Next-Generation Sequencing for Determining the Effect of Arginine on Human Dental Biofilms Using an In Situ Model
Abstract
:1. Introduction
2. Materials and Methods
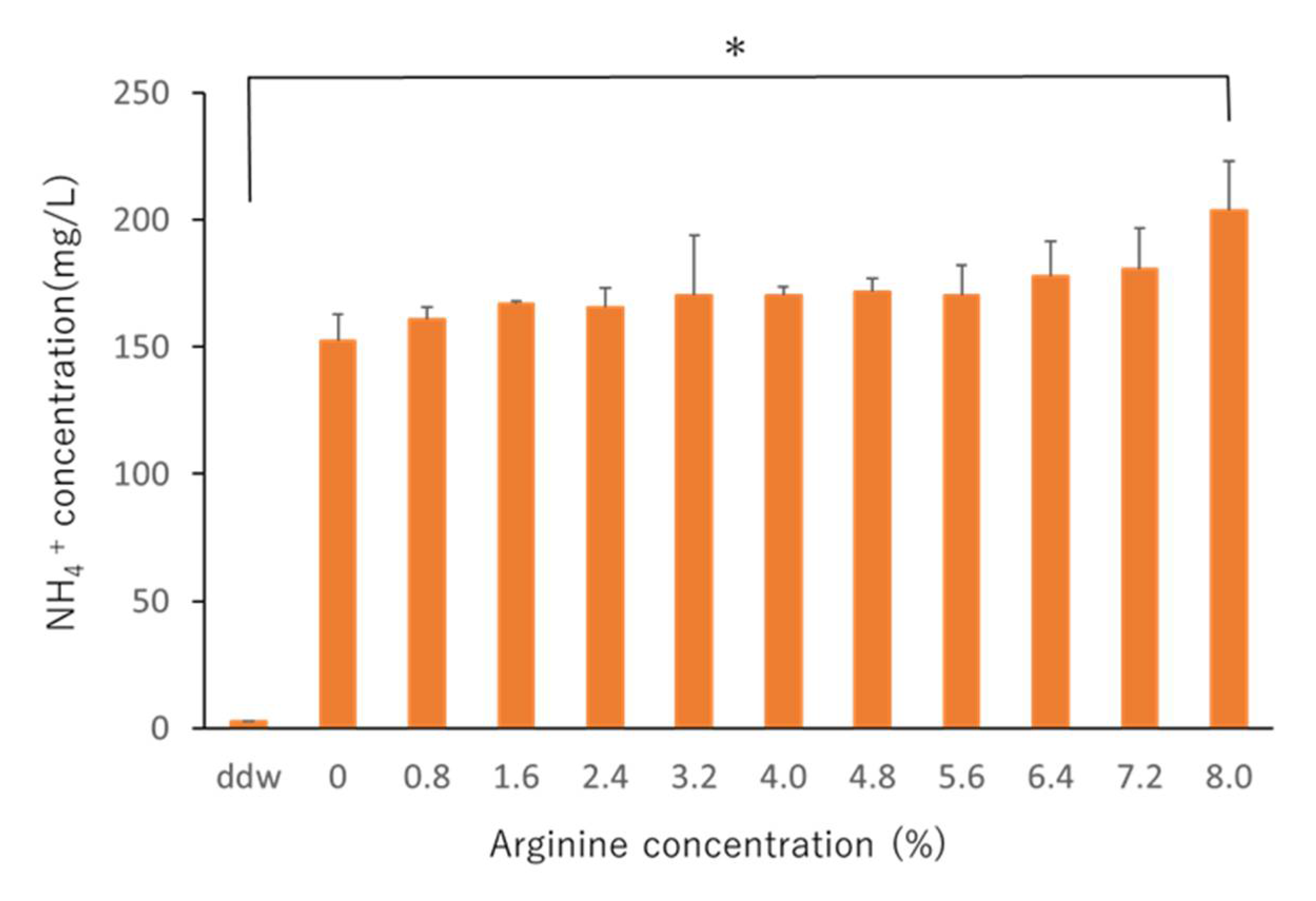
2.1. Determining the Effective Arginine Concentration In Vitro
2.2. In Situ Study
2.2.1. Study Subjects
2.2.2. In Situ Model of Biofilms
2.2.3. Experimental Protocol for Extracting Biofilms
2.2.4. Determination of Viable Counts
2.2.5. DNA Sequencing
2.3. Statistical Analysis
3. Results
3.1. Effective In Vitro Arginine Concentrations
3.2. Effect of Arginine In Situ
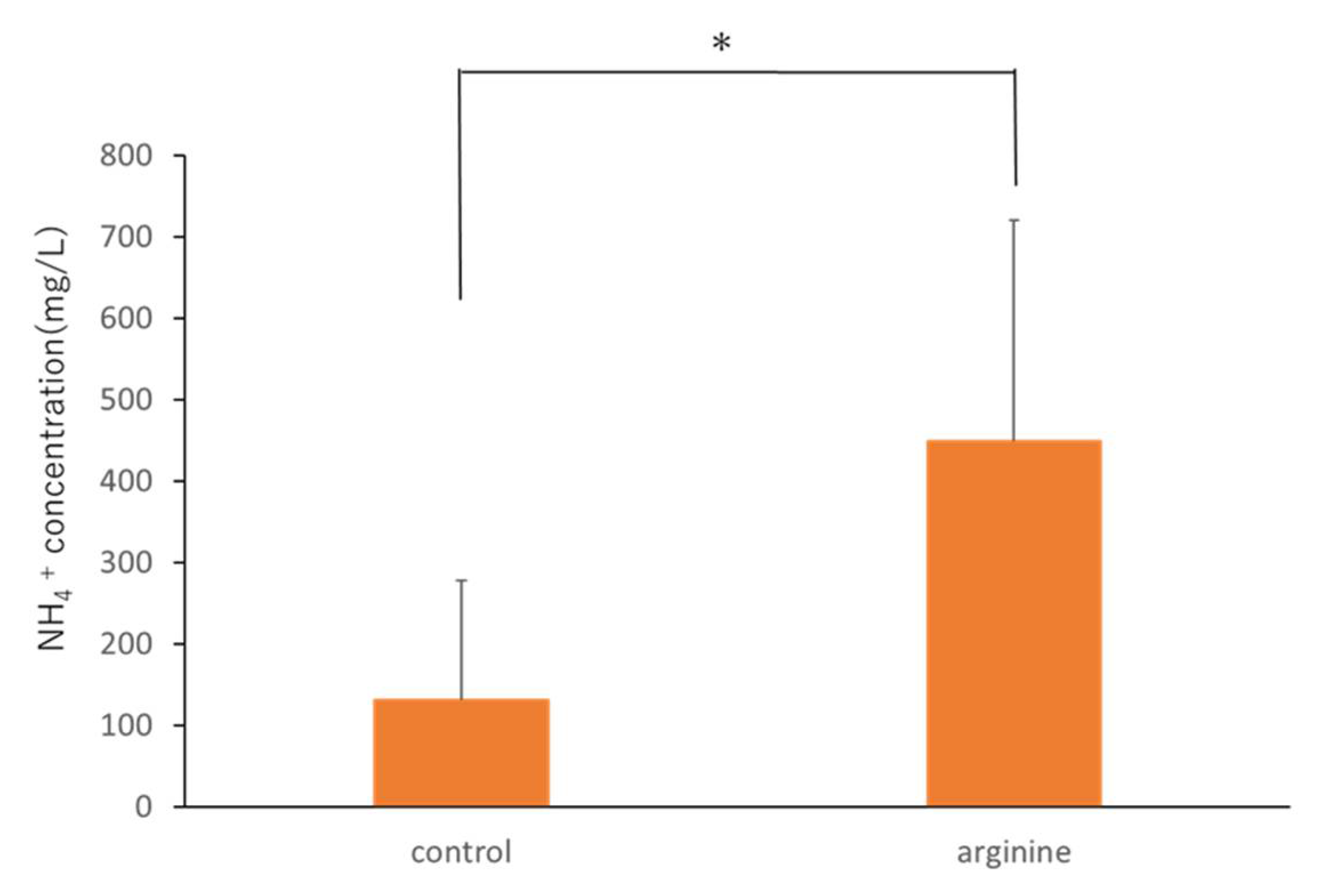
3.2.1. Effect of Arginine on the Viable Count and NH4+ Concentration In Situ
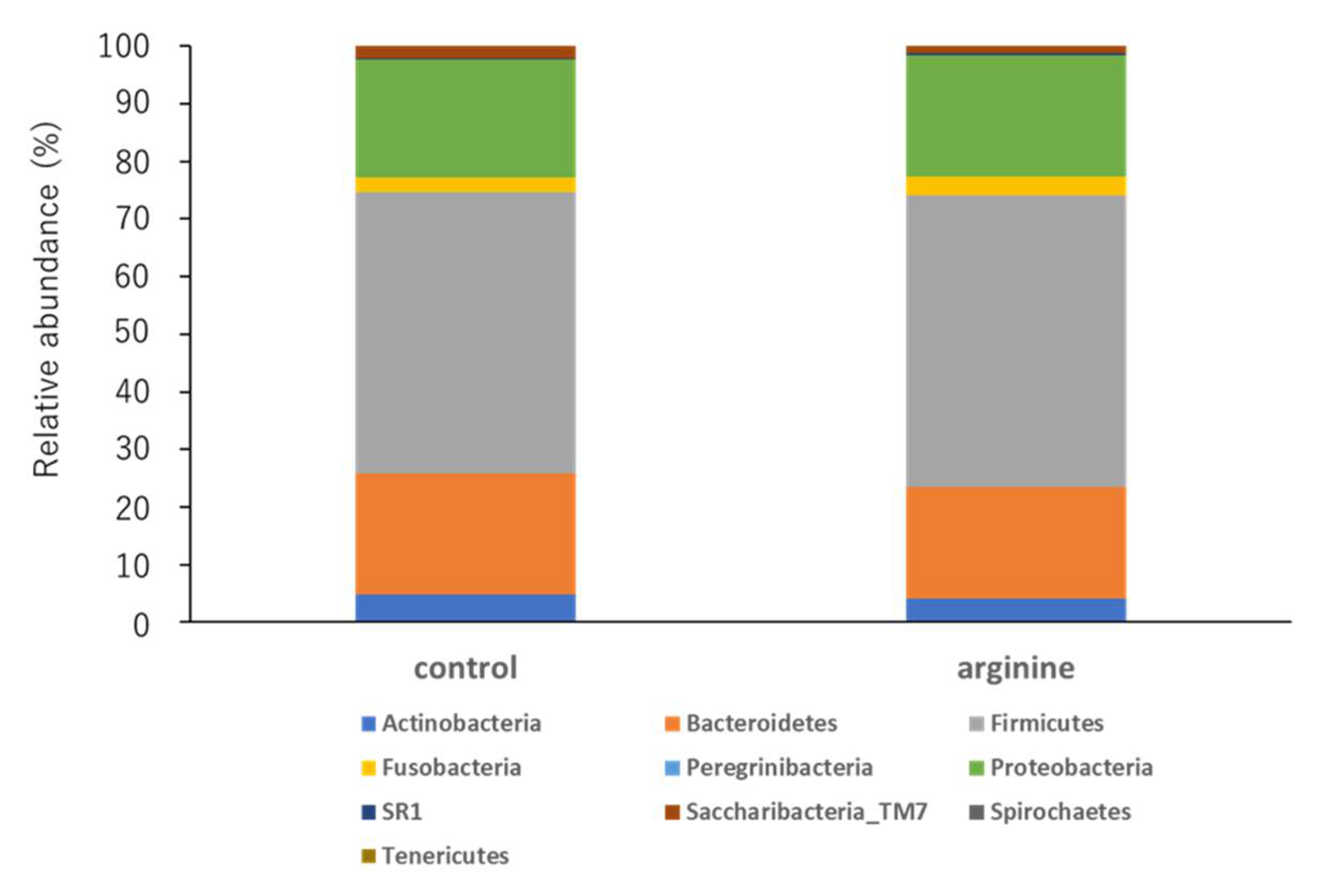
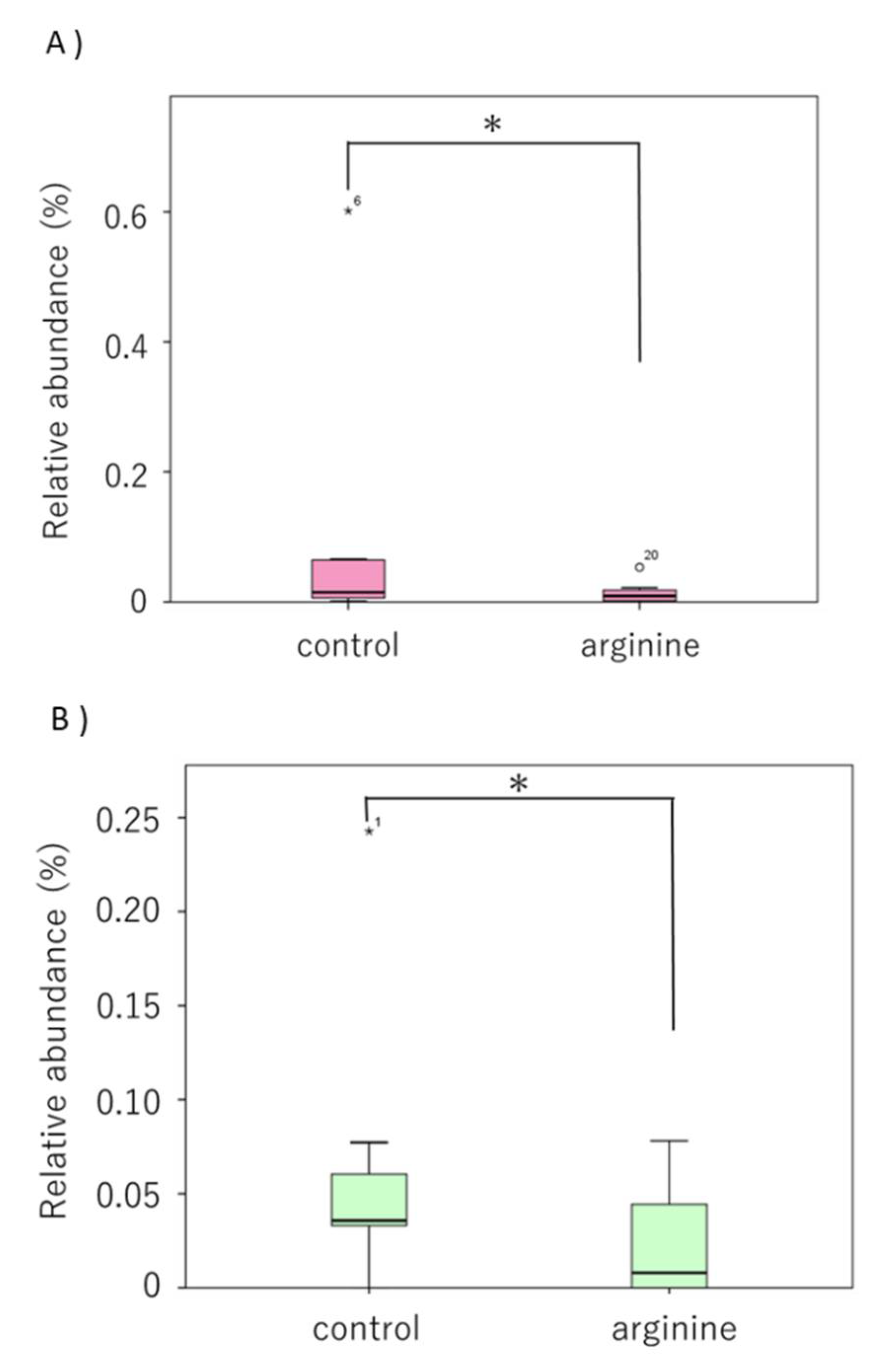
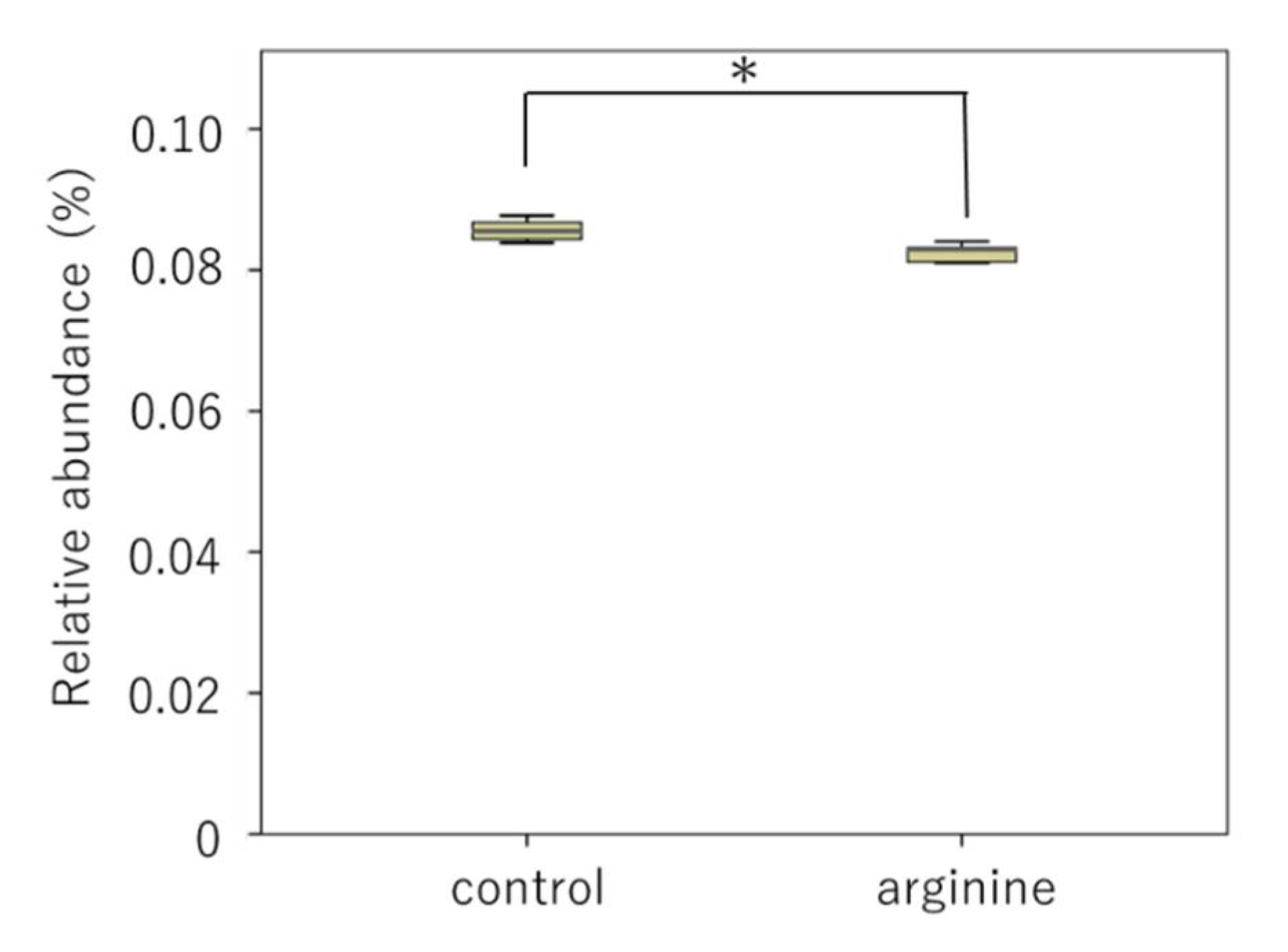
3.2.2. Effects of Arginine on the Dental Biofilm Flora and Functional Factors In Situ
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroes, I.; Lepp, P.W.; Relman, D.A. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 1999, 96, 14547–14552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ten Cate, J.M.; Klis, F.M.; Pereira-Cenci, T.; Crielaard, W.; de Groot, P.W. Molecular and cellular mechanisms that lead to Candida biofilmformation. J. Dent. Res. 2009, 88, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Dmytrenko, G.; Tana, L.; Cachau, M.V.; Bravo, M.; Gonzalez, S.; Correa, F.; Fernandez-Solari, J.; De Laurentiis, A. Presence of Trichomonas spp. in oral ulcerations of a patient with kidney transplant. A case report. J. Clin. Exp. Dent. 2020, 12, e1201. [Google Scholar] [CrossRef] [PubMed]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the oral microbiota in health: Mechanisms that prevent dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef]
- Marsh, P.D. Are dental diseases examples of ecological catastrophes? Microbiology 2003, 149, 279–294. [Google Scholar] [CrossRef]
- Marsh, P.D. Dental plaque as a biofilm and amicrobial community–implications for health and disease. BMC Oral Health 2006, 15, 6–12. [Google Scholar] [CrossRef] [Green Version]
- Darveau, R.P.; Hajishengallis, G.; Curtis, M.A. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 2012, 91, 816–820. [Google Scholar] [CrossRef] [Green Version]
- Kolenbrander, P.E.; Palmer, R.J., Jr.; Rickard, A.H.; Jakubovics, N.S.; Chalmers, N.I.; Diaz, P.I. Bacterial interactions and successions during plaque development. Periodontology 2000 2006, 42, 47–79. [Google Scholar] [CrossRef]
- Ho, M.H.; Lamont, R.J.; Xie, H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci. Rep. 2017, 7, 1413. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Wake, N.; Asahi, Y.; Noiri, Y.; Hayashi, M.; Motooka, D.; Nakamura, S.; Gotoh, K.; Miura, J.; Machi, H.; Iida, T.; et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ Biofilms Microbiomes 2016, 2, 16018. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.E.; Moore, L.V. The bacteria of periodontal diseases. Periodontology 2000 1994, 5, 66–77. [Google Scholar] [CrossRef]
- Linden, G.J.; Lyons, A.; Scannapieco, F.A. Periodontal systemic associations: Review of the evidence. J. Peridontol. 2013, 84, S8–S19. [Google Scholar] [CrossRef]
- Asahi, Y.; Noiri, Y.; Igarashi, J.; Asai, H.; Suga, H.; Ebisu, S. Effects of N-acyl homoserine lactone analogues on Porphyromonas gingivalis biofilm formation. J. Periodontal. Res. 2010, 45, 255–261. [Google Scholar] [CrossRef]
- Maezono, H.; Noiri, Y.; Asahi, Y.; Yamaguchi, M.; Yamamoto, R.; Izutani, N.; Azakami, H.; Ebisu, S. Antibiofilm effects of azithromycin and erythromycin on Porphyromonas gingivalis. Antimicrob. Agents Chemother. 2011, 55, 5887–5892. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Burne, R.A.; Zeng, L.; Ahn, S.J.; Palmer, S.R.; Liu, Y.; Lefebure, T.; Stanhope, M.J.; Nascimento, M.M. Progress dissecting the oral microbiome in caries and health. Adv. Dent. Res. 2012, 24, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Lavender, S.; Woo, J.; Guo, L.; Shi, W.; Kilpatrick-Liverman, L.; Gimzewski, J.K. Nanoscale characterization of effect of L-arginine on Streptococcus mutans biofilm adhesion by atomic force microscopy. Microbiology 2014, 160, 1466–1473. [Google Scholar] [CrossRef] [PubMed]
- Koopman, J.E.; Röling, W.F.; Buijs, M.J.; Sissons, C.H.; Jacob, M.; Keijser, B.J.; Crielaard, W.; Zaura, E. Stability and resilience of oral microcosms toward acidification and Candida outgrowth by arginine supplementation. Microb. Ecol. 2015, 69, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lin, X.; Wang, B.Y.; Wu, J.; Lamont, R.J. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 2007, 153, 3228–3234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wuyckhuyse, B.C.; Perinpanayagam, H.E.; Bevacqua, D.; Raubertas, R.F.; Billings, R.J.; Bowen, W.H.; Tabak, L.A. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J. Dent. Res. 1995, 74, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Kraivaphan, P.; Amornchat, C.; Triratana, T.; Mateo, L.R.; Ellwood, R.; Cummins, D.; DeVizio, W.; Zhang, Y.P. Two-year caries clinical study of the efficacy of novel dentifrices containing 1.5% arginine, an insoluble calcium compound and 1450 ppm fluoride. Caries Res. 2013, 47, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Otten, M.P.; Busscher, H.J.; van der Mei, H.C.; van Hoogmoed, C.G.; Abbas, F. Acute and substantive action of antimicrobial toothpastes and mouthrinses on oral biofilm in vitro. Eur. J. Oral Sci. 2011, 119, 151–155. [Google Scholar] [CrossRef] [Green Version]
- Dehghani, S.; Rezaee, A.; Hosseinkhani, S. Effect of alternating electrical current on denitrifying bacteria in a microbial electrochemical system: Biofilm viability and ATP assessment. Environ. Sci. Pollut. Res. Int. 2018, 25, 33591–33598. [Google Scholar] [CrossRef]
- Aagaard, K.; Petrosino, J.; Keitel, W.; Watson, M.; Katancik, J.; Garcia, N.; Patel, S.; Cutting, M.; Madden, T.; Hamilton, H.; et al. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J. 2013, 27, 1012–1022. [Google Scholar] [CrossRef]
- Beyth, N.; Yudovin-Farber, I.; Perez-Davidi, M.; Domb, A.J.; Weiss, E.I. Polyethyleneimine nanoparticles incorporated into resin composite cause cell death and trigger biofilm stress in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 22038–22043. [Google Scholar] [CrossRef] [Green Version]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesion. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdullah, N.; Al-Marzooq, F.; Mohamad, S.; Abd Rahman, N.; Chi Ngo, H.; Perera Samaranayake, L. Intraoral appliances for in situ oral biofilm growth: A systematic review. J. Oral Microbiol. 2019, 11, 1647757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, P.D. In sickness and in health-what does the oral microbiome mean to us? An ecological perspective. Adv. Dent. Res. 2018, 29, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Hale, J.D.; Heng, N.C.; Tagg, J.R. Developing oral probiotics from Streptococcus salivarius. Future Microbiol. 2012, 7, 1355–1371. [Google Scholar] [CrossRef]
- Sissons, C.H.; Hancock, E.M.; Perinpanayagam, H.E.; Cutress, T.W. The bacteria responsible for ureolysis in artificial dental plaque. Arch. Oral Biol. 1988, 33, 727–733. [Google Scholar] [CrossRef]
- Diaz, P.I.; Chalmers, N.I.; Rickard, A.H.; Kong, C.; Milburn, C.L.; Palmer, R.J., Jr.; Kolenbrander, P.E. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 2006, 72, 2837–2848. [Google Scholar] [CrossRef] [Green Version]
- Lang, N.P.; Cumming, B.R.; Löe, H. Toothbrushing frequency as it relates to plaque development and gingival health. J. Periodontol. 1973, 44, 396–405. [Google Scholar] [CrossRef]
- Belda-Ferre, P.; Alcaraz, L.D.; Cabrera-Rubio, R.; Romero, H.; Simón-Soro, A.; Pignatelli, M.; Mira, A. The oral metagenome in health and disease. ISME J. 2012, 6, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Aas, J.A.; Griffen, A.L.; Dardis, S.R.; Lee, A.M.; Olsen, I.; Dewhirst, F.E.; Leys, E.J.; Paster, B.J. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008, 46, 1407–1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mansell, J.; Gourtsoyannis, Y.; Draz, N.; Buchanan, R. Infective endocarditis due to Atopobium vaginae: A rare association between genital infection and endocarditis of the tricuspid valve. BMJ Case Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Hamma, T.; Ferré-D’Amaré, A.R. Pseudouridine synthases. Chem. Biol. 2006, 13, 1125–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rintala-Dempsey, A.C.; Kothe, U. Eukaryotic stand-alone pseudouridine synthases—RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017, 14, 1185–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuriki, N.; Asahi, Y.; Sotozono, M.; Machi, H.; Noiri, Y.; Hayashi, M.; Ebisu, S. Next-Generation Sequencing for Determining the Effect of Arginine on Human Dental Biofilms Using an In Situ Model. Pharmacy 2021, 9, 18. https://doi.org/10.3390/pharmacy9010018
Kuriki N, Asahi Y, Sotozono M, Machi H, Noiri Y, Hayashi M, Ebisu S. Next-Generation Sequencing for Determining the Effect of Arginine on Human Dental Biofilms Using an In Situ Model. Pharmacy. 2021; 9(1):18. https://doi.org/10.3390/pharmacy9010018
Chicago/Turabian StyleKuriki, Nanako, Yoko Asahi, Maki Sotozono, Hiroyuki Machi, Yuichiro Noiri, Mikako Hayashi, and Shigeyuki Ebisu. 2021. "Next-Generation Sequencing for Determining the Effect of Arginine on Human Dental Biofilms Using an In Situ Model" Pharmacy 9, no. 1: 18. https://doi.org/10.3390/pharmacy9010018
APA StyleKuriki, N., Asahi, Y., Sotozono, M., Machi, H., Noiri, Y., Hayashi, M., & Ebisu, S. (2021). Next-Generation Sequencing for Determining the Effect of Arginine on Human Dental Biofilms Using an In Situ Model. Pharmacy, 9(1), 18. https://doi.org/10.3390/pharmacy9010018




