Roles of Micropillar Topography and Surface Energy on Cancer Cell Dynamics
Abstract
1. Introduction
2. Materials and Methods
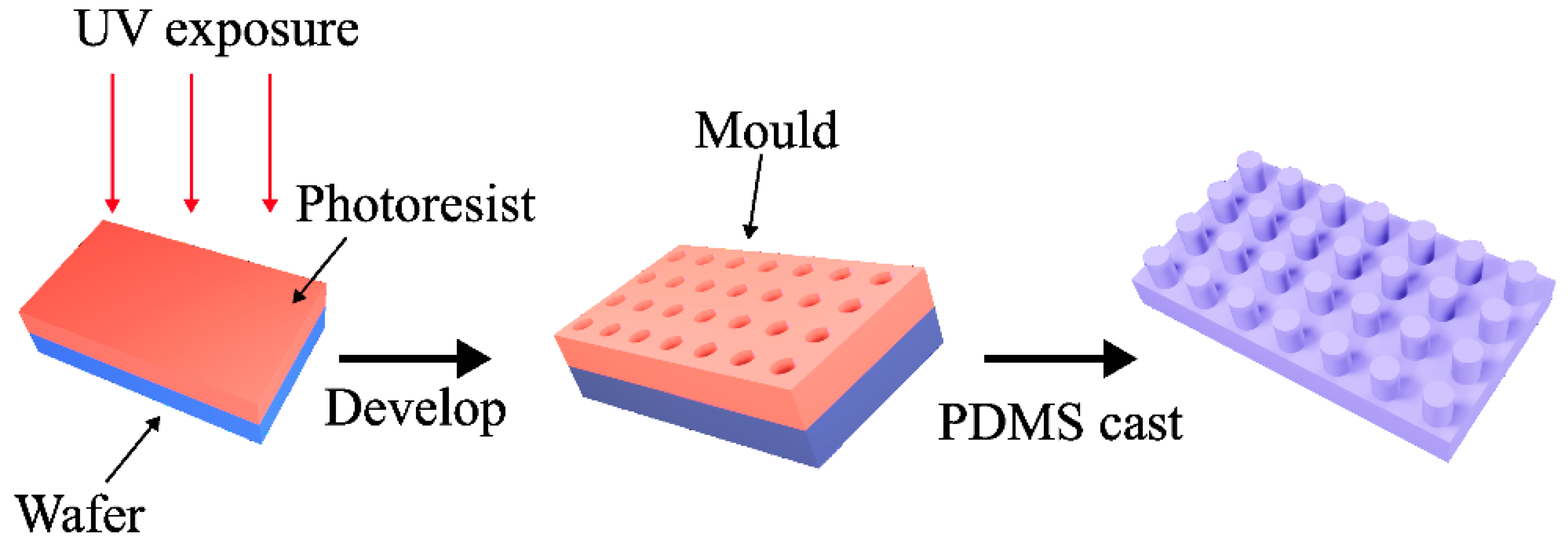
2.1. Fabrication of the Mould
2.2. Cell Culture
2.3. Quantification and Detection of MDA MB-231
2.4. Statistical Analysis
2.5. Immunofluorescence Staining
2.6. Fluorescence Microscopy
2.7. Contact Angle Measurement
2.8. Surface Free Energy Measurement
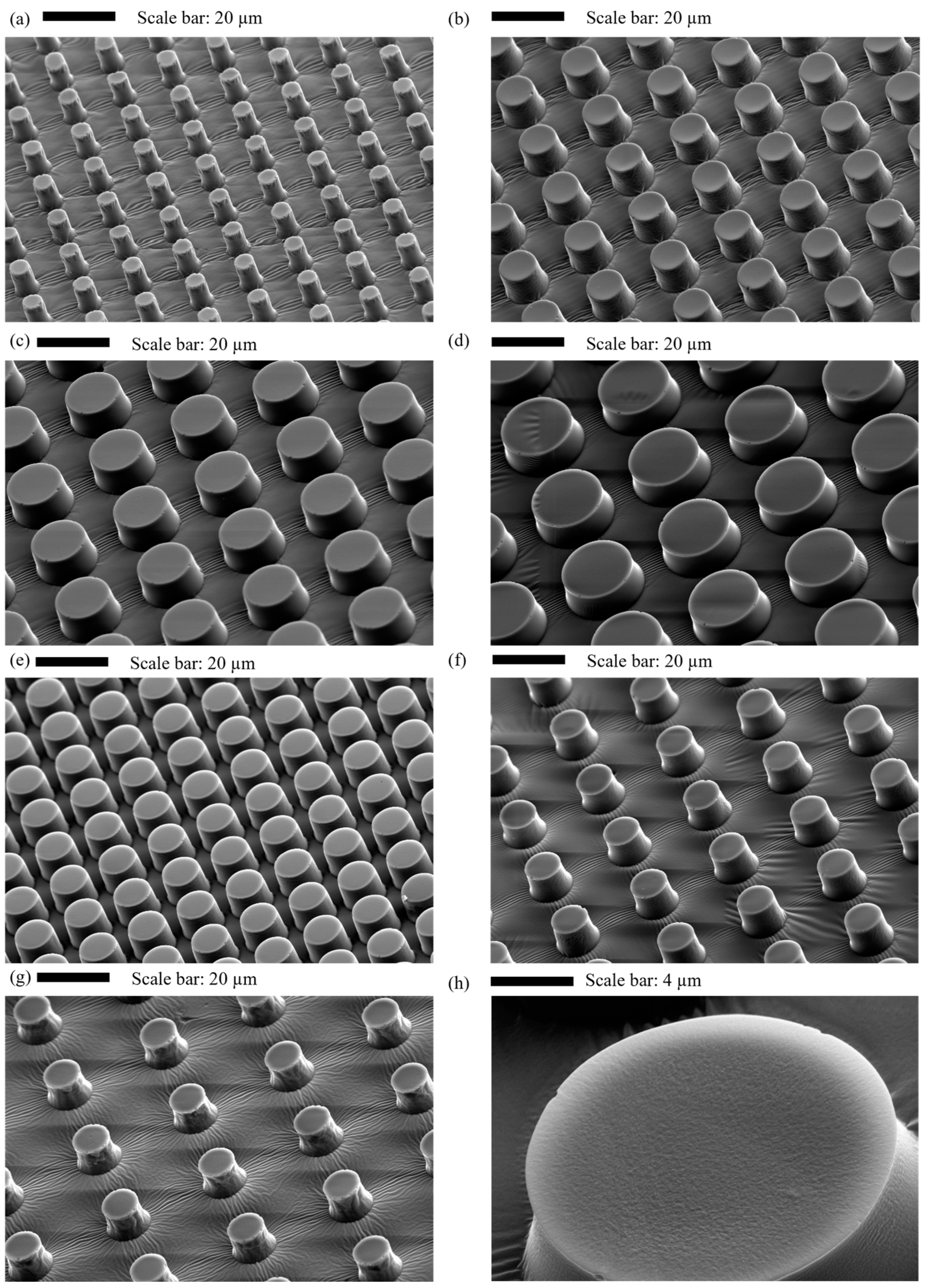
2.9. Characterisation of Micropillar Surfaces
3. Results and Discussion
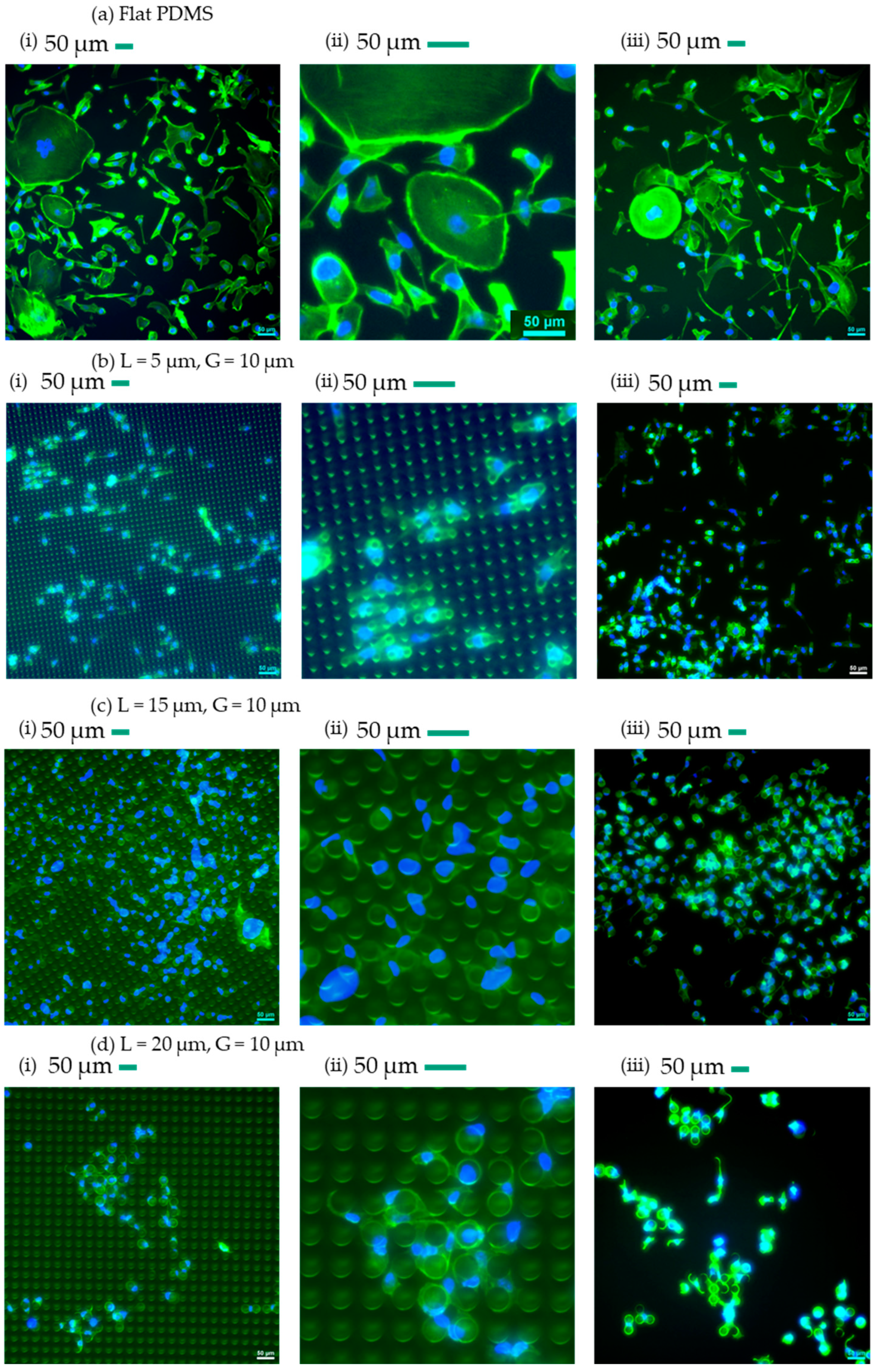
3.1. Qualitative Analysis of Cells’ Behaviour on Micropillars
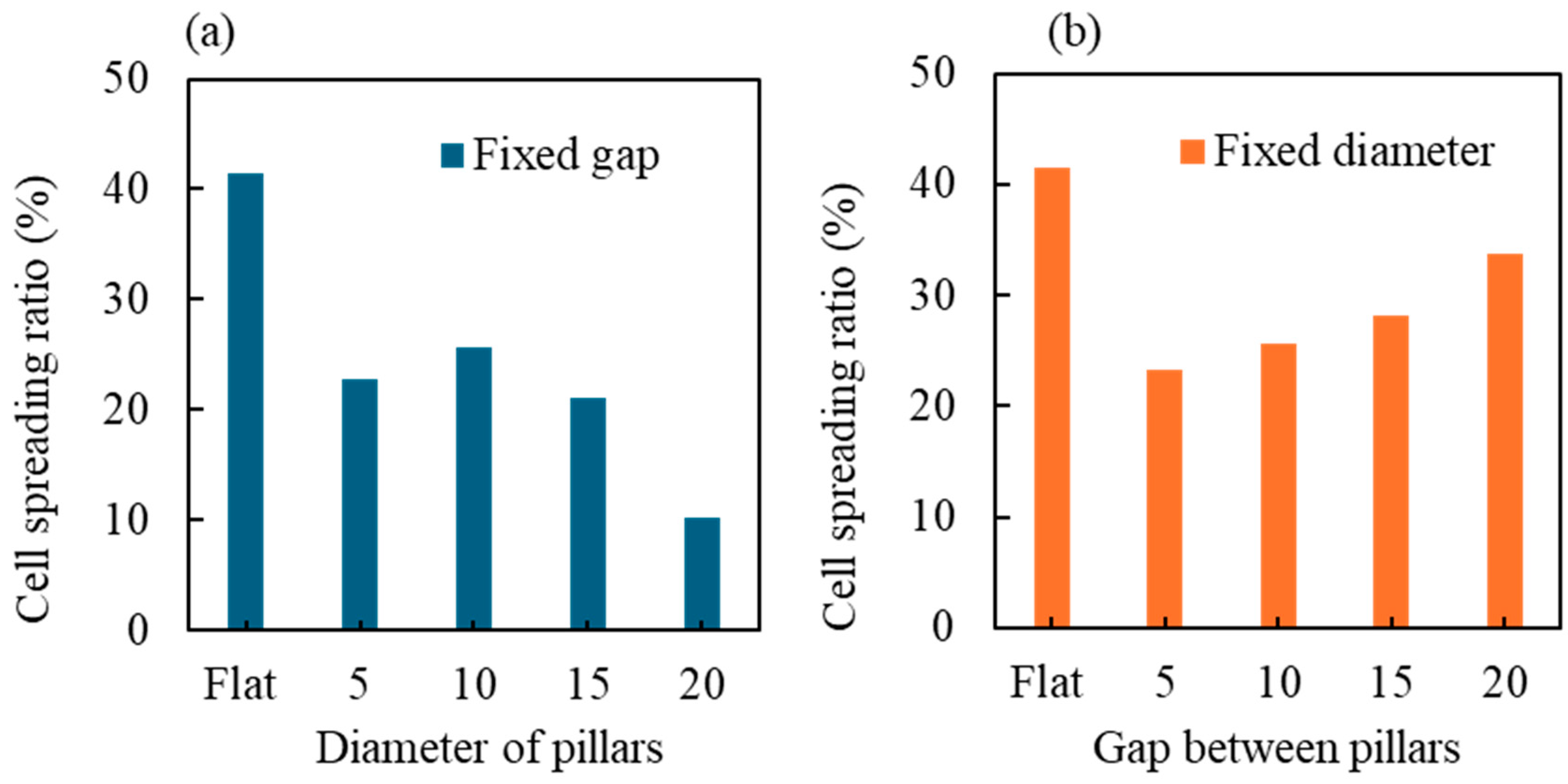
3.2. Quantitative Analysis of Cells’ Behaviour on Micropillars
3.3. Relationship of Cells’ Behaviour and Normalised Surface Free Energy of Micropillar Surfaces
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alves, N.M.; Pashkuleva, I.; Reis, R.L.; Mano, J.F. Controlling cell behavior through the design of polymer surfaces. Small 2010, 6, 2208–2220. [Google Scholar] [CrossRef] [PubMed]
- Saw, T.B.; Jain, S.; Ladoux, B.; Lim, C.T. Mechanobiology of collective cell migration. Cell. Mol. Bioeng. 2015, 8, 3–13. [Google Scholar] [CrossRef]
- Xie, W.; Wei, X.; Kang, H.; Jiang, H.; Chu, Z.; Lin, Y.; Hou, Y.; Wei, Q. Static and dynamic: Evolving biomaterial mechanical properties to control cellular mechanotransduction. Adv. Sci. 2023, 10, 2204594. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, R.A.; Ran, M.; Wang, Y.; Yan, J.; Zhang, Y.; Rosenholm, J.M.; Zhang, H. A pH/temperature responsive nanocomposite for chemo-photothermal synergistic cancer therapy. Smart Mater. Med. 2023, 4, 199–211. [Google Scholar] [CrossRef]
- Girard, P.P.; Cavalcanti-Adam, E.A.; Kemkemer, R.; Spatz, J.P. Cellular chemomechanics at interfaces: Sensing, integration and response. Soft Matter 2007, 3, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Weiss, P. Nerve patterns: The mechanisms of nerve growth. Growth 1941, 5, 163–203. [Google Scholar]
- Harrison, R.G. The reaction of embryonic cells to solid structures. J. Exp. Zool. 1914, 17, 521–544. [Google Scholar] [CrossRef]
- Stoker, M.; O’Neill, C.; Berryman, S.; Waxman, V. Anchorage and growth regulation in normal and virus-transformed cells. Int. J. Cancer 1968, 3, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Azioune, A.; Storch, M.; Bornens, M.; Théry, M.; Piel, M. Simple and rapid process for single cell micro-patterning. Lab A Chip 2009, 9, 1640–1642. [Google Scholar] [CrossRef]
- Walker, G.M.; Zeringue, H.C.; Beebe, D.J. Microenvironment design considerations for cellular scale studies. Lab A Chip 2004, 4, 91–97. [Google Scholar] [CrossRef]
- Craighead, H.G.; James, C.; Turner, A. Chemical and topographical patterning for directed cell attachment. Curr. Opin. Solid State Mater. Sci. 2001, 5, 177–184. [Google Scholar] [CrossRef]
- Kleinfeld, D.; Kahler, K.; Hockberger, P. Controlled outgrowth of dissociated neurons on patterned substrates. J. Neurosci. 1988, 8, 4098–4120. [Google Scholar] [CrossRef] [PubMed]
- Stenger, D.A.; Georger, J.H.; Dulcey, C.S.; Hickman, J.J.; Rudolph, A.S.; Nielsen, T.B.; McCort, S.M.; Calvert, J.M. Coplanar molecular assemblies of amino-and perfluorinated alkylsilanes: Characterization and geometric definition of mammalian cell adhesion and growth. J. Am. Chem. Soc. 1992, 114, 8435–8442. [Google Scholar] [CrossRef]
- Lom, B.; Healy, K.E.; Hockberger, P.E. A versatile technique for patterning biomolecules onto glass coverslips. J. Neurosci. Methods 1993, 50, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Brunette, D. Fibroblasts on micromachined substrata orient hierarchically to grooves of different dimensions. Exp. Cell Res. 1986, 164, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.; Brown, A. Alignment of fibroblasts on grooved surfaces described by a simple geometric transformation. J. Cell Sci. 1986, 83, 313–340. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.; Connolly, P.; Curtis, A.; Dow, J.; Wilkinson, C. Topographical control of cell behaviour: I. Simple step cues. Development 1987, 99, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.; Connolly, P.; Curtis, A.; Dow, J.; Wilkinson, C. Topographical control of cell behaviour: II. Multiple grooved substrata. Development 1990, 108, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Nikkhah, M.; Strobl, J.S.; Agah, M. Attachment and response of human fibroblast and breast cancer cells to three dimensional silicon microstructures of different geometries. Biomed. Microdevices 2009, 11, 429–441. [Google Scholar] [CrossRef]
- Bettinger, C.J.; Orrick, B.; Misra, A.; Langer, R.; Borenstein, J.T. Microfabrication of poly (glycerol–sebacate) for contact guidance applications. Biomaterials 2006, 27, 2558–2565. [Google Scholar] [CrossRef]
- Turner, A.; Dowell, N.; Turner, S.; Kam, L.; Isaacson, M.; Turner, J.; Craighead, H.; Shain, W. Attachment of astroglial cells to microfabricated pillar arrays of different geometries. J. Biomed. Mater. Res. 2000, 51, 430–441. [Google Scholar] [CrossRef]
- Charest, J.L.; García, A.J.; King, W.P. Myoblast alignment and differentiation on cell culture substrates with microscale topography and model chemistries. Biomaterials 2007, 28, 2202–2210. [Google Scholar] [CrossRef] [PubMed]
- Dusseiller, M.R.; Schlaepfer, D.; Koch, M.; Kroschewski, R.; Textor, M. An inverted microcontact printing method on topographically structured polystyrene chips for arrayed micro-3-D culturing of single cells. Biomaterials 2005, 26, 5917–5925. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Andersson, A.-S.; Sutherland, D.; Petronis, S.; Kasemo, B.; Thomsen, P. Response of rat osteoblast-like cells to microstructured model surfaces in vitro. Biomaterials 2003, 24, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Sun, C.; Li, S.; Zhang, X. A microfabricated platform probing cytoskeleton dynamics using multidirectional topographical cues. Biomed. Microdevices 2007, 9, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Rovensky, Y.A.; Slavnaja, I.; Vasiliev, J.M. Behaviour of fibroblast-like cells on grooved surfaces. Exp. Cell Res. 1971, 65, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Von Recum, A.; Van Kooten, T. The influence of micro-topography on cellular response and the implications for silicone implants. J. Biomater. Sci. Polym. Ed. 1996, 7, 181–198. [Google Scholar] [CrossRef]
- Clark, P. Cell behaviour on micropatterned surfaces. Biosens. Bioelectron. 1994, 9, 657–661. [Google Scholar] [CrossRef]
- Curtis, A.S.; Wilkinson, C.D. Reactions of cells to topography. J. Biomater. Sci. Polym. Ed. 1998, 9, 1313–1329. [Google Scholar] [CrossRef]
- van Kooten, T.G.; Whitesides, J.F.; von Recum, A.F. Influence of silicone (PDMS) surface texture on human skin fibroblast proliferation as determined by cell cycle analysis. J. Biomed. Mater. Res. 1998, 43, 1–14. [Google Scholar] [CrossRef]
- Walboomers, X.; Monaghan, W.; Curtis, A.; Jansen, J. Attachment of fibroblasts on smooth and microgrooved polystyrene. J. Biomed. Mater. Res. 1999, 46, 212–220. [Google Scholar] [CrossRef]
- Milner, K.R.; Siedlecki, C.A. Submicron poly (L-lactic acid) pillars affect fibroblast adhesion and proliferation. J. Biomed. Mater. Res. Part A 2007, 82, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, C.; Riehle, M.; Wood, M.; Gallagher, J.; Curtis, A. The use of materials patterned on a nano-and micro-metric scale in cellular engineering. Mater. Sci. Eng. C 2002, 19, 263–269. [Google Scholar] [CrossRef]
- Antmen, E.; Demirci, U.; Hasirci, V. Amplification of nuclear deformation of breast cancer cells by seeding on micropatterned surfaces to better distinguish their malignancies. Colloids Surf. B Biointerfaces 2019, 183, 110402. [Google Scholar] [CrossRef] [PubMed]
- Jungbauer, S.; Kemkemer, R.; Gruler, H.; Kaufmann, D.; Spatz, J.P. Cell Shape Normalization, Dendrite Orientation, and Melanin Production of Normal and Genetically Altered (Haploinsufficient NF1)-Melanocytes by Microstructured Substrate Interactions. ChemPhysChem 2004, 5, 85–92. [Google Scholar] [CrossRef]
- Arima, Y.; Iwata, H. Effect of wettability and surface functional groups on protein adsorption and cell adhesion using well-defined mixed self-assembled monolayers. Biomaterials 2007, 28, 3074–3082. [Google Scholar] [CrossRef] [PubMed]
- Benoit, D.S.; Schwartz, M.P.; Durney, A.R.; Anseth, K.S. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat. Mater. 2008, 7, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.Y.; Shaughnessy, M.C.; Zhou, Z.; Noh, H.; Vogler, E.A.; Donahue, H.J. Surface energy effects on osteoblast spatial growth and mineralization. Biomaterials 2008, 29, 1776–1784. [Google Scholar] [CrossRef]
- Vu, H.H.; Nguyen, N.T.; Kashaninejad, N. Re-Entrant Microstructures for Robust Liquid Repellent Surfaces. Adv. Mater. Technol. 2023, 8, 2370023. [Google Scholar] [CrossRef]
- Alves, N.M.; Shi, J.; Oramas, E.; Santos, J.L.; Tomás, H.; Mano, J.F. Bioinspired superhydrophobic poly (L-lactic acid) surfaces control bone marrow derived cells adhesion and proliferation. J. Biomed. Mater. Res. Part A 2009, 91, 480–488. [Google Scholar] [CrossRef]
- Song, W.; Veiga, D.D.; Custódio, C.A.; Mano, J.F. Bioinspired degradable substrates with extreme wettability properties. Adv. Mater. 2009, 21, 1830–1834. [Google Scholar] [CrossRef]
- Xu, C.; Yang, F.; Wang, S.; Ramakrishna, S. In vitro study of human vascular endothelial cell function on materials with various surface roughness. J. Biomed. Mater. Res. Part A 2004, 71, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Owen, G.R.; Jackson, J.; Chehroudi, B.; Burt, H.; Brunette, D. A PLGA membrane controlling cell behaviour for promoting tissue regeneration. Biomaterials 2005, 26, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- Majhy, B.; Priyadarshini, P.; Sen, A. Effect of surface energy and roughness on cell adhesion and growth–facile surface modification for enhanced cell culture. RSC Adv. 2021, 11, 15467–15476. [Google Scholar] [CrossRef]
- Sheidaei, Z.; Akbarzadeh, P.; Kashaninejad, N. Advances in numerical approaches for microfluidic cell analysis platforms. J. Sci. Adv. Mater. Devices 2020, 5, 295–307. [Google Scholar] [CrossRef]
- Kashaninejad, N.; Nguyen, N.-T.; Chan, W.K. Eccentricity effects of microhole arrays on drag reduction efficiency of microchannels with a hydrophobic wall. Phys. Fluids 2012, 24, 112004. [Google Scholar] [CrossRef]
- Kashaninejad, N.; Chan, W.K.; Nguyen, N.-T. Eccentricity effect of micropatterned surface on contact angle. Langmuir 2012, 28, 4793–4799. [Google Scholar] [CrossRef]
- Moghadas, H.; Saidi, M.S.; Kashaninejad, N.; Nguyen, N.-T. A high-performance polydimethylsiloxane electrospun membrane for cell culture in lab-on-a-chip. Biomicrofluidics 2018, 12, 024117. [Google Scholar] [CrossRef]
- Yadav, S.; Kashaninejad, N.; Nguyen, N.-T. RhoA and Rac1 in Liver Cancer Cells: Induction of Overexpression Using Mechanical Stimulation. Micromachines 2020, 11, 729. [Google Scholar] [CrossRef]
- Taghibakhshi, A.; Barisam, M.; Saidi, M.S.; Kashaninejad, N.; Nguyen, N.-T. Three-Dimensional Modeling of Avascular Tumor Growth in Both Static and Dynamic Culture Platforms. Micromachines 2019, 10, 580. [Google Scholar] [CrossRef]
- Vu, H.H.; Nguyen, N.-T.; Nguyen, N.-K.; Luu, C.H.; Hettiarachchi, S.; Kashaninejad, N. Tunable wettability with stretchable microstructured surfaces. Adv. Eng. Mater. 2023, 25, 2300821. [Google Scholar] [CrossRef]
- Annamalai, M.; Gopinadhan, K.; Han, S.A.; Saha, S.; Park, H.J.; Cho, E.B.; Kumar, B.; Patra, A.; Kim, S.-W.; Venkatesan, T. Surface energy and wettability of van der Waals structures. Nanoscale 2016, 8, 5764–5770. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.K.; Wendt, R. Estimation of the surface free energy of polymers. J. Appl. Polym. Sci. 1969, 13, 1741–1747. [Google Scholar] [CrossRef]
- Kaelble, D. Dispersion-polar surface tension properties of organic solids. J. Adhes. 1970, 2, 66–81. [Google Scholar] [CrossRef]
- Shaker, M.; Salahinejad, E. A combined criterion of surface free energy and roughness to predict the wettability of non-ideal low-energy surfaces. Prog. Org. Coat. 2018, 119, 123–126. [Google Scholar] [CrossRef]
- Liu, W.; Sun, Q.; Zheng, Z.L.; Gao, Y.T.; Zhu, G.Y.; Wei, Q.; Xu, J.Z.; Li, Z.M.; Zhao, C.S. Topographic cues guiding cell polarization via distinct cellular mechanosensing pathways. Small 2022, 18, 2104328. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Kang, Z.; Sun, Q.; Zhang, M.; Wang, P.; Li, Y.; Li, J.; Su, B.; Wei, Q. Microtubule Assists Actomyosin to Regulate Cell Nuclear Mechanics and Chromatin Accessibility. Research 2023, 6, 0054. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, T.; Nakajima, A.; Shimada, N.; Sawai, S. Tissue self-organization based on collective cell migration by contact activation of locomotion and chemotaxis. Proc. Natl. Acad. Sci. USA 2019, 116, 4291–4296. [Google Scholar] [CrossRef]
- Yanakieva, I.; Erzberger, A.; Matejčić, M.; Modes, C.D.; Norden, C. Cell and tissue morphology determine actin-dependent nuclear migration mechanisms in neuroepithelia. J. Cell Biol. 2019, 218, 3272–3289. [Google Scholar] [CrossRef]
- Sun, Q.; Pei, F.; Zhang, M.; Zhang, B.; Jin, Y.; Zhao, Z.; Wei, Q. Curved nanofiber network induces cellular bridge formation to promote stem cell mechanotransduction. Adv. Sci. 2023, 10, 2204479. [Google Scholar] [CrossRef]
- Masuda, Y.; Inami, W.; Miyakawa, A.; Kawata, Y. Cell culture on hydrophilicity-controlled silicon nitride surfaces. World J. Microbiol. Biotechnol. 2015, 31, 1977–1982. [Google Scholar] [CrossRef] [PubMed]
- TruongVo, T.; Kennedy, R.; Chen, H.; Chen, A.; Berndt, A.; Agarwal, M.; Zhu, L.; Nakshatri, H.; Wallace, J.; Na, S. Microfluidic channel for characterizing normal and breast cancer cells. J. Micromech. Microeng. 2017, 27, 035017. [Google Scholar] [CrossRef]
- Kim, Y.G.; Lim, N.; Kim, J.; Kim, C.; Lee, J.; Kwon, K.-H. Study on the surface energy characteristics of polydimethylsiloxane (PDMS) films modified by C4F8/O2/Ar plasma treatment. Appl. Surf. Sci. 2019, 477, 198–203. [Google Scholar] [CrossRef]
- Bettinger, C.; Langer, R.; Borenstein, J. Engineering substrate microand nanotopography to control cell function. Angew. Chem. Int. Ed. Engl. 2009, 48, 5406–5415. [Google Scholar] [CrossRef]

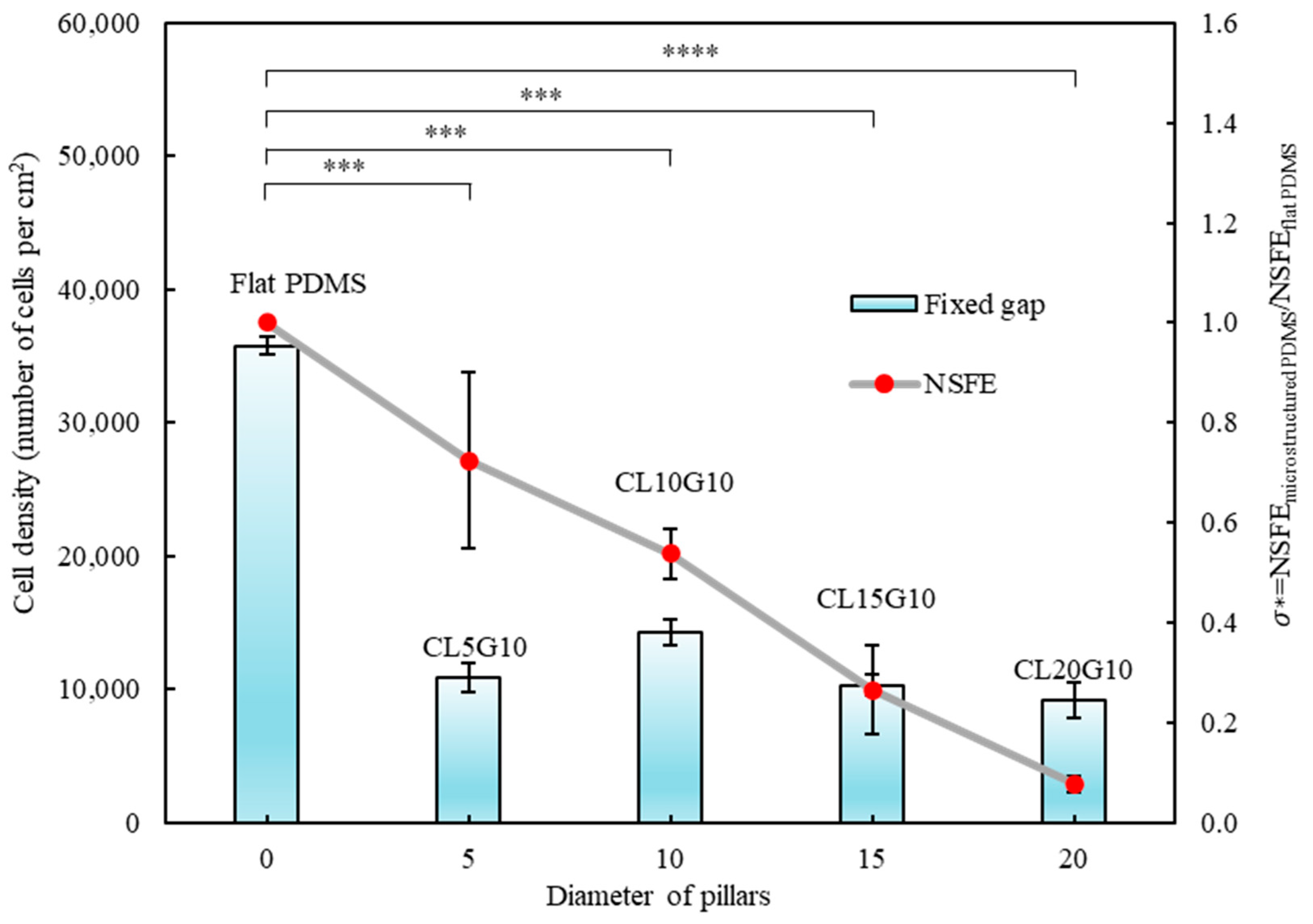
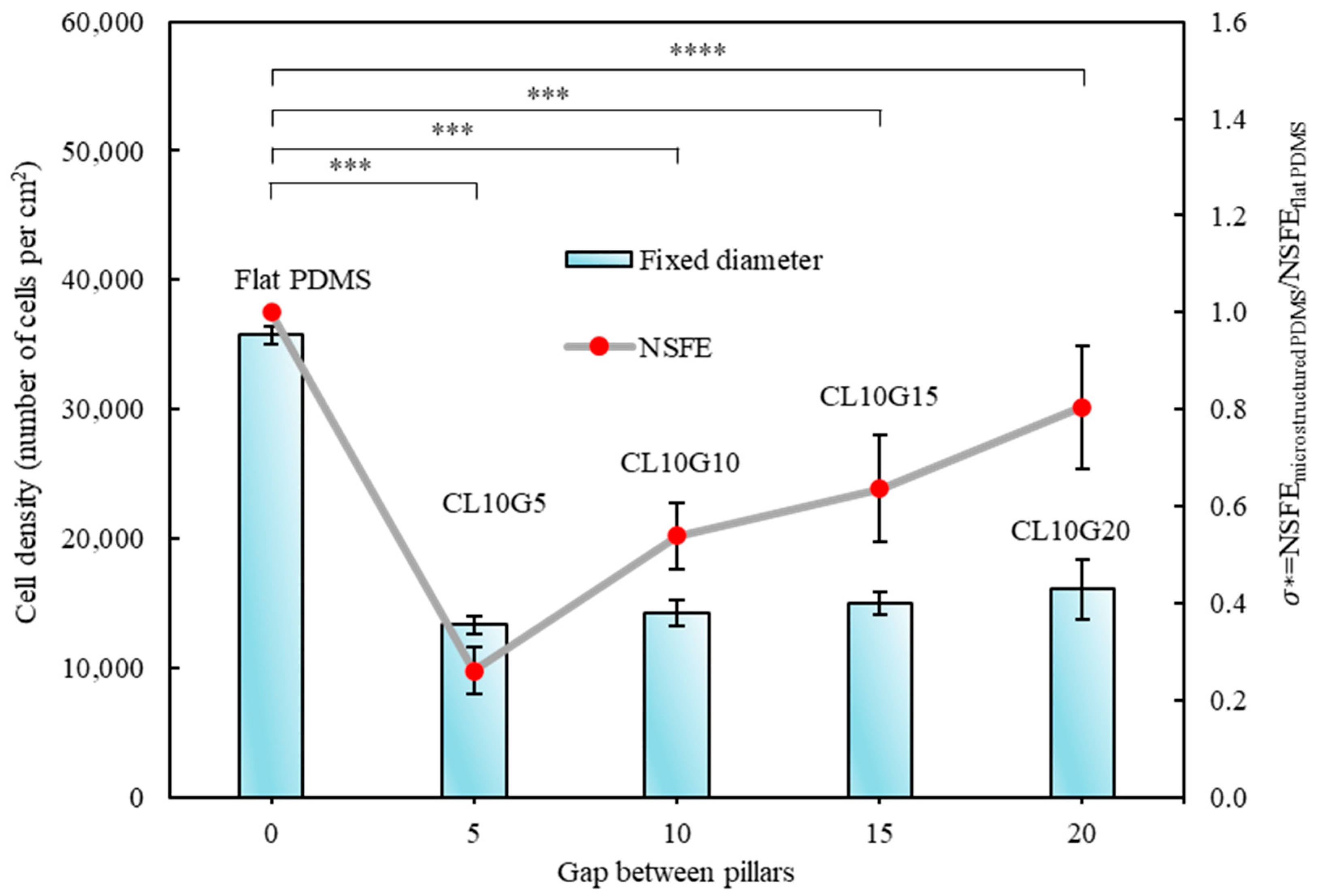
| L (µm) | G (µm) | Symbol |
|---|---|---|
| 5 | 10 | CL5G10 |
| 10 | 10 | CL10G10 |
| 15 | 10 | CL15G10 |
| 20 | 10 | CL20G10 |
| 10 | 5 | CL10G5 |
| 10 | 15 | CL10G15 |
| 10 | 20 | CL10G20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, H.H.; Nguyen, N.-T.; Yadav, S.; Nguyen, T.T.H.; Kashaninejad, N. Roles of Micropillar Topography and Surface Energy on Cancer Cell Dynamics. Technologies 2024, 12, 130. https://doi.org/10.3390/technologies12080130
Vu HH, Nguyen N-T, Yadav S, Nguyen TTH, Kashaninejad N. Roles of Micropillar Topography and Surface Energy on Cancer Cell Dynamics. Technologies. 2024; 12(8):130. https://doi.org/10.3390/technologies12080130
Chicago/Turabian StyleVu, Hoang Huy, Nam-Trung Nguyen, Sharda Yadav, Thi Thanh Ha Nguyen, and Navid Kashaninejad. 2024. "Roles of Micropillar Topography and Surface Energy on Cancer Cell Dynamics" Technologies 12, no. 8: 130. https://doi.org/10.3390/technologies12080130
APA StyleVu, H. H., Nguyen, N.-T., Yadav, S., Nguyen, T. T. H., & Kashaninejad, N. (2024). Roles of Micropillar Topography and Surface Energy on Cancer Cell Dynamics. Technologies, 12(8), 130. https://doi.org/10.3390/technologies12080130









