Paralogues of Mmp11 and Timp4 Interact during the Development of the Myotendinous Junction in the Zebrafish Embryo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Care and Spawning
2.2. Immunostaining and Microscopy
2.3. Construction of EMMA–Mmp11a Expression Vector
2.4. Microinjection and Heat Shock
2.5. Embryo Homogenization, SDS-PAGE, and Immunoblotting
2.6. Furin Inhibition Treatment
2.7. Yeast Two-Hybrid Assay
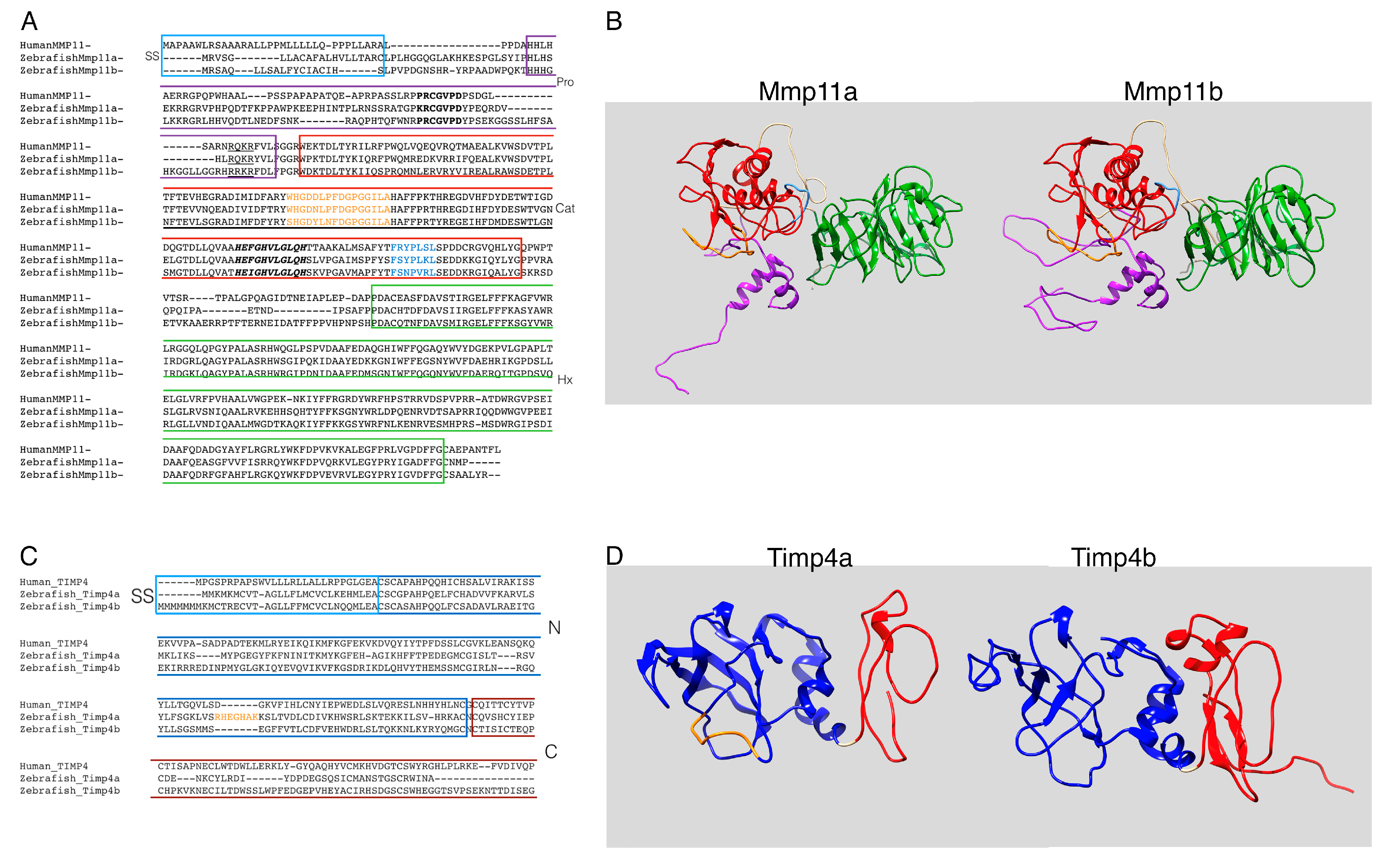
2.8. In silico Protein Modeling
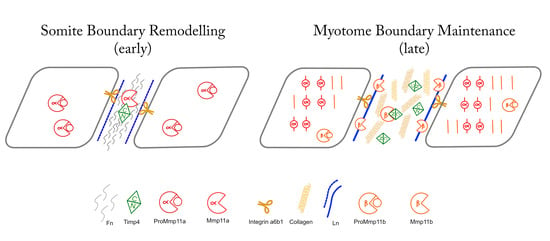
3. Results
3.1. Zebrafish Mmp11 and Timp4 Sequences Exhbit Both Conserved and Divergent Features
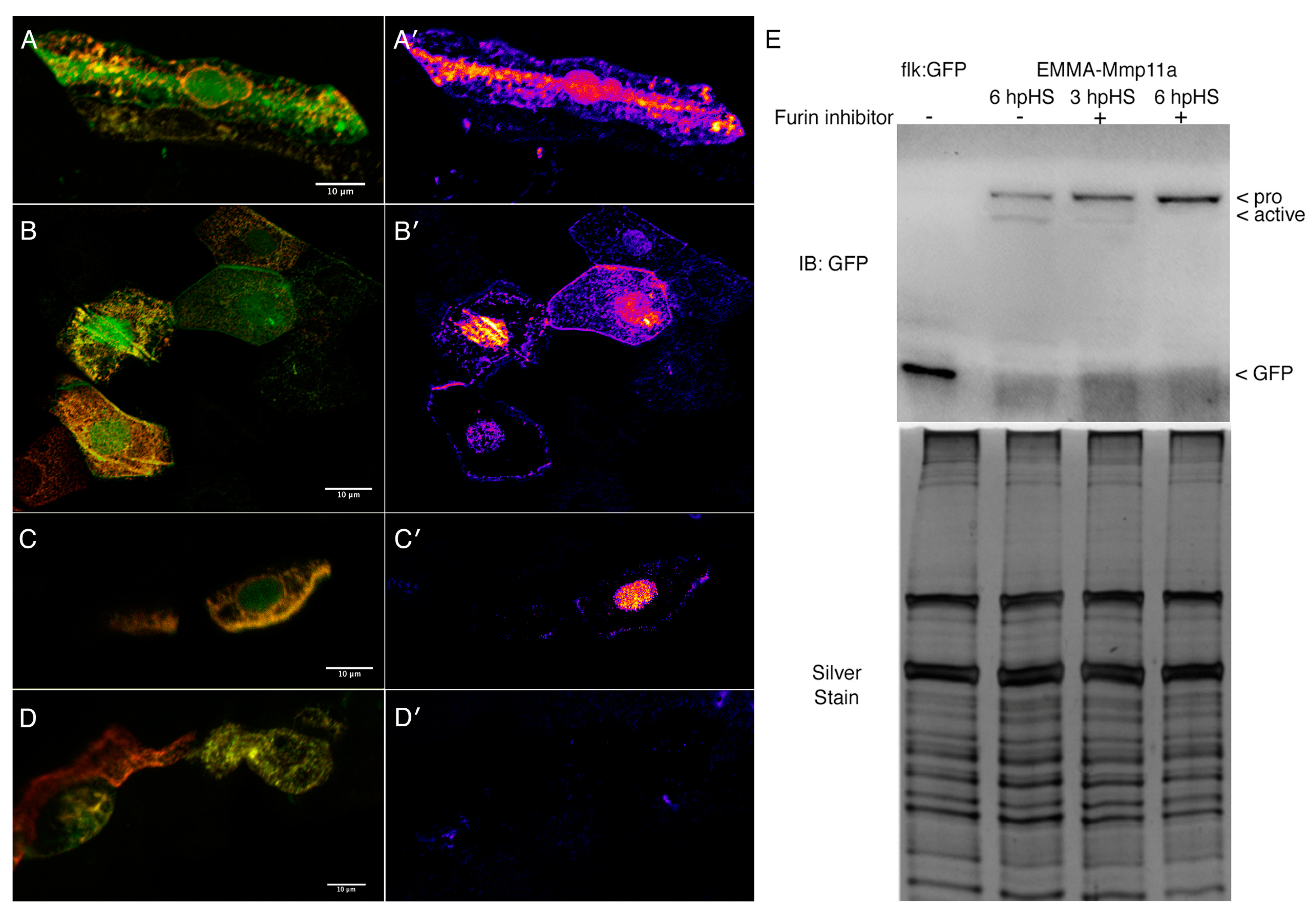
3.2. The Propeptide of Mmp11a is Removed by Furin Proprotein Convertase
3.3. Mmp11 and Timp4 Proteins Exhibit Dynamic Patterns of Accumulation in the MTJ and Muscle Cells
3.4. Mmp11 and Timp4 Proteins Interact in Specific Ways
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Goody, M.F.; Carter, E.V.; Kilroy, E.A.; Maves, L.; Henry, C.A. “Muscling” Throughout Life: Integrating Studies of Muscle Development, Homeostasis, and Disease in Zebrafish. Curr. Top. Dev. Biol. 2017, 124, 197–234. [Google Scholar] [CrossRef] [PubMed]
- Pourquié, O. The segmentation clock: Converting embryonic time into spatial pattern. Science (New York, NY) 2003, 301, 328–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yabe, T.; Takada, S. Molecular mechanism for cyclic generation of somites: Lessons from mice and zebrafish. Dev. Growth Differ. 2016, 58, 31–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMillen, P.; Chatti, V.; Jülich, D.; Holley, S.A. A Sawtooth Pattern of Cadherin 2 Stability Mechanically Regulates Somite Morphogenesis. Curr. Biol. 2016, 26, 542–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snow, C.; Henry, C. Dynamic formation of microenvironments at the myotendinous junction correlates with muscle fiber morphogenesis in zebrafish. Gene Expr. Patterns GEP 2008, 9, 37–42. [Google Scholar] [CrossRef] [Green Version]
- Charvet, B.; Malbouyres, M.; Pagnon-Minot, A.; Ruggiero, F.; Le Guellec, D. Development of the zebrafish myoseptum with emphasis on the myotendinous junction. Cell Tissue Res. 2011, 346, 439–449. [Google Scholar] [CrossRef]
- Wood, A.J.; Currie, P.D. Development Aspects of Zebrafish Myotendinous Junction: A Model System for Understanding Muscle Basement Membrane Formation and Failure. Curr. Pathobiol. Rep. 2017, 5, 1–9. [Google Scholar] [CrossRef]
- Stamenkovic, I. Extracellular matrix remodelling: The role of matrix metalloproteinases. J. Pathol. 2003, 200, 448–464. [Google Scholar] [CrossRef]
- Pedersen, M.E.; Vuong, T.T.; Rønning, S.B.; Kolset, S.O. Matrix metalloproteinases in fish biology and matrix turnover. Matrix Biol. 2015, 44–46, 86–93. [Google Scholar] [CrossRef]
- Apte, S.S.; Parks, W.C. Metalloproteinases: A parade of functions in matrix biology and an outlook for the future. Matrix Biol. J. Int. Soc. Matrix Biol. 2015. [Google Scholar] [CrossRef]
- Murphy, G. Matrix Metalloproteinases. In Encyclopedia of Cell Biology; Elsevier: Amsterdam, The Netherlands, 2016; pp. 621–629. [Google Scholar] [CrossRef]
- Jenkins, M.H.; Alrowaished, S.S.; Goody, M.F.; Crawford, B.D.; Henry, C.A. Laminin and Matrix metalloproteinase 11 regulate Fibronectin levels in the zebrafish myotendinous junction. Skeletal Muscle 2016, 6, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basset, P.; Bellocq, J.P.; Wolf, C.; Stoll, I.; Hutin, P.; Limacher, J.M.; Podhajcer, O.L.; Chenard, M.P.; Rio, M.C.; Chambon, P. A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature 1990, 348, 699–704. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, S.; Guo, J.; Zhou, L.; You, L.; Zhang, T.; Zhao, Y. Insights into the distinct roles of MMP-11 in tumor biology and future therapeutics (Review). Int. J. Oncol. 2016, 48, 1783–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damjanovski, S.; Ishizuya-Oka, A.; Shi, Y.B. Spatial and temporal regulation of collagenases-3, -4, and stromelysin -3 implicates distinct functions in apoptosis and tissue remodeling during frog metamorphosis. Cell Res. 1999, 9, 91–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizuya-Oka, A.; Li, Q.; Amano, T.; Damjanovski, S.; Ueda, S.; Shi, Y.B. Requirement for matrix metalloproteinase stromelysin-3 in cell migration and apoptosis during tissue remodeling in Xenopus laevis. J. Cell Biol. 2000, 150, 1177–1188. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Ishizuya-Oka, A.; Buchholz, D.R.; Amano, T.; Matsuda, H.; Shi, Y.-B. A causative role of stromelysin-3 in extracellular matrix remodeling and epithelial apoptosis during intestinal metamorphosis in Xenopus laevis. J. Biol. Chem. 2005, 280, 27856–27865. [Google Scholar] [CrossRef] [Green Version]
- Amano, T.; Fu, L.; Marshak, A.; Kwak, O.; Shi, Y.-B. Spatio-temporal regulation and cleavage by matrix metalloproteinase stromelysin-3 implicate a role for laminin receptor in intestinal remodeling during Xenopus laevis metamorphosis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2005, 234, 190–200. [Google Scholar] [CrossRef]
- Mathew, S.; Fu, L.; Hasebe, T.; Ishizuya-Oka, A.; Shi, Y.-B. Tissue-dependent induction of apoptosis by matrix metalloproteinase stromelysin-3 during amphibian metamorphosis. Birth Defects Res. Part C Embryo Today Rev. 2010, 90, 55–66. [Google Scholar] [CrossRef] [Green Version]
- Masson, R.; Lefebvre, O.; Noël Fahime, M.E.; Chenard, M.P.; Wendling, C. In vivo evidence that the stromelysin-3 metalloproteinase contributes in a paracrine manner to epithelial cell malignancy. J. Cell Biol. 1998, 140, 1535–1541. [Google Scholar] [CrossRef]
- Wyatt, R.A.; Keow, J.Y.; Harris, N.D.; Haché, C.A.; Li, D.H.; Crawford, B.D. The Zebrafish Embryo: A Powerful Model System for Investigating Matrix Remodeling. Zebrafish 2009, 6, 347–354. [Google Scholar] [CrossRef]
- Crawford, B.D.; Pilgrim, D.B. Ontogeny and regulation of matrix metalloproteinase activity in the zebrafish embryo by in vitro and in vivo zymography. Dev. Biol. 2005, 286, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Keow, J.Y.; Herrmann, K.M.; Crawford, B.D. Differential in vivo zymography: A method for observing matrix metalloproteinase activity in the zebrafish embryo. Matrix Biol. 2011, 30, 169–177. [Google Scholar] [CrossRef]
- Keow, J.Y.; Pond, E.D.; Cisar, J.S.; Cravatt, B.F.; Crawford, B.D. Activity-based labeling of matrix metalloproteinases in living vertebrate embryos. PLoS ONE 2012, 7, e43434. [Google Scholar] [CrossRef]
- Jeffrey, E.J.; Crawford, B.D. The epitope-mediated MMP activation assay: Detection and quantification of the activation of Mmp2 in vivo in the zebrafish embryo. Histochem. Cell Biol. 2018, 149, 277–286. [Google Scholar] [CrossRef]
- Maves, L. Recent advances using zebrafish animal models for muscle disease drug discovery. Expert Opin. Drug Discov. 2014, 9, 1033–1045. [Google Scholar] [CrossRef] [Green Version]
- Small, C.; Crawford, B. Matrix metalloproteinases in neural development: A phylogenetically diverse perspective. Neural Regen. Res. 2016, 11, 357–362. [Google Scholar] [CrossRef]
- Li, M.; Hromowyk, K.J.; Amacher, S.L.; Currie, P.D. Muscular dystrophy modeling in zebrafish. In The Zebrafish—Disease Models and Chemical Screens; Elsevier: Amsterdam, The Netherlands, 2017; Volume 138, pp. 347–380. [Google Scholar] [CrossRef]
- Luderman, L.N.; Unlu, G.; Knapik, E.W. Zebrafish Developmental Models of Skeletal Diseases. Curr. Top. Dev. Biol. 2017, 124, 81–124. [Google Scholar] [CrossRef]
- Coffey, E.; Pasquarella, M.; Goody, M.; Henry, C. Ethanol Exposure Causes Muscle Degeneration in Zebrafish. J. Dev. Biol. 2018, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Keow, J.Y.; Crawford, B.D. Investigating Matrix Metalloproteinase Regulation in its Biological Context; Detecting MMP Activity In Vivo. In Matrix Metalloproteinases; Oshiro, N., Miyagi, E., Eds.; Nova Science Publishers: New York, NY, USA, 2012; pp. 151–169. [Google Scholar]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. In Matrix Metalloproteinases and Tissue Remodeling in Health and Disease: Cardiovascular Remodeling, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2017; Volume 147, pp. 1–73. [Google Scholar] [CrossRef] [Green Version]
- Ra, H.-J.; Parks, W.C. Control of matrix metalloproteinase catalytic activity. Matrix Biol. J. Int. Soc. Matrix Biol. 2007, 26, 587–596. [Google Scholar] [CrossRef] [Green Version]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015. [Google Scholar] [CrossRef]
- Viappiani, S.; Nicolescu, A.C.; Holt, A.; Sawicki, G.; Crawford, B.D.; Leon, H.; van Mulligen, T.; Schulz, R. Activation and modulation of 72kDa matrix metalloproteinase-2 by peroxynitrite and glutathione. Biochem. Pharmacol. 2009, 77, 826–834. [Google Scholar] [CrossRef]
- Hadler-Olsen, E.; Fadnes, B.; Sylte, I.; Uhlin-Hansen, L.; Winberg, J.-O. Regulation of matrix metalloproteinase activity in health and disease. FEBS J. 2011, 278, 28–45. [Google Scholar] [CrossRef]
- Piperi, C.; GPapavassiliou, A. Molecular Mechanisms Regulating Matrix Metalloproteinases. Curr. Top. Med. Chem. 2012, 12, 1095–1112. [Google Scholar] [CrossRef]
- Pei, D.; Weiss, S.J. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature 1995, 375, 244–247. [Google Scholar] [CrossRef]
- Shi, Y.-B.; Fu, L.; Hasebe, T.; Ishizuya-Oka, A. Regulation of extracellular matrix remodeling and cell fate determination by matrix metalloproteinase stromelysin-3 during thyroid hormone-dependent post-embryonic development. Pharmacol. Ther. 2007, 116, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011, 12, 233. [Google Scholar] [CrossRef]
- Arpino, V.; Brock, M.; Gill, S.E. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 2015. [Google Scholar] [CrossRef]
- Jackson, H.W.; Defamie, V.; Waterhouse, P.; Khokha, R. TIMPs: Versatile extracellular regulators in cancer. Nat. Rev. Cancer 2016, 17, 1–16. [Google Scholar] [CrossRef]
- Doucet, A.; Overall, C.M. Protease proteomics: Revealing protease in vivo functions using systems biology approaches. Mol. Asp. Med. 2008, 29, 339–358. [Google Scholar] [CrossRef]
- Solis, N.; Overall, C.M. Identification of Protease Cleavage Sites and Substrates in Cancer by Carboxy-TAILS (C-TAILS). In Matrix Metalloproteinase Protocols; Clark, I.M., Ed.; Springer: New York, NY, USA, 2018; Volume 1731, pp. 15–28. [Google Scholar] [CrossRef]
- Cauwe, B.; Opdenakker, G. Intracellular substrate cleavage: A novel dimension in the biochemistry, biology and pathology of matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 2010, 45, 351–423. [Google Scholar] [CrossRef]
- DeCoux, A.; Lindsey, M.L.; Villarreal, F.; García, R.A.; Schulz, R. Myocardial matrix metalloproteinase-2: Inside out and upside down. J. Mol. Cell. Cardiol. 2014, 77, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Mustafa, A.; Yerzhan, A.; Merzhakupova, D.; Yerlan, P.; Orakov, A.N.; Wang, X.; Huang, Y.; Miao, L. Nuclear matrix metalloproteinases: Functions resemble the evolution from the intracellular to the extracellular compartment. Cell Death Discov. 2017, 3, 17036. [Google Scholar] [CrossRef] [Green Version]
- Jobin, P.G.; Butler, G.S.; Overall, C.M. New intracellular activities of matrix metalloproteinases shine in the moonlight. BBA Mol. Cell Res. 2017, 1864, 2043–2055. [Google Scholar] [CrossRef]
- Fallata, A.M.; Wyatt, R.A.; Levesque, J.M.; Dufour, A.; Overall, C.M.; Crawford, B.D. Intracellular localization in zebrafish muscle and conserved sequence features suggest roles for Gelatinase A in sarcomere maintenance. Biomedicines 2019, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Murphy, G.; Segain, J.P.; O’Shea, M.; Cockett, M.; Ioannou, C.; Lefebvre, O. The 28-kDa N-terminal domain of mouse stromelysin-3 has the general properties of a weak metalloproteinase. J. Biol. Chem. 1993, 268, 15435–15441. [Google Scholar]
- Noël Santavicca, M.; Stoll, I.; L’Hoir, C.; Staub, A.; Murphy, G. Identification of structural determinants controlling human and mouse stromelysin-3 proteolytic activities. J. Biol. Chem. 1995, 270, 22866–22872. [Google Scholar] [CrossRef] [Green Version]
- Matziari, M.; Dive, V.; Yiotakis, A. Matrix metalloproteinase 11 (MMP-11; stromelysin-3) and synthetic inhibitors. Med. Res. Rev. 1995, 27, 528–552. [Google Scholar] [CrossRef]
- Pan, W.; Arnone, M.; Kendall, M.; Grafstrom, R.H.; Seitz, S.P.; Wasserman, Z.R.; Albright, C.F. Identification of Peptide Substrates for Human MMP-11 (Stromelysin-3) Using Phage Display. J. Biol. Chem. 2003, 278, 27820–27827. [Google Scholar] [CrossRef] [Green Version]
- Mathew, S.; Fu, L.; Fiorentino, M.; Matsuda, H.; Das, B.; Shi, Y.-B. Differential regulation of cell type-specific apoptosis by stromelysin-3: A potential mechanism via the cleavage of the laminin receptor during tail resorption in Xenopus laevis. J. Biol. Chem. 2009, 284, 18545–18556. [Google Scholar] [CrossRef] [Green Version]
- Amano, T.; Kwak, O.; Fu, L.; Marshak, A.; Shi, Y.-B. The matrix metalloproteinase stromelysin-3 cleaves laminin receptor at two distinct sites between the transmembrane domain and laminin binding sequence within the extracellular domain. Cell Res. 2005, 15, 150–159. [Google Scholar] [CrossRef]
- Fiorentino, M.; Fu, L.; Shi, Y.-B. Mutational analysis of the cleavage of the cancer-associated laminin receptor by stromelysin-3 reveals the contribution of flanking sequences to site recognition and cleavage efficiency. Int. J. Mol. Med. 2009, 23, 389–397. [Google Scholar]
- Kwan, J.A.; Schulze, C.J.; Wang, W.; Leon, H.; Sariahmetoglu, M.; Sung, M.; Sawicka, J.; Sims, D.E.; Sawicki, G.; Schulz, R. Matrix metalloproteinase-2 (MMP-2) is present in the nucleus of cardiac myocytes and is capable of cleaving poly (ADP-ribose) polymerase (PARP) in vitro. FASEB J. 2004, 18, 690–692. [Google Scholar] [CrossRef]
- Sariahmetoglu, M.; Crawford, B.D.; Leon, H.; Sawicka, J.; Li, L.; Ballermann, B.J.; Holmes, C.; Berthiaume, L.G.; Holt, A.; Sawicki, G. Regulation of matrix metalloproteinase-2 (MMP-2) activity by phosphorylation. FASEB J. 2007, 21, 2486–2495. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.A.M.; Chow, A.K.; Kandasamy, A.D.; Fan, X.; West, L.J.; Crawford, B.D.; Simmen, T.; Schulz, R. Mechanisms of cytosolic targeting of matrix metalloproteinase-2. J. Cell. Physiol. 2012, 227, 3397–3404. [Google Scholar] [CrossRef]
- Meyer, B.S.; Rademann, J. Extra- and intracellular imaging of human matrix metalloprotease 11 (hMMP-11) with a cell-penetrating FRET substrate. J. Biol. Chem. 2012, 287, 37857–37867. [Google Scholar] [CrossRef] [Green Version]
- Hadler-Olsen, E.; Solli, A.I.; Hafstad, A.; Winberg, J.-O.; Uhlin-Hansen, L. Intracellular MMP-2 activity in skeletal muscle is associated with type II fibers. J. Cell. Physiol. 2015, 230, 160–169. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Punj, V.; Kim, J.-M.; Lee, S.; Ulmer, T.S.; Lu, W. MMP-9 facilitates selective proteolysis of the histone H3 tail at genes necessary for proficient osteoclastogenesis. Genes Dev. 2016, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Maskos, K.; Bode, W. Structural basis of matrix metalloproteinases and tissue inhibitors of metalloproteinases. Mol. Biotechnol. 2003, 25, 241–266. [Google Scholar] [CrossRef]
- Overall, C.M.; Butler, G.S. Protease yoga: Extreme flexibility of a matrix metalloproteinase. Structure (London, England: 1993) 2007, 15, 1159–1161. [Google Scholar] [CrossRef] [Green Version]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. BBA Mol. Cell Res. 2010, 1803, 55–71. [Google Scholar] [CrossRef] [Green Version]
- Pei, D.; Majmudar, G.; Weiss, S.J. Hydrolytic inactivation of a breast carcinoma cell-derived serpin by human stromelysin-3. J. Biol. Chem. 1994, 269, 25849–25855. [Google Scholar]
- Bigg, H.F.; Shi, Y.E.; Liu, Y.E.; Steffensen, B.; Overall, C.M. Specific, high affinity binding of tissue inhibitor of metalloproteinases-4 (TIMP-4) to the COOH-terminal hemopexin-like domain of human gelatinase A. TIMP-4 binds progelatinase A and the COOH-terminal domain in a similar manner to TIMP-2. J. Biol. Chem. 1997, 272, 15496–15500. [Google Scholar] [CrossRef] [Green Version]
- Bigg, H.F.; Morrison, C.J.; Butler, G.S.; Bogoyevitch, M.A.; Wang, Z.; Soloway, P.D.; Overall, C.M. Tissue inhibitor of metalloproteinases-4 inhibits but does not support the activation of gelatinase A via efficient inhibition of membrane type 1-matrix metalloproteinase. Cancer Res. 2001, 61, 3610–3618. [Google Scholar]
- Taylor, J.S.; Braasch, I.; Frickey, T.; Meyer, A.; Van de Peer, Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003, 13, 382–390. [Google Scholar] [CrossRef] [Green Version]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Klein-Szanto, A.J.; Bassi, D.E. Proprotein convertase inhibition: Paralyzing the cell’s master switches. Biochem. Pharmacol. 2017, 140, 8–15. [Google Scholar] [CrossRef]
- Wang, W.; Schulze, C.J.; Suarez-Pinzon, W.L.; Dyck, J.R.B.; Sawicki, G.; Schulz, R. Intracellular action of matrix metalloproteinase-2 accounts for acute myocardial ischemia and reperfusion injury. Circulation 2002, 106, 1543–1549. [Google Scholar] [CrossRef] [Green Version]
- Yeghiazaryan, M.; Zybura-Broda, K.; Cabaj, A.; Włodarczyk, J.; Sławińska, U.; Rylski, M.; Wilczynski, G.M. Fine-structural distribution of MMP-2 and MMP-9 activities in the rat skeletal muscle upon training: A study by high-resolution in situ zymography. Histochem. Cell Biol. 2012, 138, 75–87. [Google Scholar] [CrossRef] [Green Version]
- Marchant, D.J.; Bellac, C.L.; Moraes, T.J.; Wadsworth, S.J.; Dufour, A.; Butler, G.S. A new transcriptional role for matrix metalloproteinase-12 in antiviral immunity. Nat. Med. 2014, 20, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Baghirova, S.; Hughes, B.G.; Poirier, M.; Kondo, M.Y.; Schulz, R. Nuclear matrix metalloproteinase-2 in the cardiomyocyte and the ischemic-reperfused heart. J. Mol. Cell. Cardiol. 2016, 94, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Lamb, G.D.; Murphy, R.M. Distribution and activation of matrix metalloproteinase-2 in skeletal muscle fibers. Am. J. Physiol. Cell Physiol. 2019, 122, C613–C625. [Google Scholar] [CrossRef]
- Luo, D.; Mari, B.; Stoll, I.; Anglard, P. Alternative splicing and promoter usage generates an intracellular stromelysin 3 isoform directly translated as an active matrix metalloproteinase. J. Biol. Chem. 2002, 277, 25527–25536. [Google Scholar] [CrossRef] [Green Version]
- Kosugi, S.; Hasebe, M.; Entani, T.; Takayama, S.; Tomita, M.; Yanagawa, H. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 2008, 15, 940–949. [Google Scholar] [CrossRef] [Green Version]
- Sawicki, G. Intracellular Regulation of Matrix Metalloproteinase-2 Activity: New Strategies in Treatment and Protection of Heart Subjected to Oxidative Stress. Scientifica 2013, 2013, 130451. [Google Scholar] [CrossRef] [Green Version]
- Kaur, N.; Kumar, B.V.S.; Mahajan, K.; Singh, S. Expression and characterization of tissue inhibitor of metalloproteinase 4 from complex canine mammary carcinomas. J. Immunoass. Immunochem. 2016, 37, 515–526. [Google Scholar] [CrossRef]
- Iyer, R.P.; Patterson, N.L.; Fields, G.B.; Lindsey, M.L. The history of matrix metalloproteinases: Milestones, myths, and misperceptions. AJP Heart Circ. Physiol. 2012, 303, H919–H930. [Google Scholar] [CrossRef] [Green Version]
- Rohani, M.G.; Parks, W.C. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015. [Google Scholar] [CrossRef]
- Linde-Medina, M.; Marcucio, R. Living tissues are more than cell clusters: The extracellular matrix as a driving force in morphogenesis. Prog. Biophys. Mol. Biol. 2018. [Google Scholar] [CrossRef]
- Dzamba, B.J.; DeSimone, D.W. Extracellular Matrix (ECM) and the Sculpting of Embryonic Tissues. Curr. Top. Dev. Biol. 2018, 130, 245–274. [Google Scholar] [CrossRef]





| Timp4a-N | Timp4a-C | Timp4b-N | Timp4b-C | |
|---|---|---|---|---|
| Mmp11a-Cat | -- | - | -- | -- |
| Mmp11a-Hx | - | -- | -- | ++ |
| Mmp11b-Cat | + | -- | ++ | -- |
| Mmp11b-Hx | - | - | - | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matchett, E.F.; Wang, S.; Crawford, B.D. Paralogues of Mmp11 and Timp4 Interact during the Development of the Myotendinous Junction in the Zebrafish Embryo. J. Dev. Biol. 2019, 7, 22. https://doi.org/10.3390/jdb7040022
Matchett EF, Wang S, Crawford BD. Paralogues of Mmp11 and Timp4 Interact during the Development of the Myotendinous Junction in the Zebrafish Embryo. Journal of Developmental Biology. 2019; 7(4):22. https://doi.org/10.3390/jdb7040022
Chicago/Turabian StyleMatchett, Emma F., Shuaijin Wang, and Bryan D. Crawford. 2019. "Paralogues of Mmp11 and Timp4 Interact during the Development of the Myotendinous Junction in the Zebrafish Embryo" Journal of Developmental Biology 7, no. 4: 22. https://doi.org/10.3390/jdb7040022
APA StyleMatchett, E. F., Wang, S., & Crawford, B. D. (2019). Paralogues of Mmp11 and Timp4 Interact during the Development of the Myotendinous Junction in the Zebrafish Embryo. Journal of Developmental Biology, 7(4), 22. https://doi.org/10.3390/jdb7040022