A Novel Mutation in Cse1l Disrupts Brain and Eye Development with Specific Effects on Pax6 Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Husbandry
2.2. ENU Mutagenesis
2.3. Cse1lnull CRISPR Design
2.4. Weight
2.5. Histology
2.6. Skeletal Preparations
2.7. Exome Analysis
2.8. RNA-Seq
2.9. RNAscope
2.10. Immunofluorescence (Ascl1, Gsx2, Nkx2.1, Olig2, Pax6)
2.11. Immunofluorescence (PHH3, CC3, Tbr2)
2.12. Immunofluorescence (Lhx2, Pax6)
2.13. Immunofluorescence Quantification
2.14. Western Immunoblotting
3. Results
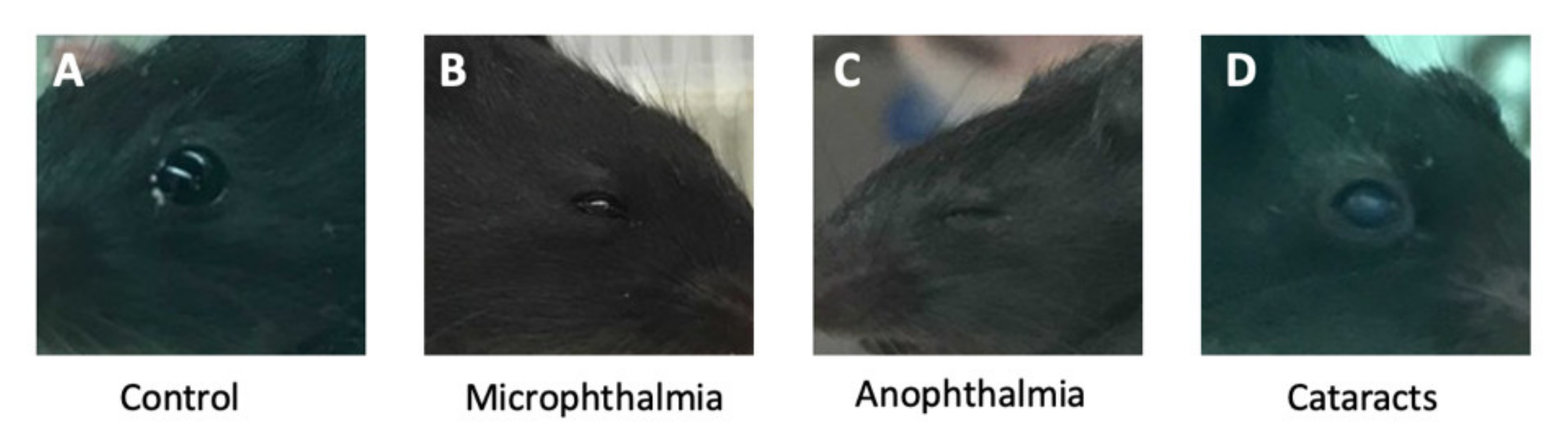
3.1. Anteater Mutants Display Variable, Incompletely Penetrant Organogenesis Phenotypes
3.2. The Anteater Phenotype Is Caused by a Variant in Cse1l
3.3. Cse1l Expression
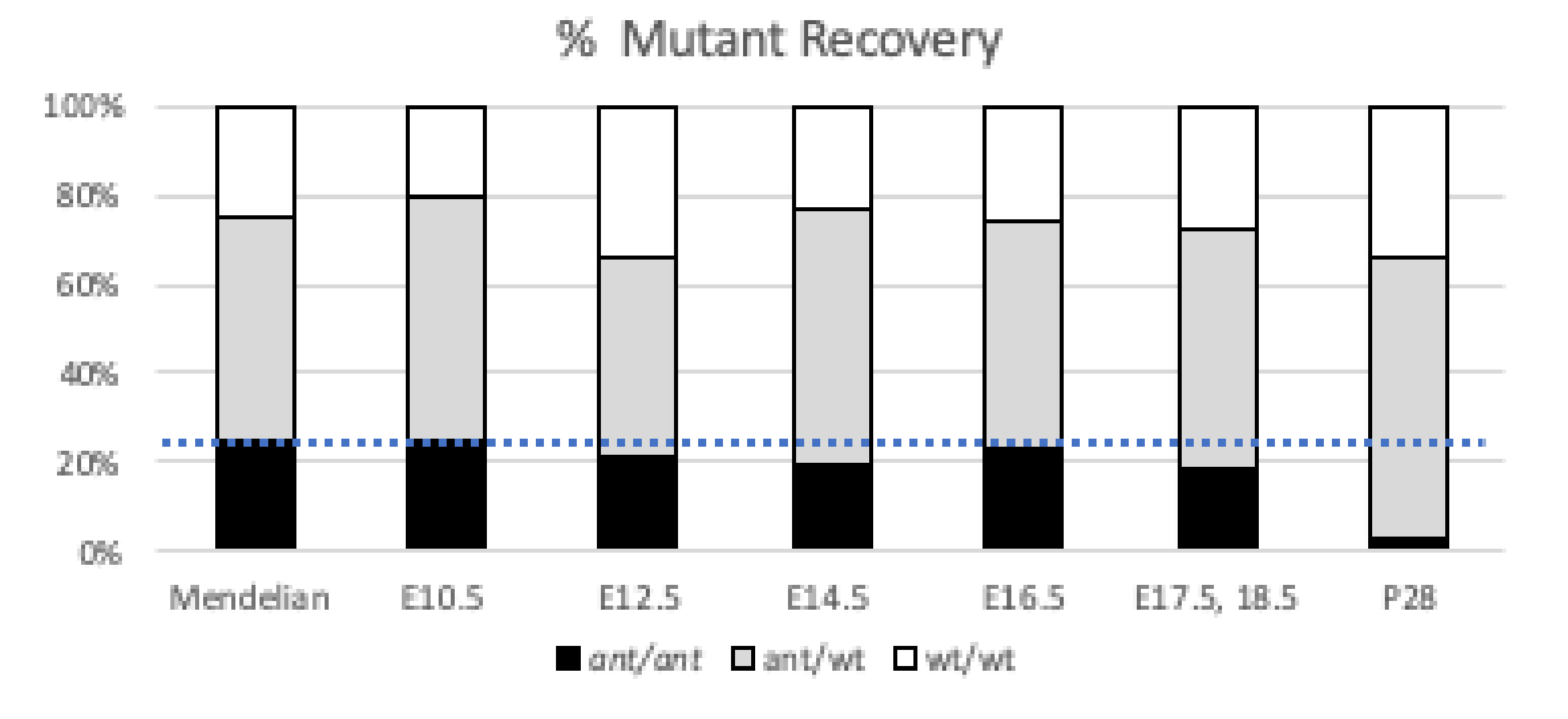
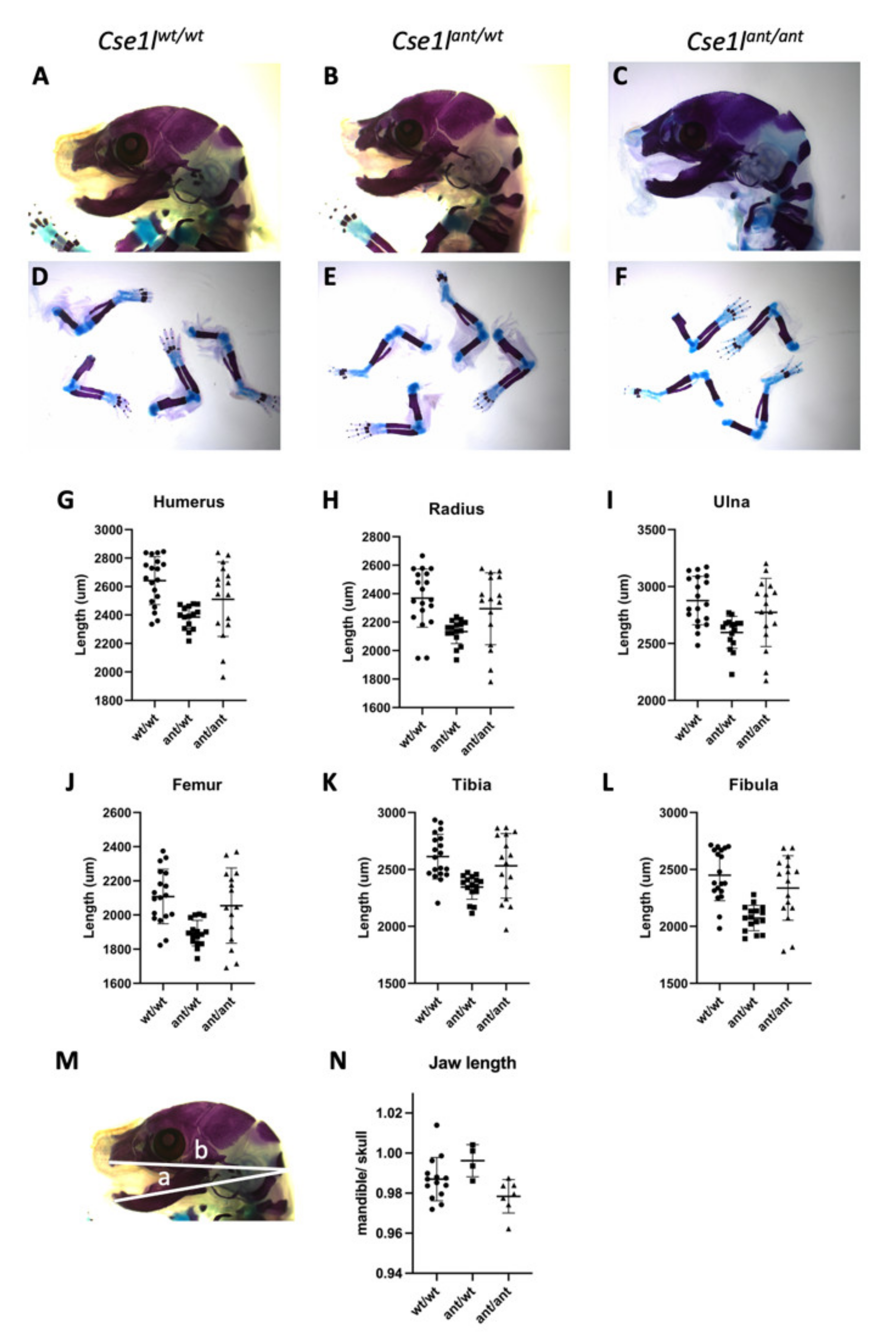
3.4. Anteater Mutants Do Not Survive Past Birth and Have a Variable Phenotype
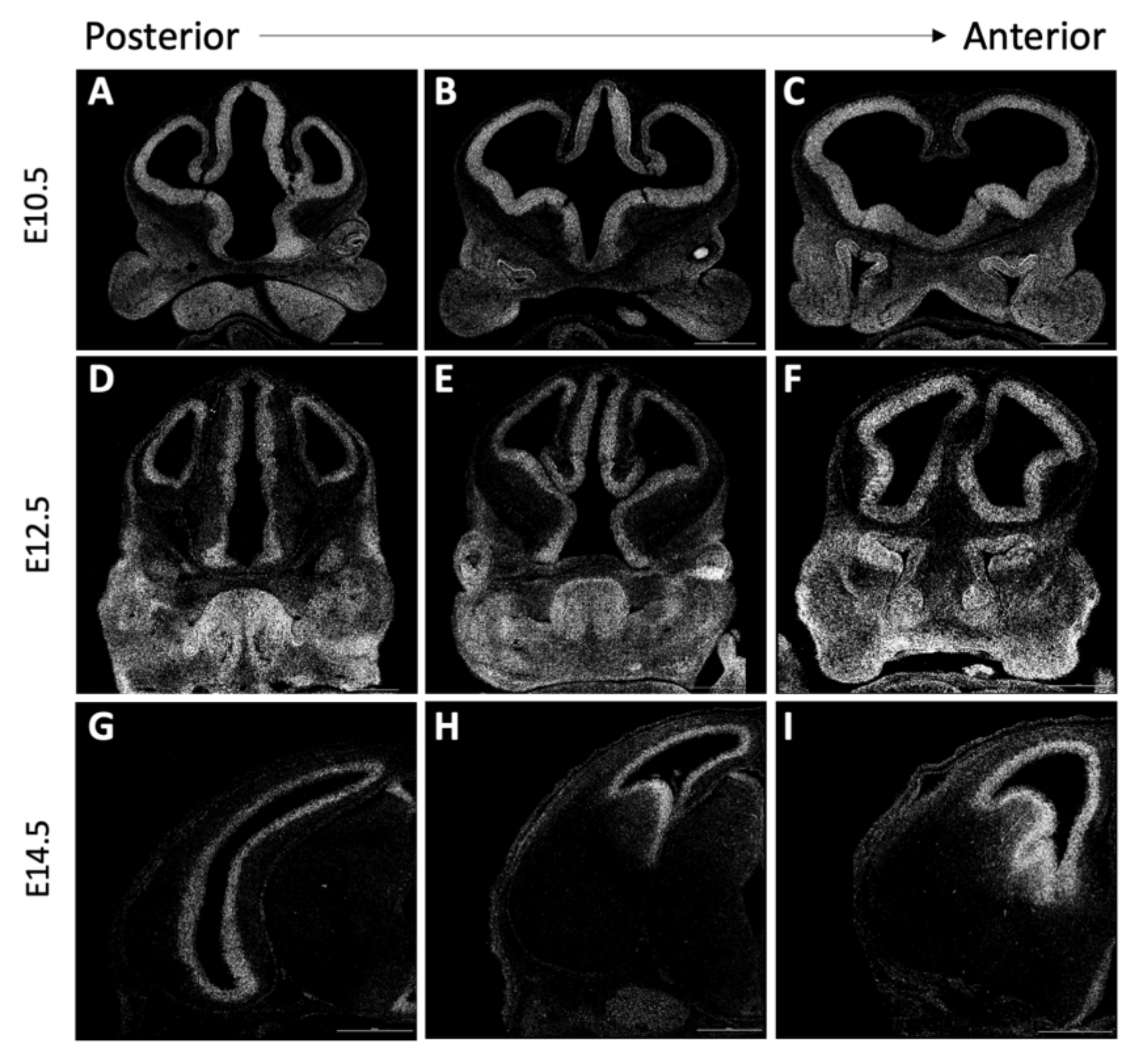
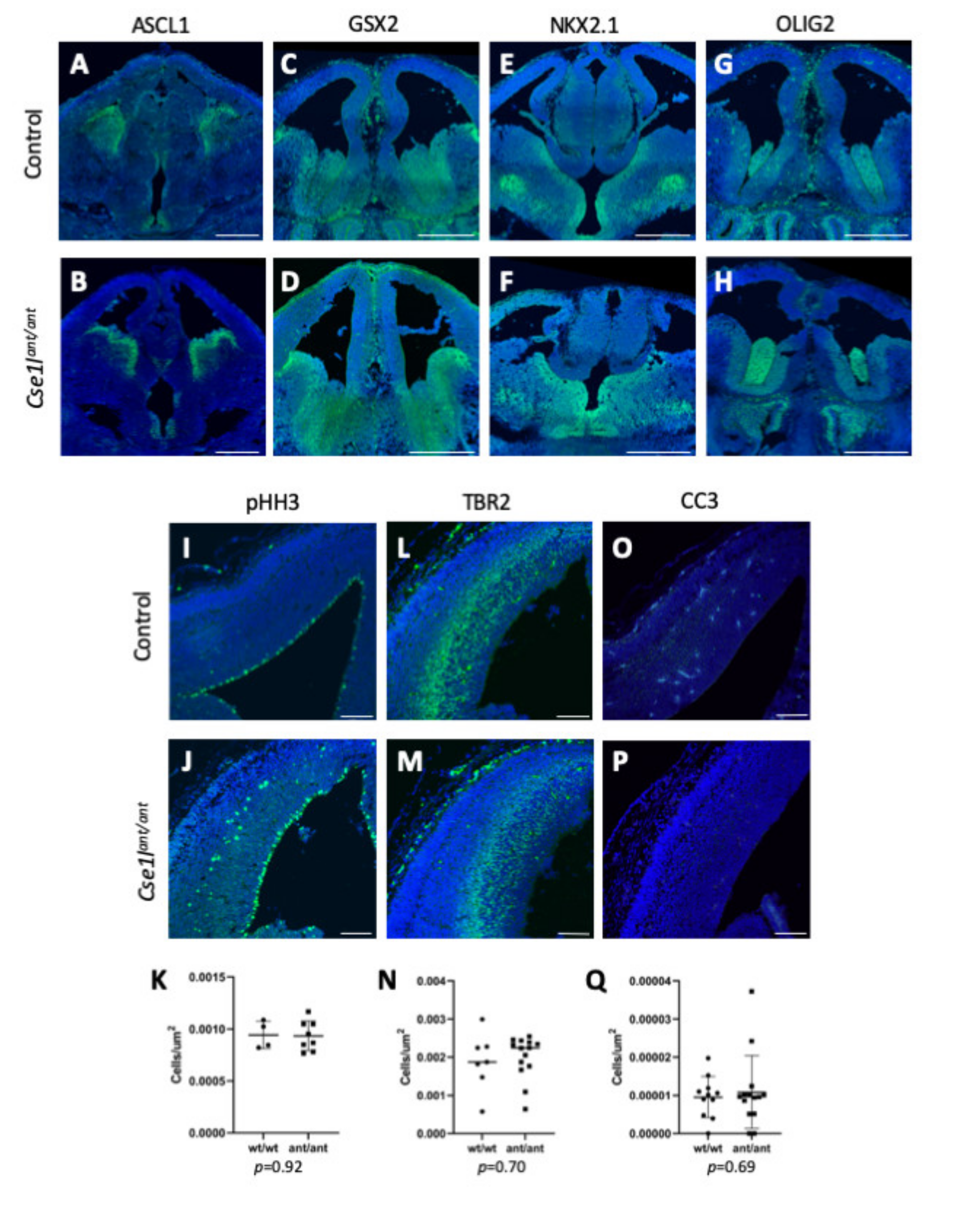
3.5. Forebrain Patterning Is Not Consistently Perturbed in Anteater Mutants
3.6. Cse1l Is Required for Proper Pax6 Regulation in the Eye and Brain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brinkmann, U.; Gallo, M.; Pastan, I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc. Natl. Acad. Sci. USA 1995, 92, 10427–10431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherf, U.; Pastan, I.; Willingham, M.C.; Brinkmann, U. The human CAS protein which is homologous to the CSE1 yeast chromosome segregation gene product is associated with microtubules and mitotic spindle. Proc. Natl. Acad. Sci. USA 1996, 93, 2670–2674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, S.A.; Gerace, L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell 1991, 66, 837–847. [Google Scholar] [CrossRef]
- Görlich, D.; Vogel, F.; Mills, A.D.; Hartmann, E.; Laskey, R.A. Distinct functions for the two importin subunits in nuclear protein import. Nat. Cell Biol. 1995, 377, 246–248. [Google Scholar] [CrossRef]
- Gorlich, D.; Pante, N.; Kutay, U.; Aebi, U.; Bischoff, F.R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996, 15, 5584–5594. [Google Scholar] [CrossRef] [Green Version]
- Chi, N.C.; Adam, S.A. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol. Biol. Cell 1997, 8, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Kutay, U.; Bischoff, F.; Kostka, S.; Kraft, R.; Görlich, D. Export of Importin α from the Nucleus Is Mediated by a Specific Nuclear Transport Factor. Cell 1997, 90, 1061–1071. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, Y.; Saiwaki, T.; Yamashita, J.; Yasuda, Y.; Kotera, I.; Shibata, S.; Shigeta, M.; Hiraoka, Y.; Haraguchi, T.; Yoneda, Y. Cellular stresses induce the nuclear accumulation of importin α and cause a conventional nuclear import block. J. Cell Biol. 2004, 165, 617–623. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, Y.; Miyamoto, Y.; Yamashiro, T.; Asally, M.; Masui, A.; Wong, C.; Loveland, K.L.; Yoneda, Y. Nuclear retention of importin α coordinates cell fate through changes in gene expression. EMBO J. 2011, 31, 83–94. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Wei, J.; Xu, K.; Sun, X.; Zhang, H.; Xiong, C. CSE1L participates in regulating cell mitosis in human seminoma. Cell Prolif. 2018, 52, e12549. [Google Scholar] [CrossRef]
- Lee, W.R.; Shen, S.C.; Wu, P.R.; Chou, C.L.; Shih, Y.H.; Yeh, C.M.; Jiang, M.C. CSE1L Links cAMP/PKA and Ras/ERK pathways and regulates the expressions and phosphorylations of ERK1/2, CREB, and MITF in melanoma cells. Mol. Carcinog. 2016, 55, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-F.; Lin, S.-H.; Chen, H.-C.; Tai, C.-J.; Chang, C.-C.; Li, L.-T.; Yeh, C.-M.; Yeh, K.-T.; Chen, Y.-C.; Hsu, T.-H.; et al. CSE1L, a Novel Microvesicle Membrane Protein, Mediates Ras-Triggered Microvesicle Generation and Metastasis of Tumor Cells. Mol. Med. 2012, 18, 1269–1280. [Google Scholar] [CrossRef]
- Okimoto, S.; Sun, J.; Fukuto, A.; Horikoshi, Y.; Matsuda, S.; Matsuda, T.; Ikura, M.; Ikura, T.; Machida, S.; Kurumizaka, H.; et al. hCAS/CSE1L regulates RAD51 distribution and focus formation for homologous recombinational repair. Genes Cells 2015, 20, 681–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.-E.; Jiang, X.; Du, F.; Wang, X. PHAPI, CAS, and Hsp70 Promote Apoptosome Formation by Preventing Apaf-1 Aggregation and Enhancing Nucleotide Exchange on Apaf-1. Mol. Cell 2008, 30, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Ohkubo, S.; Tatsuno, I.; Prives, C. hCAS/CSE1L Associates with Chromatin and Regulates Expression of Select p53 Target Genes. Cell 2007, 130, 638–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Q.; Li, X.; Wang, C.-Z.; Xu, S.; Yuan, G.; Shao, W.; Liu, B.; Zheng, Y.; Wang, H.; Lei, X.; et al. Roles of the CSE1L-mediated nuclear import pathway in epigenetic silencing. Proc. Natl. Acad. Sci. USA 2018, 115, E4013–E4022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M.; Okumura, N.; Kose, S.; Takao, T.; Imamoto, N. Identification of cargo proteins specific for importin-beta with importin-alpha applying a stable isotope labeling by amino acids in cell culture (SILAC)-based in vitro transport system. J. Biol. Chem. 2013, 288, 24540–24549. [Google Scholar] [CrossRef] [Green Version]
- Wellmann, A.; Krenacs, L.; Fest, T.; Scherf, U.; Pastan, I.; Raffeld, M.; Brinkmann, U. Localization of the cell proliferation and apoptosis-associated CAS protein in lymphoid neoplasms. Am. J. Pathol. 1997, 150, 25–30. [Google Scholar]
- Zhou, Y.; Zhou, Q.; Li, L.; Gu, Y.; Sun, X.; Wang, S.; Wang, X. Functional regulation of human trophoblast cells by CSE1L. J. Matern. Neonatal Med. 2021, 34, 1598–1605. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, J.; Fok, K.L.; Tsang, L.L.; Ye, M.; Liu, J.; Chen, H. CD147 Induces Epithelial-to-Mesenchymal Transition by Disassembling Cellular Apoptosis Susceptibility Protein/E-Cadherin/beta-Catenin Complex in Human Endometriosis. Am. J. Pathol. 2018, 188, 1597–1607. [Google Scholar] [CrossRef] [Green Version]
- Bera, T.K.; Bera, J.; Brinkmann, U.; Tessarollo, L.; Pastan, I. Cse1l Is Essential for Early Embryonic Growth and Development. Mol. Cell. Biol. 2001, 21, 7020–7024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stottmann, R.; Beier, D.R. ENU Mutagenesis in the Mouse. Curr. Protoc. Hum. Genet. 2014, 82, 15.4.1–15.4.10. [Google Scholar] [PubMed] [Green Version]
- Stottmann, R.W.; Moran, J.; Turbedoan, A.; Driver, E.; Kelley, M.W.; Beier, D.R. Focusing Forward Genetics: A Tripartite ENU Screen for Neurodevelopmental Mutations in the Mouse. Genetics 2011, 188, 615–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toresson, H.; Potter, S.; Campbell, K. Genetic control of dorsal-ventral identity in the telencephalon: Opposing roles for Pax6 and Gsh2. Dev. 2000, 127, 4361–4371. [Google Scholar] [CrossRef]
- Kakrana, A.; Yang, A.; Anand, D.; Djordjevic, D.; Ramachandruni, D.; Singh, A.; Huang, H.; Ho, J.W.K.; Lachke, S. iSyTE 2.0: A database for expression-based gene discovery in the eye. Nucleic Acids Res. 2017, 46, D875–D885. [Google Scholar] [CrossRef] [PubMed]
- Anand, D.; Kakrana, A.; Siddam, A.D.; Huang, H.; Saadi, I.; Lachke, S.A. RNA sequencing-based transcriptomic profiles of embryonic lens development for cataract gene discovery. Qual. Life Res. 2018, 137, 941–954. [Google Scholar] [CrossRef]
- Aryal, S.; Anand, D.; Hernandez, F.G.; Weatherbee, B.; Huang, H.; Reddy, A.P.; Wilmarth, P.A.; David, L.L.; Lachke, S.A. MS/MS in silico subtraction-based proteomic profiling as an approach to facilitate disease gene discovery: Application to lens development and cataract. Qual. Life Res. 2020, 139, 151–184. [Google Scholar] [CrossRef]
- Castro, D.S.; Martynoga, B.; Parras, C.; Ramesh, V.; Pacary, E.; Johnston, C.; Drechsel, D.; Lebel-Potter, M.; Garcia, L.G.; Hunt, C.; et al. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011, 25, 930–945. [Google Scholar] [CrossRef] [Green Version]
- Yun, K.; Potter, S.; Rubenstein, J. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 2001, 128, 193–205. [Google Scholar] [CrossRef]
- Sussel, L.; Marin, O.; Kimura, S.; Rubenstein, J.L. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: Evidence for a transformation of the pallidum into the striatum. Development 1999, 126, 3359–3370. [Google Scholar] [CrossRef]
- Nery, S.; Wichterle, H.; Fishell, G. Sonic hedgehog contributes to oligodendrocyte specification in the mammalian forebrain. Development 2001, 128, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Gurley, L.R.; D’Anna, J.A.; Barham, S.S.; Deaven, L.L.; Tobey, R.A. Histone Phosphorylation and Chromatin Structure during Mitosis in Chinese Hamster Cells. J. Biol. Inorg. Chem. 1978, 84, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Englund, C.; Fink, A.; Lau, C.; Pham, D.; Daza, R.A.M.; Bulfone, A.; Kowalczyk, T.; Hevner, R.F. Pax6, Tbr2, and Tbr1 Are Expressed Sequentially by Radial Glia, Intermediate Progenitor Cells, and Postmitotic Neurons in Developing Neocortex. J. Neurosci. 2005, 25, 247–251. [Google Scholar] [CrossRef] [Green Version]
- Hill, R.E.; Favor, J.; Hogan, B.L.M.; Ton, C.C.T.; Saunders, G.F.; Hanson, I.; Prosser, J.; Jordan, T.; Hastie, N.D.; van Heyningen, V. Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nat. Cell Biol. 1991, 354, 522–525. [Google Scholar] [CrossRef]
- Schmahl, W.; Knoedlseder, M.; Favor, J.; Davidson, D. Defects of neuronal migration and the pathogenesis of cortical malformations are associated with Small eye (Sey) in the mouse, a point mutation at the Pax-6-locus. Acta Neuropathol. 1993, 86, 126–135. [Google Scholar] [CrossRef]
- Porter, F.D.; Drago, J.; Xu, Y.; Cheema, S.S.; Wassif, C.; Huang, S.P.; Lee, E.; Grinberg, A.; Massalas, J.S.; Bodine, D.; et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 1997, 124, 2935–2944. [Google Scholar] [CrossRef]
- Jevtić, P.; Mukherjee, R.N.; Chen, P.; Levy, D.L. Altering the levels of nuclear import factors in early Xenopus laevis embryos affects later development. PLoS ONE 2019, 14, e0215740. [Google Scholar] [CrossRef] [Green Version]
- Kodiha, M.; Tran, D.; Morogan, A.; Qian, C.; Stochaj, U. Dissecting the Signaling Events That Impact Classical Nuclear Import and Target Nuclear Transport Factors. PLoS ONE 2009, 4, e8420. [Google Scholar] [CrossRef]
- Bharti, K.; Gasper, M.; Ou, J.; Brucato, M.; Clore-Gronenborn, K.; Pickel, J.; Arnheiter, H. A Regulatory Loop Involving PAX6, MITF, and WNT Signaling Controls Retinal Pigment Epithelium Development. PLoS Genet. 2012, 8, e1002757. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yuan, S.; Liu, J.; Wang, Y.; Zhang, Y.; Chen, X.; Si, W. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J. Cell. Physiol. 2020, 235, 2071–2079. [Google Scholar] [CrossRef]
- Widlund, H.; Fisher, D. Microphthalamia-associated transcription factor: A critical regulator of pigment cell development and survival. Oncogene 2003, 22, 3035–3041. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-S.; Peng, C.; Guo, Y.; Li, Y. CSE1L promotes proliferation and migration in oral cancer through positively regulating MITF. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5429–5435. [Google Scholar]
- Jiang, M.-C. CAS (CSE1L) signaling pathway in tumor progression and its potential as a biomarker and target for targeted therapy. Tumor Biol. 2016, 37, 13077–13090. [Google Scholar] [CrossRef] [PubMed]
- Raviv, S.; Bharti, K.; Rencus-Lazar, S.; Cohen-Tayar, Y.; Schyr, R.; Evantal, N.; Meshorer, E.; Zilberberg, A.; Idelson, M.; Reubinoff, B.; et al. PAX6 Regulates Melanogenesis in the Retinal Pigmented Epithelium through Feed-Forward Regulatory Interactions with MITF. PLoS Genet. 2014, 10, e1004360. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Li, H.; Chen, Y.; Yang, J.; Chen, H.; Arnheiter, H.; Hou, L. The transcription factor MITF in RPE function and dysfunction. Prog. Retin. Eye Res. 2019, 73, 100766. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Gómez, A.; Rubio, M.J.; Català-Mora, J.; Zanón-Moreno, V.; Lopez, M.; Hernández, C.; Yoshida, S.; Nakama, T.; Ishikawa, K.; et al. DNA Methylomes Reveal Biological Networks Involved in Human Eye Development, Functions and Associated Disorders. Sci. Rep. 2017, 7, 11762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, R.; Mishra, R. Interaction of Pax6 with SPARC and p53 in Brain of Mice Indicates Smad3 Dependent Auto-regulation. J. Mol. Neurosci. 2010, 41, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Shubham, K.; Mishra, R. Pax6 interacts with SPARC and TGF-beta in murine eyes. Mol. Vis. 2012, 18, 951–956. [Google Scholar] [PubMed]





| Cse1lant/wt x Cse1lant/wt | wt/wt | ant/wt | ant/ant (% of Expected) | Total | Chi2 p Value |
|---|---|---|---|---|---|
| E10.5 | 10 | 27 | 12 (96%) | 50 | 0.712 |
| E12.5 | 23 | 30 | 14 (84%) | 67 | 0.207 |
| E14.5 | 50 | 125 | 41 (76%) | 216 | 0.047 |
| E16.5 | 8 | 16 | 7 (90%) | 31 | 0.953 |
| E17.5, 18.5 | 9 | 18 | 6 (73%) | 33 | 0.664 |
| P28 | 70 | 133 | 6 (11%) | 209 | 1.30 × 10−12 |
| Genotype | Mean (st. dev) | ANOVA F Statistic (p Value) | Comparison | Tukey’s Multiple Comparison-Adjusted p Value * | |
|---|---|---|---|---|---|
| Humerus | wt/wt | 2641 (168.3) | 8.01 (0.001) | wt vs. ant/wt | 0.001 |
| ant/wt | 2385 (80.94) | ant/wt vs. ant/ant | 0.156 | ||
| ant/ant | 2511 (261.2) | wt vs. ant/ant | 0.108 | ||
| Radius | wt/wt | 2369 (204.2) | 6.56 (0.003) | wt vs. ant/wt | 0.002 |
| ant/wt | 2133 (83.8) | ant/wt vs. ant/ant | 0.498 | ||
| ant/ant | 2294 (252.3) | wt vs. ant/ant | 0.059 | ||
| Ulna | wt/wt | 2877 (213.4) | 6.71 (0.003) | wt vs. ant/wt | 0.002 |
| ant/wt | 2597 (140.6) | ant/wt vs. ant/ant | 0.083 | ||
| ant/ant | 2773 (299.2) | wt vs. ant/ant | 0.369 | ||
| Femur | wt/wt | 2108 (159.7) | 7.96 (0.001) | wt vs. ant/wt | 0.001 |
| ant/wt | 1894 (75.1) | ant/wt vs. ant/ant | 0.021 | ||
| ant/ant | 2055 (220.3) | wt vs. ant/ant | 0.614 | ||
| Fibula | wt/wt | 2450 (224.5) | 13.26 (<0.0001) | wt vs. ant/wt | <0.0001 |
| ant/wt | 2073 (111.6) | ant/wt vs. ant/ant | 0.004 | ||
| ant/ant | 2337 (283.1) | wt vs. ant/ant | 0.294 | ||
| Tibia | wt/wt | 2613 (197.0) | 7.34 (0.002) | wt vs. ant/wt | 0.001 |
| ant/wt | 2346 (108.5) | ant/wt vs. ant/ant | 0.486 | ||
| ant/ant | 2531 (283.3) | wt vs. ant/ant | 0.040 | ||
| Mandible/ Skull Ratio | wt/wt | 0.987 (0.011) | 4.311 (0.026) | wt vs. ant/wt | 0.243 |
| ant/wt | 0.996 (0.008) | ant/wt vs. ant/ant | 0.022 | ||
| ant/ant | 0.978 (0.008) | wt vs. ant/ant | 0.168 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blizzard, L.E.; Menke, C.; Patel, S.D.; Waclaw, R.R.; Lachke, S.A.; Stottmann, R.W. A Novel Mutation in Cse1l Disrupts Brain and Eye Development with Specific Effects on Pax6 Expression. J. Dev. Biol. 2021, 9, 27. https://doi.org/10.3390/jdb9030027
Blizzard LE, Menke C, Patel SD, Waclaw RR, Lachke SA, Stottmann RW. A Novel Mutation in Cse1l Disrupts Brain and Eye Development with Specific Effects on Pax6 Expression. Journal of Developmental Biology. 2021; 9(3):27. https://doi.org/10.3390/jdb9030027
Chicago/Turabian StyleBlizzard, Lauren E., Chelsea Menke, Shaili D. Patel, Ronald R. Waclaw, Salil A. Lachke, and Rolf W. Stottmann. 2021. "A Novel Mutation in Cse1l Disrupts Brain and Eye Development with Specific Effects on Pax6 Expression" Journal of Developmental Biology 9, no. 3: 27. https://doi.org/10.3390/jdb9030027
APA StyleBlizzard, L. E., Menke, C., Patel, S. D., Waclaw, R. R., Lachke, S. A., & Stottmann, R. W. (2021). A Novel Mutation in Cse1l Disrupts Brain and Eye Development with Specific Effects on Pax6 Expression. Journal of Developmental Biology, 9(3), 27. https://doi.org/10.3390/jdb9030027







