on sex recognition and the problem of sexual selection in Drosophila. J. Anim. Behav. 1915, 5, 351–366. [Google Scholar] [CrossRef]
- Thisse, B.; Thisse, C. Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission. 2004. Available online: http://zfin.org (accessed on 1 June 2022).
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, 315–320. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Titus, M.B.; Wright, E.G.; Bono, J.M.; Poliakon, A.K.; Goldstein, B.R.; Super, M.K.; Young, L.A.; Manaj, M.; Litchford, M.; Reist, N.E.; et al. The conserved alternative splicing factor caper regulates neuromuscular phenotypes during development and aging. Dev. Biol. 2021, 473, 15–32. [Google Scholar] [CrossRef]
- Vessey, J.P.; Schoderboeck, L.; Gingl, E.; Luzi, E.; Riefler, J.; Di Leva, F.; Karra, D.; Thomas, S.; Kiebler, M.A.; Macchi, P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc. Natl. Acad. Sci. USA 2010, 107, 3222–3227. [Google Scholar] [CrossRef]
- Wang, C.; Dickinson, L.K.; Lehmann, R. Genetics of nanos localization in Drosophila. Dev. Dyn. 1994, 199, 103–115. [Google Scholar] [CrossRef]
- Wessels, H.H.; Imami, K.; Baltz, A.G.; Kolinski, M.; Beldovskaya, A.; Selbach, M.; Small, S.; Ohler, U.; Landthaler, M. The mRNA-bound proteome of the early fly embryo. Genome Res. 2016, 26, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Qiu, S.; Robertson, R.M. The white gene controls copulation success in Drosophila melanogaster. Sci. Rep. 2017, 7, 7712. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Brechbiel, J.L.; Gavis, E.R. Dynein-dependent transport of nanos RNA in Drosophila sensory neurons requires Rumpelstiltskin and the germ plasm organizer Oskar. J. Neurosci. 2013, 33, 14791–14800. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Neumüller, R.A.; Buckner, M.; Ayers, K.; Li, H.; Hu, Y.; Yang-Zhou, D.; Pan, L.; Wang, X.; Kelley, C.; et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell 2014, 28, 459–473. [Google Scholar] [CrossRef]
- Ye, B.; Petritsch, C.; Clark, I.E.; Gavis, E.R.; Jan, L.Y.; Jan, Y.N. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr. Biol. 2004, 14, 314–321. [Google Scholar] [CrossRef]


| Males | Females | |||||||
|---|---|---|---|---|---|---|---|---|
| yw | caper−/− | RNAi Control | caper RNAi | yw | caper−/− | RNAi Control | caper RNAi | |
| Larval Death | 3(63) | 5(46) | 0(46) | 3(43) | 0(59) | 9(50) | 2(53) | 2(49) |
| Pupal Death | 6(60) | 8(41) | 2(46) | 1(40) | 8(59) | 3(41) | 4(51) | 1(47) |
| Female Genotype | ||||||
|---|---|---|---|---|---|---|
| yw | caper−/− | yw; nosGal4 | nosGal4; UAScaperRNAi | Totals | ||
| Male Genotype | yw | 6 | 0 | 2 | 4 | 270 |
| caper−/− | 60 | 39 | 38 | 29 | 270 | |
| Totals | 150 | 150 | 120 | 120 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tixtha, E.J.; Super, M.K.; Titus, M.B.; Bono, J.M.; Olesnicky, E.C. Roles for the RNA-Binding Protein Caper in Reproductive Output in Drosophila melanogaster. J. Dev. Biol. 2023, 11, 2. https://doi.org/10.3390/jdb11010002
Tixtha EJ, Super MK, Titus MB, Bono JM, Olesnicky EC. Roles for the RNA-Binding Protein Caper in Reproductive Output in Drosophila melanogaster. Journal of Developmental Biology. 2023; 11(1):2. https://doi.org/10.3390/jdb11010002
Chicago/Turabian StyleTixtha, Erika J., Meg K. Super, M. Brandon Titus, Jeremy M. Bono, and Eugenia C. Olesnicky. 2023. "Roles for the RNA-Binding Protein Caper in Reproductive Output in Drosophila melanogaster" Journal of Developmental Biology 11, no. 1: 2. https://doi.org/10.3390/jdb11010002
APA StyleTixtha, E. J., Super, M. K., Titus, M. B., Bono, J. M., & Olesnicky, E. C. (2023). Roles for the RNA-Binding Protein Caper in Reproductive Output in Drosophila melanogaster. Journal of Developmental Biology, 11(1), 2. https://doi.org/10.3390/jdb11010002