C. elegans Apical Extracellular Matrices Shape Epithelia
Abstract
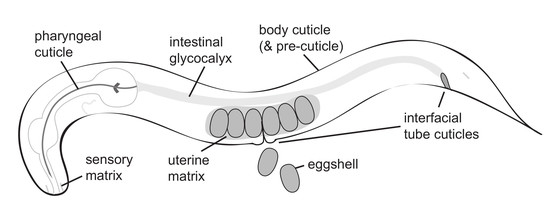
:1. Introduction
1.1. Apical Extracellular Matrix
1.2. Caenorhabditis elegans as a Model Organism for Studying Apical ECMs
2. The Eggshell
2.1. Building the Eggshell
2.2. The Eggshell Shapes Early Development
3. Pre-Cuticular aECMs
3.1. The Epidermal Sheath Elongates the Embryo
3.2. A Luminal Precuticle Shapes the Narrow Excretory Duct and Pore Lumens
3.3. Pre-Cuticular and Sensory aECMs Anchor the Pharynx and Sensory Organs to the Epidermis
3.4. The Vulva Lumen Is Shaped by a Multi-Layered Pre-Cuticular aECM
3.5. The Pre-Cuticle Patterns the Cuticle
4. Collagen-Based Cuticles and the Molt Cycle
4.1. Epidermal Cuticle Structure and Function
4.2. Alae and Annuli
4.3. Epidermal Cuticles Maintain Body Length and Girth
4.4. Cuticles of Interfacial Tubes
4.5. Regulation of the Molt Process
4.6. The Cuticle Changes during Aging
5. Chitin-Based Pharyngeal Cuticle
6. aECMs of Internal Epithelia
6.1. The Gut
6.2. The Uterus
6.3. The Excretory Canal Cell
7. Outstanding Questions Regarding aECM
7.1. How Are aECM Components Trafficked to Apical Cell Surfaces?
7.2. How Are aECMs Anchored to Cell Surfaces?
7.3. How Are aECMs Assembled and Disassembled?
7.4. What Is the Contribution of Individual aECM Components in Shaping Cell Surfaces?
Funding
Acknowledgments
Conflicts of Interest
References
- Luschnig, S.; Uv, A. Luminal matrices: An inside view on organ morphogenesis. Exp. Cell Res. 2014, 321, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Gil, J. Structure of pulmonary surfactant membranes and films: The role of proteins and lipid-protein interactions. Biochim. Biophys. Acta 2008, 1778, 1676–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarbell, J.M.; Cancel, L.M. The glycocalyx and its significance in human medicine. J. Intern. Med. 2016, 280, 97–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walma, D.A.C.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147. [Google Scholar] [CrossRef] [PubMed]
- Jayadev, R.; Sherwood, D.R. Basement membranes. Curr. Biol. 2017, 27, R207–R211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhard, W. Lung surfactant: Function and composition in the context of development and respiratory physiology. Ann. Anat. 2016, 208, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Gaudette, S.; Hughes, D.; Boller, M. The endothelial glycocalyx: Structure and function in health and critical illness. J. Vet. Emerg. Crit. Care (San Antonio) 2020. [Google Scholar] [CrossRef]
- Johansson, M.E.; Ambort, D.; Pelaseyed, T.; Schutte, A.; Gustafsson, J.K.; Ermund, A.; Hansson, G.C. Composition and functional role of the mucus layers in the intestine. Cell. Mol. Life Sci. 2011, 68, 3635–3641. [Google Scholar] [CrossRef]
- Bhattacharya, N.; Sato, W.J.; Kelly, A.; Ganguli-Indra, G.; Indra, A.K. Epidermal Lipids: Key Mediators of Atopic Dermatitis Pathogenesis. Trends Mol. Med. 2019, 25, 551–562. [Google Scholar] [CrossRef]
- Dartt, D.A. Tear lipocalin: Structure and function. Ocul. Surf. 2011, 9, 126–138. [Google Scholar] [CrossRef] [Green Version]
- Devine, W.P.; Lubarsky, B.; Shaw, K.; Luschnig, S.; Messina, L.; Krasnow, M.A. Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc. Natl. Acad. Sci. USA 2005, 102, 17014–17019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, I.; Chanut-Delalande, H.; Ferrer, P.; Latapie, Y.; Waltzer, L.; Affolter, M.; Plaza, S. Zona pellucida domain proteins remodel the apical compartment for localized cell shape changes. Dev. Cell 2010, 18, 64–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaza, S.; Chanut-Delalande, H.; Fernandes, I.; Wassarman, P.M.; Payre, F. From A to Z: Apical structures and zona pellucida-domain proteins. Trends Cell Biol. 2010, 20, 524–532. [Google Scholar] [CrossRef]
- Sapio, M.R.; Hilliard, M.A.; Cermola, M.; Favre, R.; Bazzicalupo, P. The Zona Pellucida domain containing proteins, CUT-1, CUT-3 and CUT-5, play essential roles in the development of the larval alae in Caenorhabditis elegans. Dev. Biol. 2005, 282, 231–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chappell, D.; Jacob, M.; Paul, O.; Rehm, M.; Welsch, U.; Stoeckelhuber, M.; Becker, B.F. The glycocalyx of the human umbilical vein endothelial cell: An impressive structure ex vivo but not in culture. Circ. Res. 2009, 104, 1313–1317. [Google Scholar] [CrossRef]
- Corsi, A.K.; Wightman, B.; Chalfie, M. A Transparent window into biology: A primer on Caenorhabditis elegans. WormBook 2015, 1–31. [Google Scholar] [CrossRef]
- Page, A.P.; Johnstone, I.L. The cuticle. WormBook 2007, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lazetic, V.; Fay, D.S. Molting in C. elegans. Worm 2017, 6, e1330246. [Google Scholar] [CrossRef] [Green Version]
- Costa, M.; Draper, B.W.; Priess, J.R. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev. Biol. 1997, 184, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Gill, H.K.; Cohen, J.D.; Ayala-Figueroa, J.; Forman-Rubinsky, R.; Poggioli, C.; Bickard, K.; Sundaram, M.V. Integrity of Narrow Epithelial Tubes in the C. elegans Excretory System Requires a Transient Luminal Matrix. PLoS Genet. 2016, 12, e1006205. [Google Scholar] [CrossRef]
- Mancuso, V.P.; Parry, J.M.; Storer, L.; Poggioli, C.; Nguyen, K.C.; Hall, D.H.; Sundaram, M.V. Extracellular leucine-rich repeat proteins are required to organize the apical extracellular matrix and maintain epithelial junction integrity in C. elegans. Development 2012, 139, 979–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priess, J.R.; Hirsh, D.I. Caenorhabditis elegans morphogenesis: The role of the cytoskeleton in elongation of the embryo. Dev. Biol. 1986, 117, 156–173. [Google Scholar] [CrossRef]
- Olson, S.K.; Bishop, J.R.; Yates, J.R.; Oegema, K.; Esko, J.D. Identification of novel chondroitin proteoglycans in Caenorhabditis elegans: Embryonic cell division depends on CPG-1 and CPG-2. J. Cell Biol. 2006, 173, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Stein, K.K.; Golden, A. The C. elegans eggshell. WormBook 2018, 2018, 1–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Foster, J.M.; Nelson, L.S.; Ma, D.; Carlow, C.K. The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Dev. Biol. 2005, 285, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Albertson, D.G.; Thomson, J.N. The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. B Biol. Sci. 1976, 275, 299–325. [Google Scholar] [CrossRef]
- Mango, S.E. The C. elegans pharynx: A model for organogenesis. WormBook 2007, 1–26. [Google Scholar] [CrossRef]
- Altun, Z.F.; Herndon, L.A.; Wolkow, C.A.; Crocker, C.; Lints, R.; Hall, D.H. (2002–2020). WormAtlas. Available online: http://www.wormatlas.org (accessed on 3 July 2020).
- Zimmerman, S.M.; Hinkson, I.V.; Elias, J.E.; Kim, S.K. Reproductive Aging Drives Protein Accumulation in the Uterus and Limits Lifespan in C. elegans. PLoS Genet. 2015, 11, e1005725. [Google Scholar] [CrossRef]
- Gonzalez, D.P.; Lamb, H.V.; Partida, D.; Wilson, Z.T.; Harrison, M.C.; Prieto, J.A.; Olson, S.K. CBD-1 organizes two independent complexes required for eggshell vitelline layer formation and egg activation in C. elegans. Dev. Biol. 2018, 442, 288–300. [Google Scholar] [CrossRef]
- Olson, S.K.; Greenan, G.; Desai, A.; Muller-Reichert, T.; Oegema, K. Hierarchical assembly of the eggshell and permeability barrier in C. elegans. J. Cell Biol. 2012, 198, 731–748. [Google Scholar] [CrossRef] [Green Version]
- Johnston, W.L.; Krizus, A.; Dennis, J.W. The eggshell is required for meiotic fidelity, polar-body extrusion and polarization of the C. elegans embryo. BMC Biol. 2006, 4, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starich, T.A.; Greenstein, D. Gap junctions deliver malonyl-CoA from soma to germline to support embryogenesis in Caenorhabditis elegans. Elife 2020, 9, e58619. [Google Scholar] [CrossRef] [PubMed]
- Rappleye, C.A.; Paredez, A.R.; Smith, C.W.; McDonald, K.L.; Aroian, R.V. The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode Caenorhabditis elegans. Genes Dev. 1999, 13, 2838–2851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Huang, L.-J.; Chen, S.-W.; Nebenfuehr, B.; Wysolmerski, B.; Wu, J.-C.; Wang, C.-W. Loss of the seipin gene perturbs eggshell formation in C. elegans. Development 2020. [Google Scholar] [CrossRef]
- Litscher, E.S.; Wassarman, P.M. Evolution, structure, and synthesis of vertebrate egg-coat proteins. Trends Dev. Biol. 2014, 8, 65–76. [Google Scholar] [PubMed]
- Krenger, R.; Burri, J.T.; Lehnert, T.; Nelson, B.J.; Gijs, M.A.M. Force microscopy of the Caenorhabditis elegans embryonic eggshell. Microsyst. Nanoeng. 2020, 6, 29. [Google Scholar] [CrossRef]
- Edgar, L.G.; McGhee, J.D. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 1988, 53, 589–599. [Google Scholar] [CrossRef]
- Johnston, W.L.; Dennis, J.W. The eggshell in the C. elegans oocyte-to-embryo transition. Genesis 2012, 50, 333–349. [Google Scholar] [CrossRef]
- Parry, J.M.; Velarde, N.V.; Lefkovith, A.J.; Zegarek, M.H.; Hang, J.S.; Ohm, J.; Singson, A. EGG-4 and EGG-5 Link Events of the Oocyte-to-Embryo Transition with Meiotic Progression in C. elegans. Curr. Biol. 2009, 19, 1752–1757. [Google Scholar] [CrossRef] [Green Version]
- Rappleye, C.A.; Tagawa, A.; Le Bot, N.; Ahringer, J.; Aroian, R.V. Involvement of fatty acid pathways and cortical interaction of the pronuclear complex in Caenorhabditis elegans embryonic polarity. BMC Dev. Biol. 2003, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Rappleye, C.A.; Tagawa, A.; Lyczak, R.; Bowerman, B.; Aroian, R.V. The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev. Cell 2002, 2, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Laufer, J.S.; Bazzicalupo, P.; Wood, W.B. Segregation of developmental potential in early embryos of Caenorhabditis elegans. Cell 1980, 19, 569–577. [Google Scholar] [CrossRef]
- Schierenberg, E. Reversal of cellular polarity and early cell-cell interaction in the embryos of Caenorhabditis elegans. Dev. Biol. 1987, 122, 452–463. [Google Scholar] [CrossRef]
- Wood, W.B.; Bergmann, D.; Florance, A. Maternal effect of low temperature on handedness determination in C. elegans embryos. Dev. Genet. 1996, 19, 222–230. [Google Scholar] [CrossRef]
- Schierenberg, E.; Junkersdorf, B. The role of eggshell and underlying vitelline membrane for normal pattern formation in the early C. elegans embryo. Roux Arch. Dev. Biol. 1992, 202, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Vuong-Brender, T.T.K.; Suman, S.K.; Labouesse, M. The apical ECM preserves embryonic integrity and distributes mechanical stress during morphogenesis. Development 2017, 144, 4336–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J.D.; Sparacio, A.P.; Belfi, A.C.; Forman-Rubinsky, R.; Hall, D.H.; Maul-Newby, H.; Sundaram, M.V. A multi-layered and dynamic apical extracellular matrix shapes the vulva lumen in Caenorhabditis elegans. Elife 2020, 9, e57874. [Google Scholar] [CrossRef] [PubMed]
- Heiman, M.G.; Shaham, S. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 2009, 137, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Kelley, M.; Yochem, J.; Krieg, M.; Calixto, A.; Heiman, M.G.; Kuzmanov, A.; Fay, D.S. FBN-1, a fibrillin-related protein, is required for resistance of the epidermis to mechanical deformation during C. elegans embryogenesis. Elife 2015, 4. [Google Scholar] [CrossRef]
- Low, I.I.C.; Williams, C.R.; Chong, M.K.; McLachlan, I.G.; Wierbowski, B.M.; Kolotuev, I.; Heiman, M.G. Morphogenesis of neurons and glia within an epithelium. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.D.; Flatt, K.M.; Schroeder, N.E.; Sundaram, M.V. Epithelial Shaping by Diverse Apical Extracellular Matrices Requires the Nidogen Domain Protein DEX-1 in Caenorhabditis elegans. Genetics 2019, 211, 185–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forman-Rubinsky, R.; Cohen, J.D.; Sundaram, M.V. Lipocalins Are Required for Apical Extracellular Matrix Organization and Remodeling in Caenorhabditis elegans. Genetics 2017, 207, 625–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong-Brender, T.T.K.; Ben Amar, M.; Pontabry, J.; Labouesse, M. The interplay of stiffness and force anisotropies drives embryo elongation. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Bercher, M.; Wahl, J.; Vogel, B.E.; Lu, C.; Hedgecock, E.M.; Hall, D.H.; Plenefisch, J.D. mua-3, a gene required for mechanical tissue integrity in Caenorhabditis elegans, encodes a novel transmembrane protein of epithelial attachment complexes. J. Cell Biol. 2001, 154, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Fotopoulos, P.; Kim, J.; Hyun, M.; Qamari, W.; Lee, I.; You, Y.J. DPY-17 and MUA-3 Interact for Connective Tissue-Like Tissue Integrity in Caenorhabditis elegans: A Model for Marfan Syndrome. G3 (Bethesda) 2015, 5, 1371–1378. [Google Scholar] [CrossRef] [Green Version]
- Hong, L.; Elbl, T.; Ward, J.; Franzini-Armstrong, C.; Rybicka, K.K.; Gatewood, B.K.; Bucher, E.A. MUP-4 is a novel transmembrane protein with functions in epithelial cell adhesion in Caenorhabditis elegans. J. Cell Biol. 2001, 154, 403–414. [Google Scholar] [CrossRef] [Green Version]
- Bokhove, M.; Jovine, L. Structure of Zona Pellucida Module Proteins. Curr. Top. Dev. Biol. 2018, 130, 413–442. [Google Scholar] [CrossRef]
- Davies, A.G.; Spike, C.A.; Shaw, J.E.; Herman, R.K. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics 1999, 153, 117–134. [Google Scholar]
- Nelson, F.K.; Albert, P.S.; Riddle, D.L. Fine structure of the Caenorhabditis elegans secretory-excretory system. J. Ultrastruct. Res. 1983, 82, 156–171. [Google Scholar] [CrossRef]
- Sundaram, M.V.; Buechner, M. The Caenorhabditis elegans Excretory System: A Model for Tubulogenesis, Cell Fate Specification, and Plasticity. Genetics 2016, 203, 35–63. [Google Scholar] [CrossRef] [Green Version]
- Pu, P.; Stone, C.E.; Burdick, J.T.; Murray, J.I.; Sundaram, M.V. The Lipocalin LPR-1 Cooperates with LIN-3/EGF Signaling To Maintain Narrow Tube Integrity in Caenorhabditis elegans. Genetics 2017, 205, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Stone, C.E.; Hall, D.H.; Sundaram, M.V. Lipocalin signaling controls unicellular tube development in the Caenorhabditis elegans excretory system. Dev. Biol. 2009, 329, 201–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, L.; Kovacevic, I.; Heiman, M.G.; Bao, Z. A multicellular rosette-mediated collective dendrite extension. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Grimbert, S.; Mastronardi, K.; Christensen, R.; Law, C.; Fay, D.; Piekny, A. Multi-tissue patterning drives anterior morphogenesis of the C. elegans embryo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Oikonomou, G.; Shaham, S. The glia of Caenorhabditis elegans. Glia 2011, 59, 1253–1263. [Google Scholar] [CrossRef] [Green Version]
- Yochem, J.; Bell, L.R.; Herman, R.K. The identities of sym-2, sym-3 and sym-4, three genes that are synthetically lethal with mec-8 in Caenorhabditis elegans. Genetics 2004, 168, 1293–1306. [Google Scholar] [CrossRef] [Green Version]
- Schouteden, C.; Serwas, D.; Palfy, M.; Dammermann, A. The ciliary transition zone functions in cell adhesion but is dispensable for axoneme assembly in C. elegans. J. Cell Biol. 2015, 210, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaux, G.; Gansmuller, A.; Hindelang, C.; Labouesse, M. CHE-14, a protein with a sterol-sensing domain, is required for apical sorting in C. elegans ectodermal epithelial cells. Curr. Biol. 2000, 10, 1098–1107. [Google Scholar] [CrossRef] [Green Version]
- Oikonomou, G.; Perens, E.A.; Lu, Y.; Shaham, S. Some, but not all, retromer components promote morphogenesis of C. elegans sensory compartments. Dev. Biol. 2012, 362, 42–49. [Google Scholar] [CrossRef] [Green Version]
- Oikonomou, G.; Perens, E.A.; Lu, Y.; Watanabe, S.; Jorgensen, E.M.; Shaham, S. Opposing activities of LIT-1/NLK and DAF-6/patched-related direct sensory compartment morphogenesis in C. elegans. PLoS Biol. 2011, 9, e1001121. [Google Scholar] [CrossRef] [Green Version]
- Perens, E.A.; Shaham, S. C. elegans daf-6 encodes a patched-related protein required for lumen formation. Dev. Cell 2005, 8, 893–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singhal, A.; Shaham, S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat. Commun. 2017, 8, 14100. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Chen, H.; Zhang, Y.; Wu, Z.; Zhang, Y.; Zhang, Y.; Wei, Q. An extracellular protein regulates patched-related/DAF-6-mediated sensory compartment formation in C. elegans. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gupta, B.P.; Hanna-Rose, W.; Sternberg, P.W. Morphogenesis of the vulva and the vulval-uterine connection. WormBook 2012, 1–20. [Google Scholar] [CrossRef]
- Sternberg, P.W.; Horvitz, H.R. The combined action of two intercellular signaling pathways specifies three cell fates during vulval induction in C. elegans. Cell 1989, 58, 679–693. [Google Scholar] [CrossRef]
- Farooqui, S.; Pellegrino, M.W.; Rimann, I.; Morf, M.K.; Müller, L.; Fröhli, E.; Hajnal, A. Coordinated lumen contraction and expansion during vulval tube morphogenesis in Caenorhabditis elegans. Dev. Cell 2012, 23, 494–506. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.Y.; Horvitz, H.R. The Caenorhabditis elegans vulval morphogenesis gene sqv-4 encodes a UDP-glucose dehydrogenase that is temporally and spatially regulated. Proc. Natl. Acad. Sci. USA 2002, 99, 14224–14229. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.Y.; Horvitz, H.R. The SQV-1 UDP-glucuronic acid decarboxylase and the SQV-7 nucleotide-sugar transporter may act in the Golgi apparatus to affect Caenorhabditis elegans vulval morphogenesis and embryonic development. Proc. Natl. Acad. Sci. USA 2002, 99, 14218–14223. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.Y.; Olson, S.K.; Brown, J.R.; Esko, J.D.; Horvitz, H.R. The Caenorhabditis elegans genes sqv-2 and sqv-6, which are required for vulval morphogenesis, encode glycosaminoglycan galactosyltransferase II and xylosyltransferase. J. Biol. Chem. 2003, 278, 11735–11738. [Google Scholar] [CrossRef] [Green Version]
- Hwang, H.Y.; Olson, S.K.; Esko, J.D.; Horvitz, H.R. Caenorhabditis elegans early embryogenesis and vulval morphogenesis require chondroitin biosynthesis. Nature 2003, 423, 439–443. [Google Scholar] [CrossRef]
- Yang, Q.; Roiz, D.; Mereu, L.; Daube, M.; Hajnal, A. The Invading Anchor Cell Induces Lateral Membrane Constriction during Vulval Lumen Morphogenesis in C. elegans. Dev. Cell 2017, 42, 271–285.e273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, D.Z.; Sternberg, P.W.; Inoue, T. Morphologically defined sub-stages of C. elegans vulval development in the fourth larval stage. BMC Dev. Biol. 2015, 15, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, T.; Hartwieg, E.; Horvitz, H.R. sqv mutants of Caenorhabditis elegans are defective in vulval epithelial invagination. Proc. Natl. Acad. Sci. USA 1999, 96, 968–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, N.; Toyoda, H.; Sano, M.; Nishiwaki, K. Chondroitin acts in the guidance of gonadal distal tip cells in C. elegans. Dev. Biol. 2006, 300, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Noborn, F.; Gomez Toledo, A.; Nasir, W.; Nilsson, J.; Dierker, T.; Kjellén, L.; Larson, G. Expanding the chondroitin glycoproteome of Caenorhabditis elegans. J. Biol. Chem. 2018, 293, 379–389. [Google Scholar] [CrossRef] [Green Version]
- George-Raizen, J.B.; Shockley, K.R.; Trojanowski, N.F.; Lamb, A.L.; Raizen, D.M. Dynamically-expressed prion-like proteins form a cuticle in the pharynx of Caenorhabditis elegans. Biol. Open 2014, 3, 1139–1149. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, G.J.; Gaidatzis, D.; Aeschimann, F.; Großhans, H. Extensive oscillatory gene expression during C. elegans larval development. Mol. Cell 2014, 53, 380–392. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, I.L.; Barry, J.D. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996, 15, 3633–3639. [Google Scholar] [CrossRef]
- Favre, R.; Cermola, M.; Nunes, C.P.; Hermann, R.; Muller, M.; Bazzicalupo, P. Immuno-cross-reactivity of CUT-1 and cuticlin epitopes between ascaris lumbricoides, Caenorhabditis elegans, and Heterorhabditis. J. Struct. Biol. 1998, 123, 1–7. [Google Scholar] [CrossRef]
- Favre, R.; Hermann, R.; Cermola, M.; Hohenberg, H.; Muller, M.; Bazzicalupo, P. Immuno-gold-labelling of CUT-1, CUT-2 and cuticlin epitopes in Caenorhabditis elegans and Heterorhabditis sp. processed by high pressure freezing and freeze-substitution. J. Submicrosc. Cytol. Pathol. 1995, 27, 341–347. [Google Scholar]
- Lewis, E.; Hunter, S.J.; Tetley, L.; Nunes, C.P.; Bazzicalupo, P.; Devaney, E. cut-1-like genes are present in the filarial nematodes, Brugia pahangi and Brugia malayi, and, as in other nematodes, code for components of the cuticle. Mol. Biochem. Parasitol. 1999, 101, 173–183. [Google Scholar] [CrossRef]
- Gravato-Nobre, M.J.; Stroud, D.; O’Rourke, D.; Darby, C.; Hodgkin, J. Glycosylation genes expressed in seam cells determine complex surface properties and bacterial adhesion to the cuticle of Caenorhabditis elegans. Genetics 2011, 187, 141–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kage-Nakadai, E.; Kobuna, H.; Kimura, M.; Gengyo-Ando, K.; Inoue, T.; Arai, H.; Mitani, S. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS ONE 2010, 5, e8857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaima, E.; Leymarie, N.; Stroud, D.; Mizanur, R.M.; Hodgkin, J.; Gravato-Nobre, M.J.; Cipollo, J.F. The Caenorhabditis elegans bus-2 mutant reveals a new class of O-glycans affecting bacterial resistance. J. Biol. Chem. 2010, 285, 17662–17672. [Google Scholar] [CrossRef] [Green Version]
- Parsons, L.M.; Mizanur, R.M.; Jankowska, E.; Hodgkin, J.; O′rourke, D.; Stroud, D.; Cipollo, J.F. Caenorhabditis elegans bacterial pathogen resistant bus-4 mutants produce altered mucins. PLoS ONE 2014, 9, e107250. [Google Scholar] [CrossRef] [Green Version]
- Partridge, F.A.; Tearle, A.W.; Gravato-Nobre, M.J.; Schafer, W.R.; Hodgkin, J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 2008, 317, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Quintin, S.; Wang, S.; Pontabry, J.; Bender, A.; Robin, F.; Hyenne, V.; Labouesse, M. Non-centrosomal epidermal microtubules act in parallel to LET-502/ROCK to promote C. elegans elongation. Development 2016, 143, 160–173. [Google Scholar] [CrossRef] [Green Version]
- Suman, S.K.; Daday, C.; Ferraro, T.; Vuong-Brender, T.; Tak, S.; Quintin, S.; Labouesse, M. The plakin domain of C. elegans VAB-10/plectin acts as a hub in a mechanotransduction pathway to promote morphogenesis. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Landmann, F.; Zahreddine, H.; Rodriguez, D.; Koch, M.; Labouesse, M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature 2011, 471, 99–103. [Google Scholar] [CrossRef]
- Cox, G.N.; Kusch, M.; DeNevi, K.; Edgar, R.S. Temporal regulation of cuticle synthesis during development of Caenorhabditis elegans. Dev. Biol. 1981, 84, 277–285. [Google Scholar] [CrossRef]
- Cox, G.N.; Kusch, M.; Edgar, R.S. Cuticle of Caenorhabditis elegans: Its isolation and partial characterization. J. Cell Biol. 1981, 90, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Shou, Q.; Butcher, R.A. Identification of a dTDP-rhamnose biosynthetic pathway that oscillates with the molting cycle in Caenorhabditis elegans. Biochem. J. 2016, 473, 1507–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turek, M.; Bringmann, H. Gene expression changes of Caenorhabditis elegans larvae during molting and sleep-like lethargus. PLoS ONE 2014, 9, e113269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monsalve, G.C.; Van Buskirk, C.; Frand, A.R. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr. Biol. 2011, 21, 2033–2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, G.N.; Staprans, S.; Edgar, R.S. The cuticle of Caenorhabditis elegans. II. Stage-specific changes in ultrastructure and protein composition during postembryonic development. Dev. Biol. 1981, 86, 456–470. [Google Scholar] [CrossRef] [Green Version]
- Blaxter, M.L. Cuticle surface proteins of wild type and mutant Caenorhabditis elegans. J. Biol. Chem. 1993, 268, 6600–6609. [Google Scholar]
- Katz, S.S.; Maybrun, C.; Maul-Newby, H.M.; Frand, A.R. Non-canonical apical constriction shapes emergent matrices in C. elegans. bioRxiv 2018, 189951. [Google Scholar] [CrossRef] [Green Version]
- Chisholm, A.D.; Xu, S. The Caenorhabditis elegans epidermis as a model skin. II: Differentiation and physiological roles. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 879–902. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, I.L. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000, 16, 21–27. [Google Scholar] [CrossRef]
- Kramer, J.M.; Cox, G.N.; Hirsh, D. Expression of the Caenorhabditis elegans collagen genes col-1 and col-2 is developmentally regulated. J. Biol. Chem. 1985, 260, 1945–1951. [Google Scholar]
- Thein, M.C.; McCormack, G.; Winter, A.D.; Johnstone, I.L.; Shoemaker, C.B.; Page, A.P. Caenorhabditis elegans exoskeleton collagen COL-19: An adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dyn. 2003, 226, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Ristoratore, F.; Cermola, M.; Nola, M.; Bazzicalupo, P.; Favre, R. Ultrastructural immuno-localization of CUT-1 and CUT-2 antigenic sites in the cuticles of the nematode Caenorhabditis elegans. J. Submicrosc. Cytol. Pathol. 1994, 26, 437–443. [Google Scholar] [PubMed]
- de Melo, J.V.; de Souza, W.; Peixoto, C.A. Ultrastructural analyses of the Caenorhabditis elegans DR 847 bli-1(n361) mutant which produces abnormal cuticle blisters. J. Submicrosc. Cytol. Pathol. 2002, 34, 291–297. [Google Scholar] [PubMed]
- de Melo, J.V.; de Souza, W.; Peixoto, C.A. Immunocytochemical and freeze-fracture characterization of the Caenorhabditis elegans DR 847 bli-1(n361) mutant which produces abnormal cuticle blisters. Cell Tissue Res. 2003, 312, 229–235. [Google Scholar] [CrossRef]
- Schultz, R.D.; Bennett, E.E.; Ellis, E.A.; Gumienny, T.L. Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans. PLoS ONE 2014, 9, e101929. [Google Scholar] [CrossRef] [Green Version]
- Schultz, R.D.; Gumienny, T.L. Visualization of Caenorhabditis elegans cuticular structures using the lipophilic vital dye DiI. J. Vis. Exp. 2012, 59, e3362. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Paik, Y.K. A potential role for fatty acid biosynthesis genes during molting and cuticle formation in Caenorhabditis elegans. BMB Rep. 2011, 44, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Van de Walle, P.; Geens, E.; Baggerman, G.; Jose Naranjo-Galindo, F.; Askjaer, P.; Schoofs, L.; Temmerman, L. CEH-60/PBX regulates vitellogenesis and cuticle permeability through intestinal interaction with UNC-62/MEIS in Caenorhabditis elegans. PLoS Biol. 2019, 17, e3000499. [Google Scholar] [CrossRef]
- Bosher, J.M.; Hahn, B.S.; Legouis, R.; Sookhareea, S.; Weimer, R.M.; Gansmuller, A.; Labouesse, M. The Caenorhabditis elegans vab-10 spectraplakin isoforms protect the epidermis against internal and external forces. J. Cell Biol. 2003, 161, 757–768. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Labouesse, M. The making of hemidesmosome structures in vivo. Dev. Dyn. 2010, 239, 1465–1476. [Google Scholar] [CrossRef]
- Gotenstein, J.R.; Koo, C.C.; Ho, T.W.; Chisholm, A.D. Genetic Suppression of Basement Membrane Defects in Caenorhabditis elegans by Gain of Function in Extracellular Matrix and Cell-Matrix Attachment Genes. Genetics 2018, 208, 1499–1512. [Google Scholar] [CrossRef] [Green Version]
- McMahon, L.; Muriel, J.M.; Roberts, B.; Quinn, M.; Johnstone, I.L. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol. Biol. Cell 2003, 14, 1366–1378. [Google Scholar] [CrossRef] [Green Version]
- Dodd, W.; Tang, L.; Lone, J.C.; Wimberly, K.; Wu, C.W.; Consalvo, C.; Choe, K.P. A Damage Sensor Associated with the Cuticle Coordinates Three Core Environmental Stress Responses in Caenorhabditis elegans. Genetics 2018, 208, 1467–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, W.; Li, L.; Li, Y.; Fu, R.; Zhu, Y.; Zhang, H. Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity 2015, 42, 309–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Qi, L.; Zhang, H. TGFβ-like DAF-7 acts as a systemic signal for autophagy regulation in C. elegans. J. Cell Biol. 2019, 218, 3998–4006. [Google Scholar] [CrossRef]
- Lassandro, F.; Sebastiano, M.; Zei, F.; Bazzicalupo, P. The role of dityrosine formation in the crosslinking of CUT-2, the product of a second cuticlin gene of Caenorhabditis elegans. Mol. Biochem. Parasitol. 1994, 65, 147–159. [Google Scholar] [CrossRef]
- Muriel, J.M.; Brannan, M.; Taylor, K.; Johnstone, I.L.; Lithgow, G.J.; Tuckwell, D. M142.2 (cut-6), a novel Caenorhabditis elegans matrix gene important for dauer body shape. Dev. Biol. 2003, 260, 339–351. [Google Scholar] [CrossRef]
- Sebastiano, M.; Lassandro, F.; Bazzicalupo, P. cut-1 a Caenorhabditis elegans gene coding for a dauer-specific noncollagenous component of the cuticle. Dev. Biol. 1991, 146, 519–530. [Google Scholar] [CrossRef]
- Flatt, K.M.; Beshers, C.; Unal, C.; Cohen, J.D.; Sundaram, M.V.; Schroeder, N.E. Epidermal Remodeling in Caenorhabditis elegans Dauers Requires the Nidogen Domain Protein DEX-1. Genetics 2019, 211, 169–183. [Google Scholar] [CrossRef] [Green Version]
- Meli, V.S.; Osuna, B.; Ruvkun, G.; Frand, A.R. MLT-10 defines a family of DUF644 and proline-rich repeat proteins involved in the molting cycle of Caenorhabditis elegans. Mol. Biol. Cell 2010, 21, 1648–1661. [Google Scholar] [CrossRef] [Green Version]
- Brenner, S. The genetics of Caenorhabditis elegans. Genetics 1974, 77, 71–94. [Google Scholar] [PubMed]
- Noble, L.M.; Miah, A.; Kaur, T.; Rockman, M.V. The Ancestral Caenorhabditis elegans Cuticle Suppresses rol-1. G3 2020, 10, 2385–2395. [Google Scholar] [CrossRef] [PubMed]
- Fernando, T.; Flibotte, S.; Xiong, S.; Yin, J.; Yzeiraj, E.; Moerman, D.G.; Savage-Dunn, C. C. elegans ADAMTS ADT-2 regulates body size by modulating TGFβ signaling and cuticle collagen organization. Dev. Biol. 2011, 352, 92–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maduzia, L.L.; Gumienny, T.L.; Zimmerman, C.M.; Wang, H.; Shetgiri, P.; Krishna, S.; Padgett, R.W. lon-1 regulates Caenorhabditis elegans body size downstream of the dbl-1 TGF beta signaling pathway. Dev. Biol. 2002, 246, 418–428. [Google Scholar] [CrossRef] [Green Version]
- Morita, K.; Chow, K.L.; Ueno, N. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-beta family. Development 1999, 126, 1337–1347. [Google Scholar]
- Nyström, J.; Shen, Z.Z.; Aili, M.; Flemming, A.J.; Leroi, A.; Tuck, S. Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics 2002, 161, 83–97. [Google Scholar]
- Suzuki, Y.; Morris, G.A.; Han, M.; Wood, W.B. A cuticle collagen encoded by the lon-3 gene may be a target of TGF-beta signaling in determining Caenorhabditis elegans body shape. Genetics 2002, 162, 1631–1639. [Google Scholar]
- Clark, J.F.; Meade, M.; Ranepura, G.; Hall, D.H.; Savage-Dunn, C. Caenorhabditis elegans DBL-1/BMP Regulates Lipid Accumulation via Interaction with Insulin Signaling. G3 (Bethesda) 2018, 8, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Savage-Dunn, C.; Padgett, R.W. The TGF-β Family in Caenorhabditis elegans. Cold Spring Harb. Perspect. Biol. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Madaan, U.; Yzeiraj, E.; Meade, M.; Clark, J.F.; Rushlow, C.A.; Savage-Dunn, C. BMP Signaling Determines Body Size via Transcriptional Regulation of Collagen Genes in Caenorhabditis elegans. Genetics 2018, 210, 1355–1367. [Google Scholar] [CrossRef] [Green Version]
- Madaan, U.; Faure, L.; Chowdhury, A.; Ahmed, S.; Ciccarelli, E.J.; Gumienny, T.L.; Savage-Dunn, C. Feedback regulation of BMP signaling by Caenorhabditis elegans cuticle collagens. Mol. Biol. Cell 2020, 31, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.M.; Nikolic-Paterson, D.J.; Lan, H.Y. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Wright, K.A.; Thomson, J.N. The buccal capsule of Caenorhabditis elegans (Nematoda: Rhabditoidea): An ultrastructural study. Can. J. Zool. 1981, 59, 1952–1961. [Google Scholar] [CrossRef]
- Drace, K.; McLaughlin, S.; Darby, C. Caenorhabditis elegans BAH-1 is a DUF23 protein expressed in seam cells and required for microbial biofilm binding to the cuticle. PLoS ONE 2009, 4, e6741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgkin, J.; Clark, L.C.; Gravato-Nobre, M.J. Worm-stars and half-worms: Novel dangers and novel defense. Worm 2014, 3, e27939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulston, J.E.; Albertson, D.G.; Thomson, J.N. The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev. Biol. 1980, 78, 542–576. [Google Scholar] [CrossRef]
- Sulston, J.E.; Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977, 56, 110–156. [Google Scholar] [CrossRef]
- Yu, R.Y.; Nguyen, C.Q.; Hall, D.H.; Chow, K.L. Expression of ram-5 in the structural cell is required for sensory ray morphogenesis in Caenorhabditis elegans male tail. EMBO J. 2000, 19, 3542–3555. [Google Scholar] [CrossRef] [Green Version]
- Soete, G.; Betist, M.C.; Korswagen, H.C. Regulation of Caenorhabditis elegans body size and male tail development by the novel gene lon-8. BMC Dev. Biol. 2007, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Baird, S.E.; Emmons, S.W. Properties of a class of genes required for ray morphogenesis in Caenorhabditis elegans. Genetics 1990, 126, 335–344. [Google Scholar]
- Kuno, K.; Baba, C.; Asaka, A.; Matsushima, C.; Matsushima, K.; Hosono, R. The Caenorhabditis elegans ADAMTS family gene adt-1 is necessary for morphogenesis of the male copulatory organs. J. Biol. Chem. 2002, 277, 12228–12236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, F.C.; Chow, K.L. A novel thioredoxin-like protein encoded by the C. elegans dpy-11 gene is required for body and sensory organ morphogenesis. Development 2002, 129, 1185–1194. [Google Scholar] [PubMed]
- Ko, F.C.; Chow, K.L. A mutation at the start codon defines the differential requirement of dpy-11 in Caenorhabditis elegans body hypodermis and male tail. Biochem. Biophys. Res. Commun. 2003, 309, 201–208. [Google Scholar] [CrossRef]
- Ko, F.C.; Chow, K.L. Mutations with sensory ray defect unmask cuticular glycoprotein antigens in Caenorhabditis elegans male tail. Dev. Growth Differ. 2000, 42, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Frand, A.R.; Russel, S.; Ruvkun, G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005, 3, e312. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, S.; Zhang, J.; Oksov, Y.; Lustigman, S. The Caenorhabditis elegans cathepsin Z-like cysteine protease, Ce-CPZ-1, has a multifunctional role during the worms’ development. J. Biol. Chem. 2004, 279, 6035–6045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepek, G.; McCormack, G.; Birnie, A.J.; Page, A.P. The astacin metalloprotease moulting enzyme NAS-36 is required for normal cuticle ecdysis in free-living and parasitic nematodes. Parasitology 2011, 138, 237–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.; Sagoh, N.; Iwasaki, H.; Inoue, H.; Takahashi, K. Metalloproteases with EGF, CUB, and thrombospondin-1 domains function in molting of Caenorhabditis elegans. Biol. Chem. 2004, 385, 565–568. [Google Scholar] [CrossRef]
- Joseph, B.B.; Wang, Y.; Edeen, P.; Lažetić, V.; Grant, B.D.; Fay, D.S. Control of clathrin-mediated endocytosis by NIMA family kinases. PLoS Genet. 2020, 16, e1008633. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.C.; Yu, H.; Liu, Y.C.; Chen, T.L.; Stern, A.; Lo, S.J.; Chiu, D.T. IDH-1 deficiency induces growth defects and metabolic alterations in GSPD-1-deficient Caenorhabditis elegans. J. Mol. Med. (Berl.) 2019, 97, 385–396. [Google Scholar] [CrossRef] [Green Version]
- Miao, R.; Li, M.; Zhang, Q.; Yang, C.; Wang, X. An ECM-to-Nucleus Signaling Pathway Activates Lysosomes for C. elegans Larval Development. Dev. Cell 2020, 52, 21–37.e25. [Google Scholar] [CrossRef] [PubMed]
- Soloviev, A.; Gallagher, J.; Marnef, A.; Kuwabara, P.E. C. elegans patched-3 is an essential gene implicated in osmoregulation and requiring an intact permease transporter domain. Dev. Biol. 2011, 351, 242–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perera, R.M.; Zoncu, R. The Lysosome as a Regulatory Hub. Annu. Rev. Cell Dev. Biol. 2016, 32, 223–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zha, J.; Ying, M.; Alexander-Floyd, J.; Gidalevitz, T. HSP-4/BiP expression in secretory cells is regulated by a developmental program and not by the unfolded protein response. PLoS Biol. 2019, 17, e3000196. [Google Scholar] [CrossRef] [Green Version]
- Essmann, C.L.; Martinez-Martinez, D.; Pryor, R.; Leung, K.Y.; Krishnan, K.B.; Lui, P.P.; Cabreiro, F. Mechanical properties measured by atomic force microscopy define health biomarkers in ageing C. elegans. Nat. Commun. 2020, 11, 1043. [Google Scholar] [CrossRef]
- Ewald, C.Y.; Landis, J.N.; Porter Abate, J.; Murphy, C.T.; Blackwell, T.K. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 2015, 519, 97–101. [Google Scholar] [CrossRef] [Green Version]
- Morikiri, Y.; Matsuta, E.; Inoue, H. The collagen-derived compound collagen tripeptide induces collagen expression and extends lifespan via a conserved p38 mitogen-activated protein kinase cascade. Biochem. Biophys. Res. Commun. 2018, 505, 1168–1173. [Google Scholar] [CrossRef]
- Mesbahi, H.; Pho, K.B.; Tench, A.J.; Leon Guerrero, V.L.; MacNeil, L.T. Cuticle Collagen Expression Is Regulated in Response to Environmental Stimuli by the GATA Transcription Factor ELT-3 in Caenorhabditis elegans. Genetics 2020, 215, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Sellegounder, D.; Liu, Y.; Wibisono, P.; Chen, C.H.; Leap, D.; Sun, J. Neuronal GPCR NPR-8 regulates C. elegans defense against pathogen infection. Sci. Adv. 2019, 5, eaaw4717. [Google Scholar] [CrossRef] [Green Version]
- Smit, R.B.; Schnabel, R.; Gaudet, J. The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function. PLoS Genet. 2008, 4, e1000222. [Google Scholar] [CrossRef] [Green Version]
- Avery, L. The genetics of feeding in Caenorhabditis elegans. Genetics 1993, 133, 897–917. [Google Scholar] [PubMed]
- Fang-Yen, C.; Avery, L.; Samuel, A.D. Two size-selective mechanisms specifically trap bacteria-sized food particles in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 20093–20096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straud, S.; Lee, I.; Song, B.; Avery, L.; You, Y.J. The jaw of the worm: GTPase-activating protein EAT-17 regulates grinder formation in Caenorhabditis elegans. Genetics 2013, 195, 115–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sparacio, A.P.; Trojanowski, N.F.; Snetselaar, K.; Nelson, M.D.; Raizen, D.M. Teething during sleep: Ultrastructural analysis of pharyngeal muscle and cuticular grinder during the molt in Caenorhabditis elegans. PLoS ONE 2020, 15, e0233059. [Google Scholar] [CrossRef]
- Stutz, K.; Kaech, A.; Aebi, M.; Künzler, M.; Hengartner, M.O. Disruption of the C. elegans Intestinal Brush Border by the Fungal Lectin CCL2 Phenocopies Dietary Lectin Toxicity in Mammals. PLoS ONE 2015, 10, e0129381. [Google Scholar] [CrossRef] [Green Version]
- Knight, C.G.; Patel, M.N.; Azevedo, R.B.; Leroi, A.M. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol. Dev. 2002, 4, 16–27. [Google Scholar] [CrossRef]
- Park, J.O.; Pan, J.; Möhrlen, F.; Schupp, M.O.; Johnsen, R.; Baillie, D.L.; Hutter, H. Characterization of the astacin family of metalloproteases in C. elegans. BMC Dev. Biol. 2010, 10, 14. [Google Scholar] [CrossRef] [Green Version]
- Bento, G.; Ogawa, A.; Sommer, R.J. Co-option of the hormone-signalling module dafachronic acid-DAF-12 in nematode evolution. Nature 2010, 466, 494–497. [Google Scholar] [CrossRef]
- Bose, N.; Ogawa, A.; von Reuss, S.H.; Yim, J.J.; Ragsdale, E.J.; Sommer, R.J.; Schroeder, F.C. Complex small-molecule architectures regulate phenotypic plasticity in a nematode. Angew. Chem. Int. Ed. Engl. 2012, 51, 12438–12443. [Google Scholar] [CrossRef] [Green Version]
- Sanghvi, G.V.; Baskaran, P.; Röseler, W.; Sieriebriennikov, B.; Rödelsperger, C.; Sommer, R.J. Life History Responses and Gene Expression Profiles of the Nematode Pristionchus pacificus Cultured on Cryptococcus Yeasts. PLoS ONE 2016, 11, e0164881. [Google Scholar] [CrossRef]
- Werner, M.S.; Sieriebriennikov, B.; Loschko, T.; Namdeo, S.; Lenuzzi, M.; Dardiry, M.; Sommer, R.J. Environmental influence on Pristionchus pacificus mouth form through different culture methods. Sci. Rep. 2017, 7, 7207. [Google Scholar] [CrossRef] [PubMed]
- Ragsdale, E.J.; Müller, M.R.; Rödelsperger, C.; Sommer, R.J. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell 2013, 155, 922–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieriebriennikov, B.; Prabh, N.; Dardiry, M.; Witte, H.; Röseler, W.; Kieninger, M.R.; Sommer, R.J. A Developmental Switch Generating Phenotypic Plasticity Is Part of a Conserved Multi-gene Locus. Cell Rep. 2018, 23, 2835–2843.e4. [Google Scholar] [CrossRef] [PubMed]
- Sieriebriennikov, B.; Sommer, R.J. Developmental Plasticity and Robustness of a Nematode Mouth-Form Polyphenism. Front. Genet. 2018, 9, 382. [Google Scholar] [CrossRef] [PubMed]
- Waterston, R.; Ainscough, R.; Anderson, K.; Berks, M.; Blair, D.; Connell, M.; A Cooper, J.; Coulson, A.; Craxton, M.; Dear, S.; et al. The genome of the nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 1993, 58, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Maduro, M.F. Gut development in C. elegans. Semin. Cell Dev. Biol. 2017, 66, 3–11. [Google Scholar] [CrossRef]
- Geisler, F.; Coch, R.A.; Richardson, C.; Goldberg, M.; Bevilacqua, C.; Prevedel, R.; Leube, R.E. Intestinal intermediate filament polypeptides in C. elegans: Common and isotype-specific contributions to intestinal ultrastructure and function. Sci. Rep. 2020, 10, 3142. [Google Scholar] [CrossRef] [Green Version]
- Schulenburg, H.; Félix, M.A. The Natural Biotic Environment of Caenorhabditis elegans. Genetics 2017, 206, 55–86. [Google Scholar] [CrossRef] [Green Version]
- Maduzia, L.L.; Yu, E.; Zhang, Y. Caenorhabditis elegans galectins LEC-6 and LEC-10 interact with similar glycoconjugates in the intestine. J. Biol. Chem. 2011, 286, 4371–4381. [Google Scholar] [CrossRef] [Green Version]
- Joshua, G.W. Functional analysis of leucine aminopeptidase in Caenorhabditis elegans. Mol. Biochem. Parasitol. 2001, 113, 223–232. [Google Scholar] [CrossRef]
- Gravato-Nobre, M.J.; Vaz, F.; Filipe, S.; Chalmers, R.; Hodgkin, J. The Invertebrate Lysozyme Effector ILYS-3 Is Systemically Activated in Response to Danger Signals and Confers Antimicrobial Protection in C. elegans. PLoS Pathog. 2016, 12, e1005826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Julien-Gau, I.; Schmidt, M.; Kurz, C.L. f57f4.4p::gfp as a fluorescent reporter for analysis of the C. elegans response to bacterial infection. Dev. Comp. Immunol. 2014, 42, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.P.; White, J.G.; Sternberg, P.W. Morphogenesis of the C. elegans hermaphrodite uterus. Development 1996, 122, 3617–3626. [Google Scholar] [PubMed]
- Kubagawa, H.M.; Watts, J.L.; Corrigan, C.; Edmonds, J.W.; Sztul, E.; Browse, J.; Miller, M.A. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat. Cell Biol. 2006, 8, 1143–1148. [Google Scholar] [CrossRef] [PubMed]
- McCaughey, J.; Stephens, D.J. ER-to-Golgi Transport: A Sizeable Problem. Trends Cell Biol. 2019, 29, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.; Clucas, C.; Johnstone, I.L. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and extracellular matrix secretion. Mol. Biol. Cell 2003, 14, 4414–4426. [Google Scholar] [CrossRef] [Green Version]
- McCaughey, J.; Stevenson, N.L.; Cross, S.; Stephens, D.J. ER-to-Golgi trafficking of procollagen in the absence of large carriers. J. Cell Biol. 2019, 218, 929–948. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Bai, M.; Barbosa, G.O.; Chen, A.; Wei, Y.; Luo, S.; Ma, D.K. Broadly conserved roles of TMEM131 family proteins in intracellular collagen assembly and secretory cargo trafficking. Sci. Adv. 2020, 6, eaay7667. [Google Scholar] [CrossRef] [Green Version]
- Birchenough, G.M.; Johansson, M.E.; Gustafsson, J.K.; Bergström, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.D.; Bermudez, J.G.; Good, M.C.; Sundaram, M.V. A C. elegans Zona Pellucida domain protein functions via its ZPc domain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hyenne, V.; Apaydin, A.; Rodriguez, D.; Spiegelhalter, C.; Hoff-Yoessle, S.; Diem, M.; Labouesse, M. RAL-1 controls multivesicular body biogenesis and exosome secretion. J. Cell Biol. 2015, 211, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolotuev, I.; Schwab, Y.; Labouesse, M. A precise and rapid mapping protocol for correlative light and electron microscopy of small invertebrate organisms. Biol. Cell 2009, 102, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Liégeois, S.; Benedetto, A.; Garnier, J.M.; Schwab, Y.; Labouesse, M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J. Cell Biol. 2006, 173, 949–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2010, 339, 269–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Surma, M.A.; Simons, K. Polarized sorting and trafficking in epithelial cells. Cell Res. 2012, 22, 793–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dernedde, J.; Weise, C.; Muller, E.C.; Hagiwara, A.; Bachmann, S.; Suzuki, M.; Scherer, H. Cupulin is a zona pellucida-like domain protein and major component of the cupula from the inner ear. PLoS ONE 2014, 9, e111917. [Google Scholar] [CrossRef]
- Devuyst, O.; Olinger, E.; Rampoldi, L. Uromodulin: From physiology to rare and complex kidney disorders. Nat. Rev. Nephrol. 2017, 13, 525–544. [Google Scholar] [CrossRef]
- Gupta, S.K. The Human Egg’s Zona Pellucida. Curr. Top. Dev. Biol. 2018, 130, 379–411. [Google Scholar] [CrossRef]
- Liaskos, C.; Rigopoulou, E.I.; Orfanidou, T.; Bogdanos, D.P.; Papandreou, C.N. CUZD1 and anti-CUZD1 antibodies as markers of cancer and inflammatory bowel diseases. Clin. Dev. Immunol. 2013, 2013, 968041. [Google Scholar] [CrossRef] [Green Version]
- McAllister, K.A.; Grogg, K.M.; Johnson, D.W.; Gallione, C.J.; Baldwin, M.A.; Jackson, C.E.; Helmbold, E.A.; Markel, D.S.; McKinnon, W.C.; Murrell, J. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat. Genet. 1994, 8, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Roggenbuck, D.; Hausdorf, G.; Martinez-Gamboa, L.; Reinhold, D.; Büttner, T.; Jungblut, P.R.; Porstmann, T.; Laass, M.W.; Henker, J.; Büning, C.; et al. Identification of GP2, the major zymogen granule membrane glycoprotein, as the autoantigen of pancreatic antibodies in Crohn’s disease. Gut 2009, 12, 1620–1628. [Google Scholar] [CrossRef] [PubMed]
- Judge, D.P.; Dietz, H.C. Marfan’s syndrome. Lancet 2005, 366, 1965–1976. [Google Scholar] [CrossRef]
- Kinsella, M.G.; Bressler, S.L.; Wight, T.N. The regulated synthesis of versican, decorin, and biglycan: Extracellular matrix proteoglycans that influence cellular phenotype. Crit. Rev. Eukaryot. Gene Expr. 2004, 14, 203–234. [Google Scholar] [CrossRef] [PubMed]
- Flower, D.R. The lipocalin protein family: Structure and function. Biochem. J. 1996, 318 Pt 1, 1–14. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cohen, J.D.; Sundaram, M.V. C. elegans Apical Extracellular Matrices Shape Epithelia. J. Dev. Biol. 2020, 8, 23. https://doi.org/10.3390/jdb8040023
Cohen JD, Sundaram MV. C. elegans Apical Extracellular Matrices Shape Epithelia. Journal of Developmental Biology. 2020; 8(4):23. https://doi.org/10.3390/jdb8040023
Chicago/Turabian StyleCohen, Jennifer D., and Meera V. Sundaram. 2020. "C. elegans Apical Extracellular Matrices Shape Epithelia" Journal of Developmental Biology 8, no. 4: 23. https://doi.org/10.3390/jdb8040023
APA StyleCohen, J. D., & Sundaram, M. V. (2020). C. elegans Apical Extracellular Matrices Shape Epithelia. Journal of Developmental Biology, 8(4), 23. https://doi.org/10.3390/jdb8040023