HOX Protein Activity Regulation by Cellular Localization
Abstract
:1. Introduction
2. HOX Protein Localization: Cell-Type and Stage-Specific Changes
3. Entering and Leaving the Nucleus
3.1. NLS, Predictions and Validations
3.2. Leaving the Nucleus
3.3. Leaving the Nucleus to Leave Cells?
3.4. Interactions with Karyopherins and Proteins Associated with Cytoplasmic Organelles
4. If Not in the Nucleus, What Do HOX Proteins Do?
5. Not to Conclude…
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NES | Nuclear Export Signal |
| NLS | Nuclear Localization Signal |
| PG | paralogue group |
| PTM | Post-Translational Modification |
| TALE | Three-Amino Acid Long Extension |
References
- Rezsohazy, R.; Saurin, A.J.; Maurel-Zaffran, C.; Graba, Y. Cellular and molecular insights into Hox protein action. Development 2015, 142, 1212–1227. [Google Scholar] [CrossRef] [Green Version]
- Gaunt, S.J. Hox cluster genes and collinearities throughout the tree of animal life. Int. J. Dev. Biol. 2018, 62, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Mallo, M.; Wellik, D.M.; Deschamps, J. Hox genes and regional patterning of the vertebrate body plan. Dev. Biol. 2010, 344, 7–15. [Google Scholar] [CrossRef] [Green Version]
- Hudry, B.; Thomas-Chollier, M.; Volovik, Y.; Duffraisse, M.; Dard, A.; Frank, D.; Technau, U.; Merabet, S. Molecular insights into the origin of the Hox-TALE patterning system. eLife 2014, 3, e01939. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Del Viso, F.; Chen, C.-Y.; Ikmi, A.; Kroesen, A.E.; Gibson, M.C. An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 2018, 361, 1377–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, H.C.; Edvardsen, R.; Maeland, A.D.; Bjordal, M.; Jensen, M.F.; Hansen, A.; Flaat, M.; Weissenbach, J.; Lehrach, H.; Wincker, P.; et al. Hox cluster disintegration with persistent anteroposterior order of expression in Oikopleura dioica. Nat. Cell Biol. 2004, 431, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Aboobaker, A.; Blaxter, M. Hox gene evolution in nematodes: Novelty conserved. Curr. Opin. Genet. Dev. 2003, 13, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Anaya, J.; D’Aniello, S.; Kuratani, S.; Garcia-Fernàndez, J. Evolution of Hox gene clusters in deuterostomes. BMC Dev. Biol. 2013, 13, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiedlmeier, B.; Santos, A.C.; Ribeiro, A.; Moncaut, N.; Lesinski, D.; Auer, H.; Kornacker, K.; Ostertag, W.; Baum, C.; Mallo, M.; et al. HOXB4’s road map to stem cell expansion. Proc. Natl. Acad. Sci. USA 2007, 104, 16952–16957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, E.M.; Thompson, A. HOX genes in normal, engineered and malignant hematopoiesis. Int. J. Dev. Biol. 2018, 62, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Gofflot, F.; Lizen, B. Emerging roles for HOX proteins in synaptogenesis. Int. J. Dev. Biol. 2018, 62, 807–818. [Google Scholar] [CrossRef]
- Philippidou, P.; Dasen, J.S. Hox genes: Choreographers in neural development, architects of circuit organization. Neuron 2013, 80, 12–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, M.; Zaffran, S. Hox genes in cardiovascular development and diseases. J. Dev. Biol. 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Tischfield, M.; Bosley, T.M.; Salih, M.A.M.; Alorainy, I.; Sener, E.C.; Nester, M.J.; Oystreck, D.T.; Chan, W.-M.; Andrews, C.; Erickson, R.P.; et al. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat. Genet. 2005, 37, 1035–1037. [Google Scholar] [CrossRef]
- Bobola, N.; Merabet, S. Homeodomain proteins in action: Similar DNA binding preferences, highly variable connectivity. Curr. Opin. Genet. Dev. 2017, 43, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Mann, R.S. To be specific or not: The critical relationship between hox and TALE proteins. Trends Genet. 2016, 32, 334–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Kumar, B.; Darland, D.C. The Hox protein conundrum: The “specifics” of DNA binding for Hox proteins and their partners. Dev. Biol. 2021, 477, 284–292. [Google Scholar] [CrossRef]
- Banreti, A.; Hudry, B.; Sass, M.; Saurin, A.; Graba, Y. Hox proteins mediate developmental and environmental control of autophagy. Dev. Cell 2014, 28, 56–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saurin, A.J.; Delfini, M.C.; Maurel-Zaffran, C.; Graba, Y. The generic facet of hox protein function. Trends Genet. 2018, 34, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor binding evokes latent differences in DNA binding specificity between hox proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, B.; Vandeputte, J.; Remacle, S.; Bergiers, I.; Simonis, N.; Twizere, J.-C.; Vidal, M.; Rezsohazy, R. Protein interactions of the transcription factor Hoxa1. BMC Dev. Biol. 2012, 12, 29. [Google Scholar] [CrossRef] [Green Version]
- Bondos, S.E.; Tan, X.-X.; Matthews, K.S. Physical and genetic interactions link hox function with diverse transcription factors and cell signaling proteins. Mol. Cell. Proteom. 2006, 5, 824–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carnesecchi, J.; Sigismondo, G.; Domsch, K.; Baader, C.E.P.; Rafiee, M.-R.; Krijgsveld, J.; Lohmann, I. Multi-level and lineage-specific interactomes of the Hox transcription factor Ubx contribute to its functional specificity. Nat. Commun. 2020, 11, 1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baëza, M.; Viala, S.; Heim, M.; Dard, A.; Hudry, B.; Duffraisse, M.; Rogulja-Ortmann, A.; Brun, C.; Merabet, S. Inhibitory activities of short linear motifs underlie Hox interactome specificity in vivo. eLife 2015, 4, e06034. [Google Scholar] [CrossRef] [PubMed]
- Rezsohazy, R. Non-transcriptional interactions of Hox proteins: Inventory, facts, and future directions. Dev. Dyn. 2014, 243, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, C.; Resetca, D.; Redel, C.; Lin, P.; MacDonald, A.S.; Ciaccio, R.; Kenney, T.M.G.; Wei, Y.; Andrews, D.W.; Sunnerhagen, M.; et al. MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 2021, 21, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Yang, X.D.; Lamb, A.; Chen, L.F. Posttranslational modifications of NF-kappaB: Another layer of regulation for NF-kappaB signaling pathway. Cell Signal 2010, 22, 1282–1290. [Google Scholar] [CrossRef] [Green Version]
- Draime, A.; Bridoux, L.; Graba, Y.; Rezsohazy, R. Post-translational modifications of HOX proteins, an underestimated issue. Int. J. Dev. Biol. 2018, 62, 733–744. [Google Scholar] [CrossRef]
- Comel, A.; Sorrentino, G.; Capaci, V.; Del Sal, G. The cytoplasmic side of p53’s oncosuppressive activities. FEBS Lett. 2014, 588, 2600–2609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, T.; Choi, K.-B.; Gilks, C.B.; Leung, P.; Auersperg, N. Cell type- and stage-specific changes in HOXA7 protein expression in human ovarian folliculogenesis: Possible role of GDF-9. Differentiation 2006, 74, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sauvegarde, C.; Paul, D.; Bridoux, L.; Jouneau, A.; Degrelle, S.; Hue, I.; Rezsohazy, R.; Donnay, I. Dynamic pattern of HOXB9 protein localization during oocyte maturation and early embryonic development in mammals. PLoS ONE 2016, 11, e0165898. [Google Scholar] [CrossRef] [PubMed]
- Komuves, L.G.; Shen, W.F.; Kwong, A.; Stelnicki, E.; Rozenfeld, S.; Oda, Y.; Blink, A.; Krishnan, K.; Lau, B.; Mauro, T.; et al. Changes in HOXB6 homeodomain protein structure and localization during human epidermal development and differentiation. Dev. Dyn. 2000, 218, 636–647. [Google Scholar] [CrossRef]
- Kömüves, L.G.; Michael, E.; Arbeit, J.M.; Ma, X.-K.; Kwong, A.; Stelnicki, E.; Rozenfeld, S.; Morimune, M.; Yu, Q.-C.; Largman, C. HOXB4 homeodomain protein is expressed in developing epidermis and skin disorders and modulates keratinocyte proliferation. Dev. Dyn. 2002, 224, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Kömüves, L.G.; Ma, X.-K.; Stelnicki, E.; Rozenfeld, S.; Oda, Y.; Largman, C. HOXB13 homeodomain protein is cytoplasmic throughout fetal skin development. Dev. Dyn. 2003, 227, 192–202. [Google Scholar] [CrossRef]
- LaRosa, G.J.; Gudas, L.J. Early retinoic acid-induced F9 teratocarcinoma stem cell gene ERA-1: Alternate splicing creates transcripts for a homeobox-containing protein and one lacking the homeobox. Mol. Cell. Biol. 1988, 8, 3906–3917. [Google Scholar] [PubMed]
- Fernandez, C.C.; Gudas, L.J. The truncated Hoxa1 protein interacts with Hoxa1 and Pbx1 in stem cells. J. Cell. Biochem. 2009, 106, 427–443. [Google Scholar] [CrossRef] [Green Version]
- Kessel, M.; Schulze, F.; Fibi, M.; Gruss, P. Primary structure and nuclear localization of a murine homeodomain protein. Proc. Natl. Acad. Sci. USA 1987, 84, 5306–5310. [Google Scholar] [CrossRef] [Green Version]
- Schulze, F.; Chowdhury, K.; Zimmer, A.; Drescher, U.; Gruss, P. The murine homeo box gene product, Hox 1.1 protein, is growth-controlled and associated with chromatin. Differentiation 1987, 36, 130–137. [Google Scholar] [CrossRef] [PubMed]
- White, R.A.; Wilcox, M. Protein products of the bithorax complex in Drosophila. Cell 1984, 39, 163–171. [Google Scholar] [CrossRef]
- Ye, W.; Lin, W.; Tartakoff, A.M.; Tao, T. Karyopherins in nuclear transport of homeodomain proteins during development. Biochim. Biophys. Acta (BBA)-Bioenerg. 2011, 1813, 1654–1662. [Google Scholar] [CrossRef]
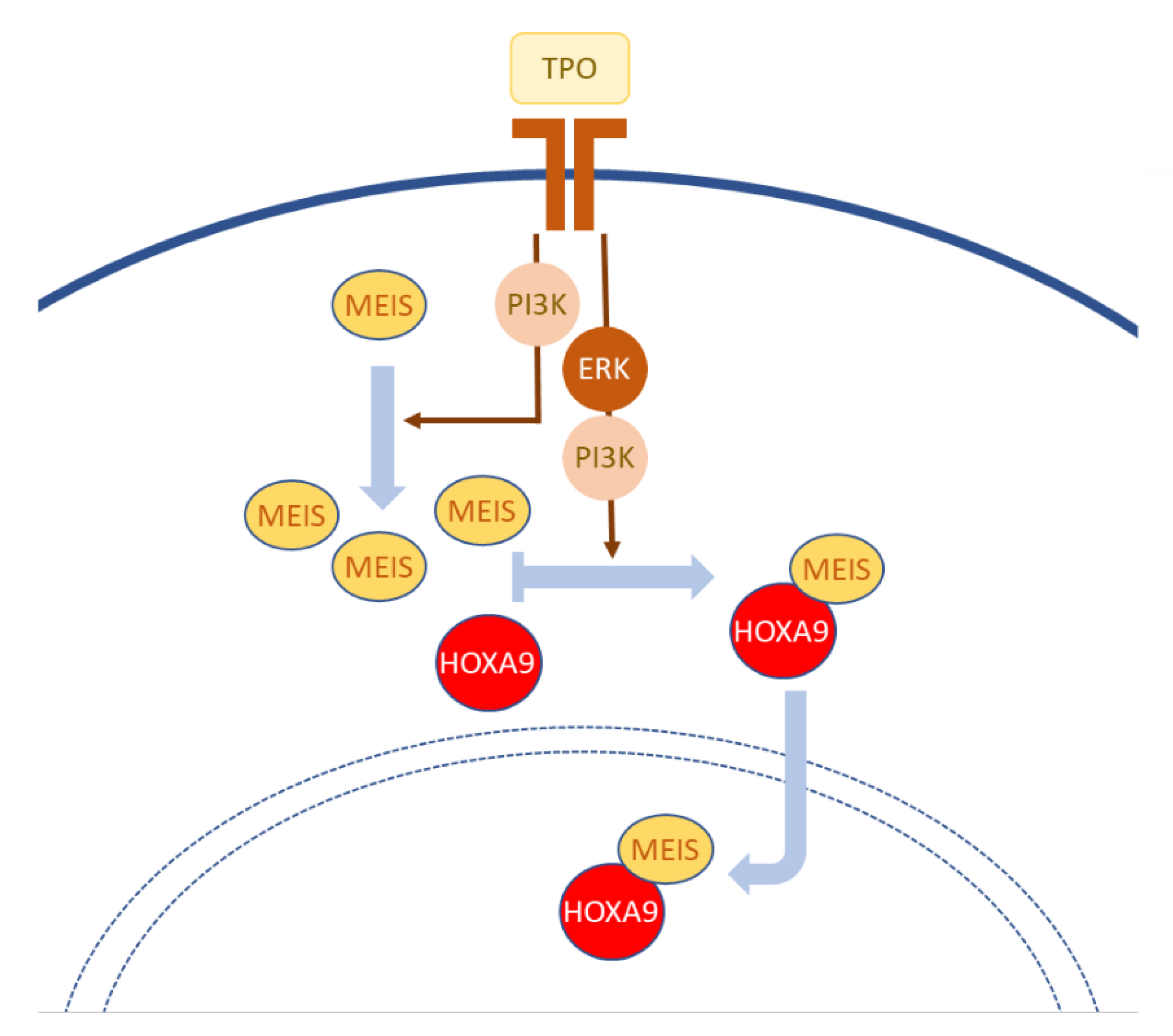
- Kirito, K.; Fox, N.; Kaushansky, K. Thrombopoietin induces HOXA9 nuclear transport in immature hematopoietic cells: Potential mechanism by which the hormone favorably affects hematopoietic stem cells. Mol. Cell. Biol. 2004, 24, 6751–6762. [Google Scholar] [CrossRef] [Green Version]
- Bridoux, L.; Bergiers, I.; Draime, A.; Halbout, M.; Deneyer, N.; Twizere, J.-C.; Rezsohazy, R. KPC2 relocalizes HOXA2 to the cytoplasm and decreases its transcriptional activity. Biochim. Biophys. Acta (BBA)-Bioenerg. 2015, 1849, 1298–1311. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Mura, S.; Yamada, K.; Sangel, P.; Hirata, S.; Maehara, K.; Kawakami, K.; Tachibana, T.; Ohkawa, Y.; Kimura, H.; et al. Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. Elife 2016, 5, e09540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosugi, S.; Hasebe, M.; Tomita, M.; Yanagawa, H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA 2009, 106, 10171–10176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffraisse, M.; Paul, R.; Carnesecchi, J.; Hudry, B.; Banreti, A.; Reboulet, J.; Ajuria, L.; Lohmann, I.; Merabet, S. Role of a versatile peptide motif controlling Hox nuclear export and autophagy in the Drosophila fat body. J. Cell Sci. 2020, 133, jcs241943. [Google Scholar] [CrossRef]
- Berthelsen, J.; Kilstrup-Nielsen, C.; Blasi, F.; Mavilio, F.; Zappavigna, V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999, 13, 946–953. [Google Scholar] [CrossRef] [Green Version]
- Mercader, N.; Leonardo, E.; Azpiazu, N.; Serrano, A.; Morata, G.; Martinez, C.; Torres, M. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature 1999, 402, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Capdevila, J.; Tsukui, T.; Esteban, C.R.; Zappavigna, V.; Belmonte, J.C.I. Control of vertebrate limb outgrowth by the proximal factor meis2 and distal antagonism of BMPs by gremlin. Mol. Cell 1999, 4, 839–849. [Google Scholar] [CrossRef]
- Deneyer, N.; Bridoux, L.; Bombled, C.; Pringels, T.; Bergiers, I.; Ruys, S.P.D.; Vertommen, D.; Twizere, J.-C.; Rezsohazy, R. HOXA2 activity regulation by cytoplasmic relocation, protein stabilization and post-translational modification. Biochim. Biophys. Acta (BBA)-Bioenerg. 2019, 1862, 194404. [Google Scholar] [CrossRef]
- Kamura, T.; Hara, T.; Matsumoto, M.; Ishida, N.; Okumura, F.; Hatakeyama, S.; Yoshida, M.; Nakayama, K.; Nakayama, K.I. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 2004, 6, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Ryan, T.; Shelton, M.; Lambert, J.-P.; Malecova, B.; Boisvenue, S.; Ruel, M.; Figeys, D.; Puri, P.L.; Skerjanc, I.S. Myosin phosphatase modulates the cardiac cell fate by regulating the subcellular localization of Nkx2.5 in a Wnt/Rho–associated protein kinase–dependent pathway. Circ. Res. 2013, 112, 257–266. [Google Scholar] [CrossRef]
- Fung, H.Y.J.; Fu, S.-C.; Chook, Y.M. Nuclear export receptor CRM1 recognizes diverse conformations in nuclear export signals. eLife 2017, 6, e23961. [Google Scholar] [CrossRef]
- La Cour, T.; Kiemer, L.; Mølgaard, A.; Gupta, R.; Skriver, K.; Brunak, S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004, 17, 527–536. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Marquis, K.; Pei, J.; Fu, S.-C.; Cağatay, T.; Grishin, N.V.; Chook, Y.M. LocNES: A computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 2015, 31, 1357–1365. [Google Scholar] [CrossRef] [Green Version]
- Chatelin, L.; Volovitch, M.; Joliot, A.H.; Perez, F.; Prochiantz, A. Transcription factor Hoxa-5 is taken up by cells in culture and conveyed to their nuclei. Mech. Dev. 1996, 55, 111–1175. [Google Scholar] [CrossRef]
- DeRossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994, 269, 10444–10450. [Google Scholar] [CrossRef]
- Sugiyama, S.; Di Nardo, A.A.; Aizawa, S.; Matsuo, I.; Volovitch, M.; Prochiantz, A.; Hensch, T.K. Experience-dependent transfer of otx2 homeoprotein into the visual cortex activates postnatal plasticity. Cell 2008, 134, 508–520. [Google Scholar] [CrossRef] [Green Version]
- Brunet, I.; Weinl, C.; Piper, M.; Trembleau, A.; Volovitch, M.; Harris, W.; Prochiantz, A.; Holt, C. The transcription factor Engrailed-2 guides retinal axons. Nat. Cell Biol. 2005, 438, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lesaffre, B.; Joliot, A.; Prochiantz, A.; Volovitch, M. Direct non-cell autonomous Pax6 activity regulates eye development in the zebrafish. Neural Dev. 2007, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Joliot, A.; Maizel, A.; Rosenberg, D.; Trembleau, A.; Dupas, S.; Volovitch, M.; Prochiantz, A. Identification of a signal sequence necessary for the unconventional secretion of Engrailed homeoprotein. Curr. Biol. 1998, 8, 856–863. [Google Scholar] [CrossRef] [Green Version]
- Dupont, E.; Prochiantz, A.; Joliot, A. Identification of a Signal Peptide for Unconventional Secretion. J. Biol. Chem. 2007, 282, 8994–9000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maizel, A.; Bensaude, O.; Prochiantz, A.; Joliot, A. A short region of its homeodomain is necessary for engrailed nuclear export and secretion. Development 1999, 126, 3183–3190. [Google Scholar] [CrossRef] [PubMed]
- Maizel, A.; Tassetto, M.; Filhol, O.; Cochet, C.; Prochiantz, A.; Joliot, A. Engrailed homeoprotein secretion is a regulated process. Development 2002, 129, 3545–3553. [Google Scholar] [CrossRef] [PubMed]
- Di Nardo, A.A.; Joliot, A.; Prochiantz, A. Homeoprotein transduction in neurodevelopment and physiopathology. Sci. Adv. 2020, 6, eabc6374. [Google Scholar] [CrossRef] [PubMed]
- Bardine, N.; Lamers, G.; Wacker, S.; Donow, C.; Knoechel, W.; Durston, A. Vertical signalling involves transmission of hox information from gastrula mesoderm to neurectoderm. PLoS ONE 2014, 9, e115208. [Google Scholar]
- Rolland, T.; Taşan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A proteome-scale map of the human interactome network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef] [Green Version]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A reference map of the human binary protein interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Aranda, B.; Blankenburg, H.; Kerrien, S.; Brinkman, F.; Ceol, A.; Chautard, E.; Dana, J.M.; Rivas, J.D.L.; Dumousseau, M.; Galeota, E.; et al. PSICQUIC and PSISCORE: Accessing and scoring molecular interactions. Nat. Methods 2011, 8, 528–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oka, M.; Mura, S.; Otani, M.; Miyamoto, Y.; Nogami, J.; Maehara, K.; Harada, A.; Tachibana, T.; Yoneda, Y.; Ohkawa, Y. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. eLife 2019, 8, e46667. [Google Scholar] [CrossRef] [PubMed]
- Saulnier, O.; Guedri-Idjouadiene, K.; Aynaud, M.-M.; Chakraborty, A.; Bruyr, J.; Pineau, J.; O’Grady, T.; Mirabeau, O.; Grossetête, S.; Galvan, B.; et al. ERG transcription factors have a splicing regulatory function involving RBFOX2 that is altered in the EWS-FLI1 oncogenic fusion. Nucleic Acids Res. 2021, 49, 5038–5056. [Google Scholar] [CrossRef] [PubMed]
- Bergiers, I.; Bridoux, L.; Nguyen, N.; Twizere, J.-C.; Rezsöhazy, R. The homeodomain transcription factor hoxa2 interacts with and promotes the proteasomal degradation of the E3 ubiquitin protein ligase RCHY1. PLoS ONE 2013, 8, e80387. [Google Scholar]
- Bridoux, L.; Deneyer, N.; Bergiers, I.; Rezsöhazy, R. Molecular analysis of the HOXA2-dependent degradation of RCHY1. PLoS ONE 2015, 10, e0141347. [Google Scholar]
- Taminiau, A.; Draime, A.; Tys, J.; Lambert, B.; Vandeputte, J.; Nguyen, N.; Renard, P.; Geerts, D.; Rezsohazy, R. HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-kappaB pathway in a transcription-independent manner. Nucleic Acids Res. 2016, 44, 7331–7349. [Google Scholar] [PubMed] [Green Version]
- Quéré, R.; Karlsson, G.; Hertwig, F.; Rissler, M.; Lindqvist, B.; Fioretos, T.; Vandenberghe, P.; Slovak, M.L.; Cammenga, J.; Karlsson, S. Smad4 binds Hoxa9 in the cytoplasm and protects primitive hematopoietic cells against nuclear activation by Hoxa9 and leukemia transformation. Blood 2011, 117, 5918–5930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draime, A.; Bridoux, L.; Belpaire, M.; Pringels, T.; Tys, J.; Rezsohazy, R. PRDM14, a putative histone methyl-transferase, interacts with and decreases the stability and activity of the HOXA1 transcription factor. Biochim. Biophys. Acta (BBA)-Bioenerg. 2018, 1861, 534–542. [Google Scholar] [CrossRef]

| HoxA | HoxB | HoxC | HoxD | ||
|---|---|---|---|---|---|
| 1 | HD | HX + HD | N-ter + HD | ||
| 2 | HD | HD | |||
| 3 | HD | HD | HD | ||
| 4 | HX | HD | HX | HD | |
| 5 | HX + HD | HX + HD | HD | ||
| 6 | HD | HD | HD | ||
| 7 | HD | HD | |||
| 8 | HD | HD | HD | ||
| 9 | HD | HD | HX | HD | HD |
| 10 | HD | HD | HD | ||
| 11 | HD | HD | HD | ||
| 12 | HD | HD | |||
| 13 | HD | HD | HD | HD | |
| lab | HX + HD | |
| pb | N-ter | |
| Dfd | HX | C-ter |
| Scr | HX + HD | |
| Antp | HD | |
| Ubx | HD extremities + HD surrounding | |
| abd-A | HD | |
| Abd-B | HD | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bridoux, L.; Gofflot, F.; Rezsohazy, R. HOX Protein Activity Regulation by Cellular Localization. J. Dev. Biol. 2021, 9, 56. https://doi.org/10.3390/jdb9040056
Bridoux L, Gofflot F, Rezsohazy R. HOX Protein Activity Regulation by Cellular Localization. Journal of Developmental Biology. 2021; 9(4):56. https://doi.org/10.3390/jdb9040056
Chicago/Turabian StyleBridoux, Laure, Françoise Gofflot, and René Rezsohazy. 2021. "HOX Protein Activity Regulation by Cellular Localization" Journal of Developmental Biology 9, no. 4: 56. https://doi.org/10.3390/jdb9040056
APA StyleBridoux, L., Gofflot, F., & Rezsohazy, R. (2021). HOX Protein Activity Regulation by Cellular Localization. Journal of Developmental Biology, 9(4), 56. https://doi.org/10.3390/jdb9040056






