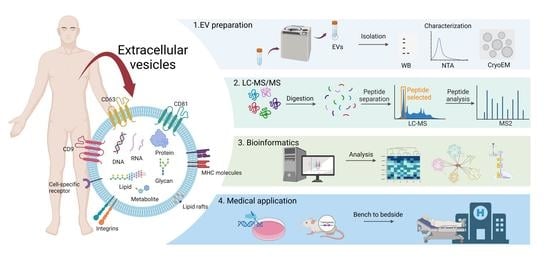
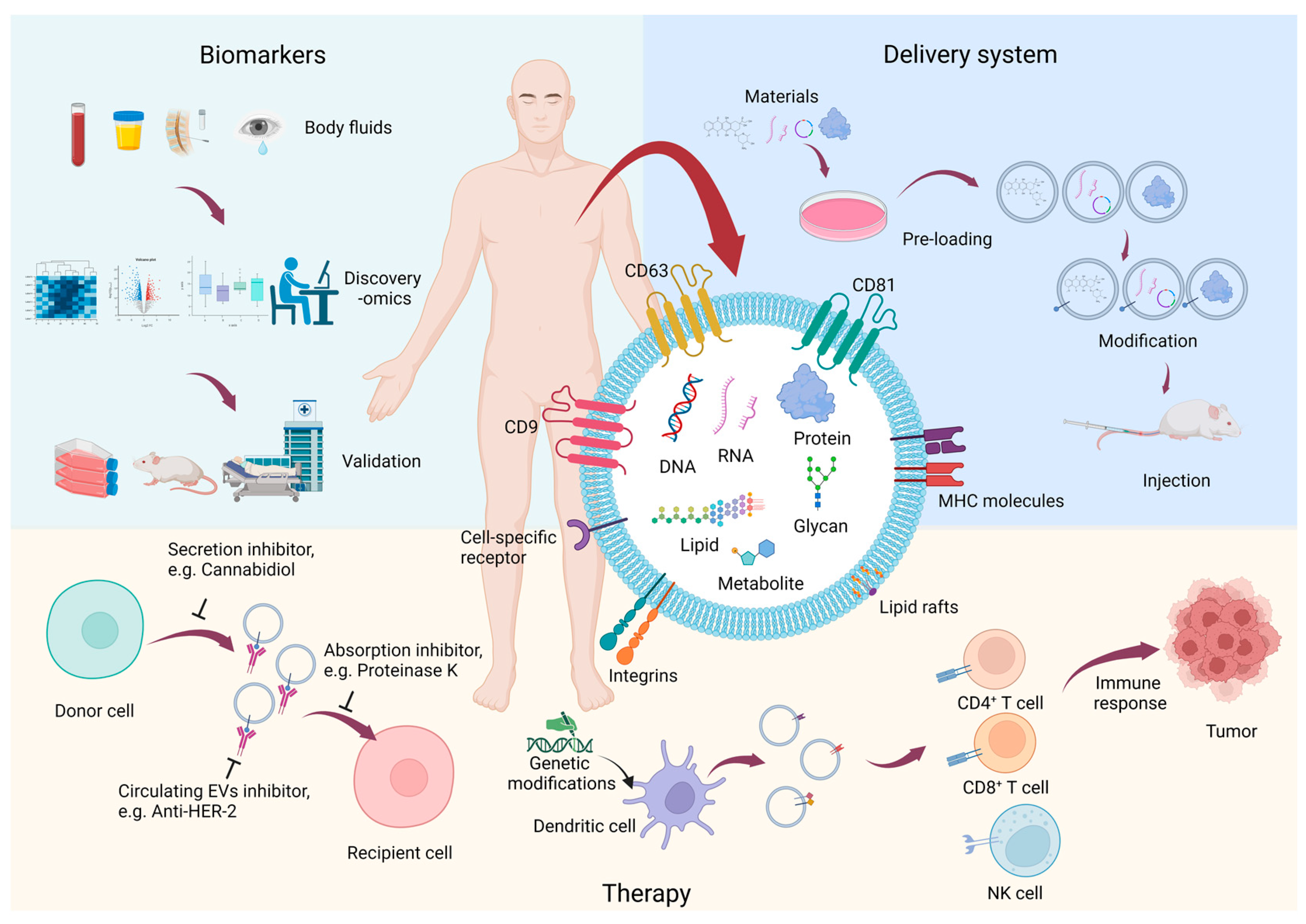
Proteomic Research of Extracellular Vesicles in Clinical Biofluid
Abstract
:1. Introduction
2. The Features of EVs
3. An Overview of the EV Biology in Disease Context
3.1. Cancer Progression
3.2. Immune Response Modulation
3.3. Neurodegenerative Disease
3.4. Viral Infection
4. Methods of EV Isolation
4.1. Conventional Approaches for the Isolation of EVs
4.2. Advanced Approaches for the Isolation of EVs
4.2.1. Asymmetric Flow Field Flow Fraction, AF4
4.2.2. Microfluidic-Based Technologies
4.2.3. Dichotomic SEC
4.2.4. Ultrafast-Isolation System, EXODUS
4.2.5. EV Enrichment Device, EVrich
4.2.6. Commercial Exosome Isolation Kits
4.2.7. Others
5. -Omics Approaches to Study EV in Clinical Biofluid
The Role of -Omics Methods in Clinical Applications of EVs
| Source | Biomarker | Isolation Method/Identification Method | Screening Method/Verification Method | Disease | Ref. |
|---|---|---|---|---|---|
| Serum | COPB2↑ | Filter column/WB, SEM | LC-MS/MS/WB, ELISA | COVID-19 | [140] |
| Plasma and serum | HSP90A↑, STIP1↑, TAGLN-2↑ | Ultrafiltration, differential centrifugation, density gradient centrifugation/TEM, NTA, WB, LVSEM | LC-MS/MS/WB | Adenomyosis | [141] |
| Plasma | PKG1↑, RALGAPA2↑, TJP2↑ | Ultracentrifugation/WB | LC-MS/MS/PRM | Breast Cancer | [142] |
| Plasma | TSPAN1↑ | Differential centrifugation, ExoQuick®/TEM, NTA, WB | LC-MS/MS/WB, ELISA | Colon Cancer | [143] |
| Serum | GCLM↓, KEL↑, APOF↑, CFB↓, PDE5A↓, ATIC↓ | Size-exclusion chromatography/TEM, WB | LC-MS/MS/NA | Colon Cancer | [43] |
| Blood | ORM1 | NA/NA | Large-scale targeted proteomics analysis/NA | Colon Cancer | [144] |
| Serum | Stratifin↑ | Size-exclusion chromatography, exoEasy kit/TEM, NTA, WB | LC-MS/MS (TMT)/ELISA | Colon Cancer | [45] |
| Serum | Annexin A3↑, A4↑, and A11↑ | Differential ultracentrifugation, density gradient centrifugation/NA | LC-MS/MS (SRM)/NA | Colon Cancer | [145] |
| Serum | TRIM3↓ | ExoQuick®/WB, TEM, NTA, | LC-MS/MS/ELISA, WB | Gastric Cancer | [146] |
| Plasma | TGFβ1↑ | Extracellular vesicles enrichment kit/TEM, NTA, WB | LC-MS/MS(TMT)/ELISA | Head and Neck Squamous Cell Carcinoma | [147] |
| Serum | AMPN↑, PIGR↑, VNN↑ | Filtration, ultracentrifugation/TEM, NTA, WB | LC-MS/MS/WB | Liver Cancer | [148] |
| Plasma | SRGN↑, TPM3↑, THBS1↑, HUWE1↑ | Density gradient flotation/TEM, NTA, WB | LC-MS/MS/WB | Lung Cancer | [149] |
| Serum | CD5L↑ | Precipitation and magnetic-based immunoaffinity/TEM, NTA, WB, DLS | MALDI-TOF-MS/WB | Lung Cancer | [150] |
| Serum | CD91↑ | MSIA monolith tips/NA | LC-MS/MS/ELISA | Lung Cancer | [151] |
| Serum | α-synuclein↑, Clusterin↑ | Immunoaffinity/SEM, NTA, WB | LC-MS/MS/electrochemiluminescence | Parkinson’s Disease | [152] |
| Serum | Syntenin-1↑ | Ultracentrifugation/NTA, EM, WB | LC-MS/MS/WB | Parkinson’s Disease | [153] |
| Plasma | IgG↑, IgM↑, C1q↑ | Immunoaffinity/flow cytometry | LC-MS/MS/NA | Systemic Lupus Erythematosus | [154] |
| Plasma | G3BP↑, TGFβ1↓ | Centrifugation/NA | LC-MS/MS/NA | Systemic Lupus Erythematosus | [155] |
| Urine | Hsp 90↑, syndecan-1↑, MARCKS↑, ZO-2↑ | Gradient density ultracentrifugation, differential ultracentrifugation/TEM, NTA, WB | LC-MS/MS(TMT/SRM/MRM)/immunohistochemical | Bladder Cancer | [156] |
| Urine | EHD4↑, EPS8L1↑, EPS8L2↑, GBP3↑, GsGTPa↑, GTPase Nras↑, MUC4↑, RAI3↑, Resistin↑ | Ultracentrifugation/WB | LC-MS/MS/WB | Bladder Cancer | [157] |
| Urine | APOA1↑, TTR↑, PIGR↑, HPX↑, AZGP1↑, CP↑ | Differential ultracentrifugation/protein concentration | LC-MS/MS (DDA)/WB | Chronic Active Antibody-Mediated Rejection | [158] |
| Urine | Calbindin↑, SNAP23↑ | Ultracentrifugation/NTA, TEM, WB | LC-MS/MS/WB | Parkinson’s Disease | [159] |
| Urine | AGP1↑ | Differential ultracentrifugation/TEM, NTA, WB | LC-MS/MS/WB | Primary Aldosteronism | [160] |
| Urine | AQP1↓, CAIX↑, CD10↓, CD147↓, CP↑, DKK4↑, DPEP1↓, MMP9↑, PODXL↑, Syntenin-1↓ | Differential centrifugation, density gradient ultracentrifugation, ultrafiltration/TEM, WB, NTA | LC-MS/MS/WB | Renal Cancer | [161] |
| Saliva | BASP1↑, NUCB2↑, PSMA7↑, PSMB7↑, TKT↑, TLN1↑, WDR1↑ | Centrifugation, exosome isolation kit/EM, WB | LC-MS/MS/WB | Inflammatory Bowel Disease/Ulcerative Colitis/Crohn’s Disease | [162] |
| Tear and Saliva | STOM↑, ANXA4↑, ANXA1↑ | Size-exclusion chromatography/NTA, flow cytometry | LC-MS/MS/NA | Primary Sjögren’s Syndrome | [163] |
| Source | Type | Biomarker | Isolation Method/Identification Method | Screening Method/Verification Method | Disease | Ref. |
|---|---|---|---|---|---|---|
| Serum | circular RNAs | Chr10q11↑, Chr1p11↑, Chr7q11↑ | exoRNeasy Midi kit, ultracentrifugation/TEM, NTA, WB | RNA Seq/RT-qPCR | Gastric Cancer | [164] |
| Plasma | circRNAs↑ | Differential centrifugation/cryo-EM, NTA | RNA-Seq/NA | Multiple Sclerosis | [165] | |
| Serum | long non-coding RNAs | HULC↑ | Ultracentrifugation/- | Microarray/RT-qPCR | Pancreatic Cancer | [166] |
| Serum | LINC00853↑ | ExoQuick/TEM, NTA, WB | RNA Seq/RT-qPCR | Hepatocellular Carcinoma | [167] | |
| Plasma | RP3-399L15.2↓, CH507-513H4.6↓ | exoRNeasy/TEM, NTA, WB | RNA Seq/RT-qPCR | Endometriosis | [168] | |
| Plasma | exLR | NFKBIA↑, NDUFB10↑, SLC7A7↑, ARPC5↑, SEPTIN9↑, etc. | Ultracentrifugation/TEM, NTA, WB | RNA Seq/RT-qPCR | Lung Cancer | [169] |
| Plasma | microRNAs | hsa-miR-106b-3p↑, hsa-miR-125a-5p↑, hsa-miR-3615↑, et al. | Ultracentrifugation/TEM, NTA, WB | RNA Seq/RT-qPCR | Lung Cancer | [170] |
| Plasma | hsa-miR-186-5p↑, hsa-miR-200c-3p↑, hsa-miR-429↑, etc. | SEC/TEM, NTA, WB | RNA Seq/RT-qPCR | Gastric Cancer | [171] | |
| Plasma | long RNAs | hsa-miR-483-5p↑ | Total Exosome Isolation Kit, differential ultracentrifugation/TEM, DLS, flowcytometry | Microarray/RT-qPCR | Adrenocortical Tumors | [172] |
| Plasma | microRNAs | microRNA-29a↑ | Differential centrifugation, density gradient centrifugation/TEM, NTA, WB | RNA Seq/ddPCR | Chronic Methamphetamine Use Disorder | [173] |
| Serum | MicroRNA-431-5p↑ | Differential centrifugation/TEM, NTA, WB | Microarray/RT-qPCR | Diabetic Retinopathy | [174] | |
| Plasma | microRNA-491-5p↑ | ExoQuick/TEM, NTA, WB | NanoString miRNAs analysis/RT-qPCR | Head and Neck Squamous Cell Carcinoma | [175] | |
| Plasma | miR-101↓ | Differential centrifugation/TEM, NTA, WB | RNA Seq/RT-qPCR | Osteosarcoma | [176] | |
| Plasma | miR-101-3p↓, miR-150-5p↑ | Precipitation/TEM, NTA, WB, ExoView | RNA Seq/RT-qPCR | Lung Cancer | [177] | |
| Plasma | miR-103a-3p↑, miR-30e-3p↓ | Ultracentrifugation/TEM, NTA, flow cytometry | OpenArray/RT-qPCR | Malignant Pleural Mesothelioma | [178] | |
| Serum | miR-122-5p↑, miR-2110↑, miR-483-5p↑; miR-370-3p↓, miR-409-3p↓, etc. | miRCURY/NA | RNA-Seq/RT-qPCR- | Atherosclerosis | [179] | |
| Serum | miR-1246↑ | SEC/TEM, NTA | Microarray/RT-qPCR | Gallbladder Cancer | [180] | |
| Plasma | miR-127-3p↓, miR-155-5p↓, miR-21-5p↓, miR-24-3p↓, let-7a-5p↓ | SEC/NA | RNA Seq/RT-qPCR | Classical Hodgkin Lymphoma | [181] | |
| Plasma | miR-134-5p↓, miR-205-5p↑, miR-409-3p↓ | SEC/TEM, NTA, WB | RNA Seq/RT-qPCR | Nasopharyngeal Carcinoma | [182] | |
| Plasma | miR-181a↑, miR-1908↑, miR-21↑, miR-486↑, miR-223↑ | ExoQuick, exoRNeasy/NA | RNA Seq/NA | Ovarian Cancer | [183] | |
| Serum | miR-181a-5p↑ | Total exosome isolation kit/TEM, NTA, WB | Microarray/RT-qPCR | Prostate Cancer | [184] | |
| Serum | miR-21-5p’(3′ addition C)↑, miR-23a-3p↑, tRF-Lys↑ | Total exosome isolation kit/TEM, NTA, WB | RNA Seq/NA | Breast Cancer | [185] | |
| Serum | miR-223↑, let-7e-5p↑, miR-486-3p↑, etc. | ExoQuick/TEM, NTA, flowcytometry | RNA Seq/RT-qPCR | Acute Rejection | [186] | |
| Plasma | miR-22-3p↑, miR-99a-5p↑, miR-151a-5p↑, miR-320b↑, miR-320d↑, etc. | ExoQuick, Exo-Spin/TEM, NTA, tunable resistive pulse sensing, WB | RNA Seq/RT-qPCR | Chronic Obstructive Pulmonary Disease | [187] | |
| Serum | miR-342-3p↑, miR-1254↓ | ExoChip/SEM, NTA, WB | NanoString miRNAs Analysis/NA | Sporadic Amyotrophic Lateral Sclerosis | [100] | |
| Plasma | miR-92b-3p↑, miR-374a-5p↑, miR-106b-3p↑ | miRCURY/NTA, TEM, WB | RNA Seq/RT-qPCR | Chronic Obstructive Pulmonary Disease | [188] | |
| Plasma | miRNA-152-3p↑, miRNA-1277-5p↑ | SEC/NTA, TEM, WB | RNA Seq/RT-qPCR | Lung Cancer | [189] | |
| Serum | miRNA-21↑ | ExoQuick/NTA, WB | miRNA array/RT-qPCR | Chronic Lung Disease | [190] | |
| Plasma | miRNAs, miR-500a-3p↑, miR-501-3p↑, miR-502-3p↑ | 3D medicine isolation reagent, polyethylene glycol-based method/NTA, SEM, WB | RNA Seq/NA | Pulmonary Ground-Glass Nodules | [191] | |
| Plasma | Let-7b-5p↑, miR-184↓, circulating miR-22-3p↓ | SEC/NTA, EM, WB | RNA Seq/RT-qPCR | Lung Cancer | [192] | |
| Plasma | let-7e↑ | Norgen plasma, serum exosome purification mini kit/WB | RNA Seq/RT-qPCR | Alzheimer’s Disease | [193] | |
| Plasma | let-7i-5p↑ | ExoQuick/TEM, NanoFCM, WB | RNA Seq/RT-qPCR | Asthma | [194] | |
| Serum | piRNAs | DQ593039↑ | Total exosome isolation reagent, exoEasy kit/TEM, NTA, WB | RNA Seq/RT-qPCR | Pulmonary Hypertension | [195] |
| CSF | microRNAs | miR-21↑ | miRCURY/TEM, NanoFCM, WB | RNA Seq/ddPCR | Leptomeningeal Metastasis | [196] |
| Urine | microRNAs | hsa-miR-193b-3p↓, hsa-miR-8485↓ | miRCURY/ExoView | miRNA Seq/NA | Acute Exercise-Induced Fatigue | [197] |
| Neurosurgical aspirate fluids | microRNAs | miR-486-3p↑ | Ultracentrifugation/TEM, NTA, WB | RNA Seq/NA | Glioblastoma | [198] |
| TDV | microRNAs | miR-203a-3p↑ | Ultracentrifugation/TEM, NTA, WB | RNA Seq/RT-qPCR | Lung Cancer | [199] |
6. The Identification of EVs
7. The Proteomic Profile Workflow of EVs in Clinical Investigation
8. Challenges for Proteomics of EVs in Clinical Investigation
9. Recent Progress and Future Directions in EV Proteomics
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Cai, J.; Han, Y.; Ren, H.; Chen, C.; He, D.; Zhou, L.; Eisner, G.M.; Asico, L.D.; Jose, P.A.; Zeng, C. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J. Mol. Cell. Biol. 2013, 5, 227–238. [Google Scholar] [CrossRef]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2016, 5, e1071008. [Google Scholar] [CrossRef]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e418. [Google Scholar] [CrossRef] [PubMed]
- Atkin-Smith, G.K.; Tixeira, R.; Paone, S.; Mathivanan, S.; Collins, C.; Liem, M.; Goodall, K.J.; Ravichandran, K.S.; Hulett, M.D.; Poon, I.K. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat. Commun. 2015, 6, 7439. [Google Scholar] [CrossRef]
- Tricarico, C.; Clancy, J.; D’Souza-Schorey, C. Biology and biogenesis of shed microvesicles. Small GTPases 2017, 8, 220–232. [Google Scholar] [CrossRef]
- Shkair, L.; Garanina, E.E.; Stott, R.J.; Foster, T.L.; Rizvanov, A.A.; Khaiboullina, S.F. Membrane Microvesicles as Potential Vaccine Candidates. Int. J. Mol. Sci. 2021, 22, 1142. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, S.; Lv, J.; Wang, X.; Afewerky, H.K.; Li, H.; Lu, Y. The emerging role of exosomes in Alzheimer’s disease. Ageing Res. Rev. 2021, 68, 101321. [Google Scholar] [CrossRef]
- ExoCarta. Available online: http://www.exocarta.org/ (accessed on 11 November 2022).
- Vesiclepedia. Available online: http://www.microvesicles.org/ (accessed on 11 November 2022).
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef]
- Schorey, J.S.; Bhatnagar, S. Exosome function: From tumor immunology to pathogen biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Huang, Z.; Huang, C.; Nice, E.C. Clinical applications of plasma proteomics and peptidomics: Towards precision medicine. Proteom. Clin. Appl. 2022, 16, e2100097. [Google Scholar] [CrossRef]
- Haney, M.J.; Klyachko, N.L.; Zhao, Y.; Gupta, R.; Plotnikova, E.G.; He, Z.; Patel, T.; Piroyan, A.; Sokolsky, M.; Kabanov, A.V.; et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J. Control. Release 2015, 207, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Panfoli, I.; Santucci, L.; Bruschi, M.; Petretto, A.; Calzia, D.; Ramenghi, L.A.; Ghiggeri, G.; Candiano, G. Microvesicles as promising biological tools for diagnosis and therapy. Expert Rev. Proteom. 2018, 15, 801–808. [Google Scholar] [CrossRef]
- Carnino, J.M.; Lee, H.; Jin, Y. Isolation and characterization of extracellular vesicles from Broncho-alveolar lavage fluid: A review and comparison of different methods. Respir. Res. 2019, 20, 240. [Google Scholar] [CrossRef]
- Mehryab, F.; Rabbani, S.; Shahhosseini, S.; Shekari, F.; Fatahi, Y.; Baharvand, H.; Haeri, A. Exosomes as a next-generation drug delivery system: An update on drug loading approaches, characterization, and clinical application challenges. Acta Biomater. 2020, 113, 42–62. [Google Scholar] [CrossRef]
- Chen, Y.; Li, G.; Liu, M.L. Microvesicles as Emerging Biomarkers and Therapeutic Targets in Cardiometabolic Diseases. Genom. Proteom. Bioinform. 2018, 16, 50–62. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Y.J.; Tang, Y.C.; Hao, X.Y.; Xu, W.J.; Xiang, D.X.; Wu, J.Y. Apoptotic bodies for advanced drug delivery and therapy. J. Control. Release 2022, 351, 394–406. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020, 5, 144. [Google Scholar] [CrossRef]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Marban, E. Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology. Annu. Rev. Physiol. 2016, 78, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Han, Y.; Zhu, Y.; Youngblood, H.A.; Almuntashiri, S.; Jones, T.W.; Wang, X.; Liu, Y.; Somanath, P.R.; Zhang, D. Nebulization of extracellular vesicles: A promising small RNA delivery approach for lung diseases. J. Control. Release 2022, 352, 556–569. [Google Scholar] [CrossRef]
- Zhang, S.; Liao, X.; Chen, S.; Qian, W.; Li, M.; Xu, Y.; Yang, M.; Li, X.; Mo, S.; Tang, M.; et al. Large Oncosome-Loaded VAPA Promotes Bone-Tropic Metastasis of Hepatocellular Carcinoma Via Formation of Osteoclastic Pre-Metastatic Niche. Adv. Sci. 2022, 9, e2201974. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.E. Extracellular vesicles in cancer therapy. Semin. Cancer Biol. 2022, 86, 296–309. [Google Scholar] [CrossRef]
- Zhou, X.; Xie, F.; Wang, L.; Zhang, L.; Zhang, S.; Fang, M.; Zhou, F. The function and clinical application of extracellular vesicles in innate immune regulation. Cell. Mol. Immunol. 2020, 17, 323–334. [Google Scholar] [CrossRef]
- Xiong, X.; Ke, X.; Wang, L.; Lin, Y.; Wang, S.; Yao, Z.; Li, K.; Luo, Y.; Liu, F.; Pan, Y.; et al. Neoantigen-based cancer vaccination using chimeric RNA-loaded dendritic cell-derived extracellular vesicles. J. Extracell. Vesicles 2022, 11, e12243. [Google Scholar] [CrossRef]
- Szczepanski, M.J.; Szajnik, M.; Welsh, A.; Whiteside, T.L.; Boyiadzis, M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica 2011, 96, 1302–1309. [Google Scholar] [CrossRef]
- Koh, Y.Q.; Ng, D.Q.; Ng, C.C.; Boey, A.; Wei, M.; Sze, S.K.; Ho, H.K.; Acharya, M.; Limoli, C.L.; Chan, A. Extracellular Vesicle Proteome of Breast Cancer Patients with and Without Cognitive Impairment Following Anthracycline-based Chemotherapy: An Exploratory Study. Biomarker. Insights 2021, 16, 11772719211018204. [Google Scholar] [CrossRef]
- Chang, L.C.; Hsu, Y.C.; Chiu, H.M.; Ueda, K.; Wu, M.S.; Kao, C.H.; Shen, T.L. Exploration of the Proteomic Landscape of Small Extracellular Vesicles in Serum as Biomarkers for Early Detection of Colorectal Neoplasia. Front. Oncol. 2021, 11, 732743. [Google Scholar] [CrossRef] [PubMed]
- Matthiesen, R.; Gameiro, P.; Henriques, A.; Bodo, C.; Moraes, M.C.; Costa-Silva, B.; Cabeçadas, J.; Gomes da Silva, M.; Beck, H.C.; Carvalho, A.S. Extracellular Vesicles in Diffuse Large B Cell Lymphoma: Characterization and Diagnostic Potential. Int. J. Mol. Sci. 2022, 23, 13327. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Pan, M.; Xiao, Y.; Ge, W. Serum Extracellular Vesicle Stratifin Is a Biomarker of Perineural Invasion in Patients With Colorectal Cancer and Predicts Worse Prognosis. Front. Oncol. 2022, 12, 912584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, T.; Du, Z.; Li, H.; Qin, W. A new integrated method for tissue extracellular vesicle enrichment and proteome profiling. RSC Adv. 2022, 12, 33409–33418. [Google Scholar] [CrossRef]
- Pane, K.; Quintavalle, C.; Nuzzo, S.; Ingenito, F.; Roscigno, G.; Affinito, A.; Scognamiglio, I.; Pattanayak, B.; Gallo, E.; Accardo, A.; et al. Comparative Proteomic Profiling of Secreted Extracellular Vesicles from Breast Fibroadenoma and Malignant Lesions: A Pilot Study. Int. J. Mol. Sci. 2022, 23, 3989. [Google Scholar] [CrossRef]
- Pachane, B.C.; Nunes, A.C.; Cataldi, T.R.; Micocci, K.C.; Moreira, B.C.; Labate, C.A.; Selistre-de-Araujo, H.S.; Altei, W.F. Small Extracellular Vesicles from Hypoxic Triple-Negative Breast Cancer Cells Induce Oxygen-Dependent Cell Invasion. Int. J. Mol. Sci. 2022, 23, 12646. [Google Scholar] [CrossRef]
- Carli, A.L.; Afshar-Sterle, S.; Rai, A.; Fang, H.; O’Keefe, R.; Tse, J.; Ferguson, F.M.; Gray, N.S.; Ernst, M.; Greening, D.W.; et al. Cancer stem cell marker DCLK1 reprograms small extracellular vesicles toward migratory phenotype in gastric cancer cells. Proteomics 2021, 21, e2000098. [Google Scholar] [CrossRef]
- Zheng, X.; Xu, K.; Zhou, B.; Chen, T.; Huang, Y.; Li, Q.; Wen, F.; Ge, W.; Wang, J.; Yu, S.; et al. A circulating extracellular vesicles-based novel screening tool for colorectal cancer revealed by shotgun and data-independent acquisition mass spectrometry. J. Extracell. Vesicles 2020, 9, 1750202. [Google Scholar] [CrossRef]
- Walbrecq, G.; Lecha, O.; Gaigneaux, A.; Fougeras, M.R.; Philippidou, D.; Margue, C.; Tetsi Nomigni, M.; Bernardin, F.; Dittmar, G.; Behrmann, I.; et al. Hypoxia-Induced Adaptations of miRNomes and Proteomes in Melanoma Cells and Their Secreted Extracellular Vesicles. Cancers 2020, 12, 692. [Google Scholar] [CrossRef]
- Tavasolian, F.; Hosseini, A.Z.; Rashidi, M.; Soudi, S.; Abdollahi, E.; Momtazi-Borojeni, A.A.; Sathyapalan, T.; Sahebkar, A. The Impact of Immune Cell-derived Exosomes on Immune Response Initiation and Immune System Function. Curr. Pharm. Des. 2021, 27, 197–205. [Google Scholar] [CrossRef]
- Hou, P.P.; Chen, H.Z. Extracellular vesicles in the tumor immune microenvironment. Cancer Lett. 2021, 516, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo, E.; Viry, E.; Morande, P.E.; Largeot, A.; Gonder, S.; Xian, F.; Ioannou, N.; Benzarti, M.; Kleine Borgmann, F.B.; Mittelbronn, M.; et al. Extracellular Vesicle Secretion by Leukemia Cells In Vivo Promotes CLL Progression by Hampering Antitumor T-cell Responses. Blood Cancer Discov. 2023, 4, 54–77. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Thaler, J.; Nieuwland, R. Extracellular Vesicles in Human Milk. Pharmaceuticals 2021, 14, 1050. [Google Scholar] [CrossRef] [PubMed]
- Jena, S.R.; Nayak, J.; Kumar, S.; Kar, S.; Samanta, L. Comparative proteome profiling of seminal components reveal impaired immune cell signalling as paternal contributors in recurrent pregnancy loss patients. Am. J. Reprod. Immunol. 2022, 89, e13613. [Google Scholar] [CrossRef] [PubMed]
- Finamore, F.; Cecchettini, A.; Ceccherini, E.; Signore, G.; Ferro, F.; Rocchiccioli, S.; Baldini, C. Characterization of Extracellular Vesicle Cargo in Sjögren’s Syndrome through a SWATH-MS Proteomics Approach. Int. J. Mol. Sci. 2021, 22, 4864. [Google Scholar] [CrossRef]
- Gerwing, M.; Kocman, V.; Stölting, M.; Helfen, A.; Masthoff, M.; Roth, J.; Barczyk-Kahlert, K.; Greune, L.; Schmidt, M.A.; Heindel, W.; et al. Tracking of Tumor Cell-Derived Extracellular Vesicles In Vivo Reveals a Specific Distribution Pattern with Consecutive Biological Effects on Target Sites of Metastasis. Mol. Imaging Biol. 2020, 22, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.F. Extracellular Vesicles and Neurodegenerative Diseases. J. Neurosci. Off. J. Soc. Neurosci. 2019, 39, 9269–9273. [Google Scholar] [CrossRef]
- You, Y.; Muraoka, S.; Jedrychowski, M.P.; Hu, J.; McQuade, A.K.; Young-Pearse, T.; Aslebagh, R.; Shaffer, S.A.; Gygi, S.P.; Blurton-Jones, M.; et al. Human neural cell type-specific extracellular vesicle proteome defines disease-related molecules associated with activated astrocytes in Alzheimer’s disease brain. J. Extracell. Vesicles 2022, 11, e12183. [Google Scholar] [CrossRef]
- Soares Martins, T.; Marçalo, R.; da Cruz e Silva, C.B.; Trindade, D.; Catita, J.; Amado, F.; Melo, T.; Rosa, I.M.; Vogelgsang, J.; Wiltfang, J.; et al. Novel Exosome Biomarker Candidates for Alzheimer’s Disease Unravelled Through Mass Spectrometry Analysis. Mol. Neurobiol. 2022, 59, 2838–2854. [Google Scholar] [CrossRef]
- Huang, Y.; Driedonks, T.A.; Cheng, L.; Rajapaksha, H.; Routenberg, D.A.; Nagaraj, R.; Redding, J.; Arab, T.; Powell, B.H.; Pletniková, O.; et al. Brain Tissue-Derived Extracellular Vesicles in Alzheimer’s Disease Display Altered Key Protein Levels Including Cell Type-Specific Markers. J. Alzheimers Dis. 2022, 90, 1057–1072. [Google Scholar] [CrossRef]
- Cai, H.; Pang, Y.; Wang, Q.; Qin, W.; Wei, C.; Li, Y.; Li, T.; Li, F.; Wang, Q.; Li, Y.; et al. Proteomic profiling of circulating plasma exosomes reveals novel biomarkers of Alzheimer’s disease. Alzheimers Res. Ther. 2022, 14, 181. [Google Scholar] [CrossRef]
- Nielsen, J.E.; Honoré, B.; Vestergård, K.; Maltesen, R.G.; Christiansen, G.; Bøge, A.U.; Kristensen, S.R.; Pedersen, S. Shotgun-based proteomics of extracellular vesicles in Alzheimer’s disease reveals biomarkers involved in immunological and coagulation pathways. Sci. Rep. 2021, 11, 18518. [Google Scholar] [CrossRef] [PubMed]
- Cohn, W.; Melnik, M.; Huang, C.; Teter, B.; Chandra, S.; Zhu, C.; McIntire, L.B.; John, V.; Gylys, K.H.; Bilousova, T. Multi-Omics Analysis of Microglial Extracellular Vesicles From Human Alzheimer’s Disease Brain Tissue Reveals Disease-Associated Signatures. Front. Pharmacol. 2021, 12, 766082. [Google Scholar] [CrossRef]
- Arioz, B.I.; Tufekci, K.U.; Olcum, M.; Durur, D.Y.; Akarlar, B.A.; Ozlu, N.; Bagriyanik, H.A.; Keskinoglu, P.; Yener, G.; Genc, S. Proteome profiling of neuron-derived exosomes in Alzheimer’s disease reveals hemoglobin as a potential biomarker. Neurosci. Lett. 2021, 755, 135914. [Google Scholar] [CrossRef]
- Muraoka, S.; Jedrychowski, M.P.; Yanamandra, K.; Ikezu, S.; Gygi, S.P.; Ikezu, T. Proteomic Profiling of Extracellular Vesicles Derived from Cerebrospinal Fluid of Alzheimer’s Disease Patients: A Pilot Study. Cells 2020, 9, 1959. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Liu, Y.; Hwang, S.; Archuleta, K.; Huang, H.; Campos, A.; Murad, R.; Piña-Crespo, J.; Xu, H.; Huang, T.Y. Trem2 deletion enhances tau dispersion and pathology through microglia exosomes. Mol. Neurodegener. 2022, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.S.; Garcia Romeu, H.; Aliyandi, A.; de Vries, M.P.; Zuhorn, I.S. DNAJB6-Containing Extracellular Vesicles as Chaperone Delivery Systems: A Proteomic Analysis. Pharmaceutics 2022, 14, 2485. [Google Scholar] [CrossRef]
- Jewett, K.A.; Thomas, R.E.; Phan, C.Q.; Lin, B.; Milstein, G.; Yu, S.; Bettcher, L.F.; Neto, F.C.; Djukovic, D.; Raftery, D.; et al. Glucocerebrosidase reduces the spread of protein aggregation in a Drosophila melanogaster model of neurodegeneration by regulating proteins trafficked by extracellular vesicles. PLoS Genet. 2021, 17, e1008859. [Google Scholar] [CrossRef]
- Vassileff, N.; Vella, L.J.; Rajapaksha, H.; Shambrook, M.; Kenari, A.N.; McLean, C.; Hill, A.F.; Cheng, L. Revealing the Proteome of Motor Cortex Derived Extracellular Vesicles Isolated from Amyotrophic Lateral Sclerosis Human Postmortem Tissues. Cells 2020, 9, 1709. [Google Scholar] [CrossRef]
- Thompson, A.G.; Gray, E.; Mäger, I.; Thézénas, M.L.; Charles, P.D.; Talbot, K.; Fischer, R.; Kessler, B.M.; Wood, M.; Turner, M.R. CSF extracellular vesicle proteomics demonstrates altered protein homeostasis in amyotrophic lateral sclerosis. Clin. Proteom. 2020, 17, 31. [Google Scholar] [CrossRef]
- Sjoqvist, S.; Otake, K. A pilot study using proximity extension assay of cerebrospinal fluid and its extracellular vesicles identifies novel amyotrophic lateral sclerosis biomarker candidates. Biochem. Biophys. Res. Commun. 2022, 613, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.T.; Alves, L.R. Extracellular Vesicles in Viral Infections: Two Sides of the Same Coin? Front. Cell. Infect. Microbiol. 2020, 10, 593170. [Google Scholar] [CrossRef] [PubMed]
- McNamara, R.P.; Dittmer, D.P. Extracellular vesicles in virus infection and pathogenesis. Curr. Opin. Virol. 2020, 44, 129–138. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, L.; Cao, P.; Li, S.; Zhang, W.; Dang, W.; Xin, S.; Jiang, M.; Xin, Y.; Li, J.; et al. Plasma Exosomal Proteomic Pattern of Epstein-Barr Virus-Associated Hemophagocytic Lymphohistiocytosis. Front. Microbiol. 2022, 13, 821311. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Kudo, K.; Higuchi, H.; Otsuka, H.; Tanaka, M.; Fukunishi, N.; Araki, T.; Takamatsu, M.; Ino, Y.; Kimura, Y.; et al. Proteomic and phospholipidomic characterization of extracellular vesicles inducing tumor microenvironment in Epstein-Barr virus-associated lymphomas. FASEB J. 2021, 35, e21505. [Google Scholar] [CrossRef] [PubMed]
- DeMarino, C.; Cowen, M.; Khatkar, P.; Cotto, B.; Branscome, H.; Kim, Y.; Sharif, S.A.; Agbottah, E.T.; Zhou, W.; Costiniuk, C.T.; et al. Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells 2022, 11, 723. [Google Scholar] [CrossRef] [PubMed]
- Falasca, K.; Lanuti, P.; Ucciferri, C.; Pieragostino, D.; Cufaro, M.C.; Bologna, G.; Federici, L.; Miscia, S.; Pontolillo, M.; Auricchio, A.; et al. Circulating extracellular vesicles as new inflammation marker in HIV infection. Aids 2020, 35, 595–604. [Google Scholar] [CrossRef]
- Pesce, E.; Manfrini, N.; Cordiglieri, C.; Santi, S.; Bandera, A.; Gobbini, A.; Gruarin, P.; Favalli, A.; Bombaci, M.; Cuomo, A.; et al. Exosomes Recovered From the Plasma of COVID-19 Patients Expose SARS-CoV-2 Spike-Derived Fragments and Contribute to the Adaptive Immune Response. Front. Immunol. 2021, 12, 785941. [Google Scholar] [CrossRef] [PubMed]
- Barberis, E.; Vanella, V.V.; Falasca, M.; Caneapero, V.; Cappellano, G.; Raineri, D.; Ghirimoldi, M.; De Giorgis, V.; Puricelli, C.; Vaschetto, R.; et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 2021, 8, 632290. [Google Scholar] [CrossRef]
- Morales-Sanfrutos, J.; Munoz, J. Unraveling the complexity of the extracellular vesicle landscape with advanced proteomics. Expert. Rev. Proteom. 2022, 19, 89–101. [Google Scholar] [CrossRef]
- Burkova, E.E.; Dmitrenok, P.S.; Bulgakov, D.V.; Vlassov, V.V.; Ryabchikova, E.I.; Nevinsky, G.A. Exosomes from human placenta purified by affinity chromatography on sepharose bearing immobilized antibodies against CD81 tetraspanin contain many peptides and small proteins. IUBMB Life 2018, 70, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Q.; Chen, J.; Chen, X.; Yang, W. Ultrasensitive electrochemiluminescence biosensor for the detection of tumor exosomes based on peptide recognition and luminol-AuNPs@g-C(3)N(4) nanoprobe signal amplification. Talanta 2021, 221, 121379. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, J.; Han, Z.; Liu, C.; Tian, F.; Xu, R.; Han, D.; Zhang, S.; Sun, J. Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation. J. Am. Chem. Soc. 2021, 143, 1290–1295. [Google Scholar] [CrossRef] [PubMed]
- Tzaridis, T.; Bachurski, D.; Liu, S.; Surmann, K.; Babatz, F.; Gesell Salazar, M.; Volker, U.; Hallek, M.; Herrlinger, U.; Vorberg, I.; et al. Extracellular Vesicle Separation Techniques Impact Results from Human Blood Samples: Considerations for Diagnostic Applications. Int. J. Mol. Sci. 2021, 22, 9211. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Yu, L.; Ma, T.; Xu, W.; Qian, H.; Sun, Y.; Shi, H. Small extracellular vesicles isolation and separation: Current techniques, pending questions and clinical applications. Theranostics 2022, 12, 6548–6575. [Google Scholar] [CrossRef]
- Askeland, A.; Borup, A.; Ostergaard, O.; Olsen, J.V.; Lund, S.M.; Christiansen, G.; Kristensen, S.R.; Heegaard, N.H.; Pedersen, S. Mass-Spectrometry Based Proteome Comparison of Extracellular Vesicle Isolation Methods: Comparison of ME-kit, Size-Exclusion Chromatography, and High-Speed Centrifugation. Biomedicines 2020, 8, 246. [Google Scholar] [CrossRef]
- Turner, N.P.; Abeysinghe, P.; Kwan Cheung, K.A.; Vaswani, K.; Logan, J.; Sadowski, P.; Mitchell, M.D. A Comparison of Blood Plasma Small Extracellular Vesicle Enrichment Strategies for Proteomic Analysis. Proteomes 2022, 10, 19. [Google Scholar] [CrossRef]
- Mussack, V.; Wittmann, G.; Pfaffl, M.W. Comparing small urinary extracellular vesicle purification methods with a view to RNA sequencing-Enabling robust and non-invasive biomarker research. Biomol. Detect. Quantif. 2019, 17, 100089. [Google Scholar] [CrossRef]
- Karimi, N.; Cvjetkovic, A.; Jang, S.C.; Crescitelli, R.; Hosseinpour Feizi, M.A.; Nieuwland, R.; Lotvall, J.; Lasser, C. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell. Mol. Life Sci. 2018, 75, 2873–2886. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, D.; Yang, H.; Shi, L.; Li, L.; Yu, C.; Wei, J.; Ding, Q. A light-activated magnetic bead strategy utilized in spatio-temporal controllable exosomes isolation. Front. Bioeng. Biotechnol. 2022, 10, 1006374. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.H.; Rau, C.S.; Wu, S.C.; Wu, Y.C.; Wu, C.J.; Tsai, C.W.; Lin, C.W.; Lu, T.H.; Hsieh, C.H. Identification and characterization of hADSC-derived exosome proteins from different isolation methods. J. Cell. Mol. Med. 2021, 25, 7436–7450. [Google Scholar] [CrossRef] [PubMed]
- Schimpf, M.E.; Caldwell, K.; Giddings, J.C. Field-Flow Fractionation Handbook; John Wiley & Sons: New York, NY, USA, 2000. [Google Scholar]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell. Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Zhang, H.; Lyden, D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc. 2019, 14, 1027–1053. [Google Scholar] [CrossRef]
- Contreras-Naranjo, J.C.; Wu, H.J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab. Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Q.; Bao, Q.Y.; Yu, S.X.; Liu, Q.; Xie, Y.; Li, X.; Liu, Y.J.; Shen, Y.H. A Novel Microfluidic Chip for Fast, Sensitive Quantification of Plasma Extracellular Vesicles as Biomarkers in Patients With Osteosarcoma. Front. Oncol. 2021, 11, 709255. [Google Scholar] [CrossRef] [PubMed]
- Lo, T.W.; Figueroa-Romero, C.; Hur, J.; Pacut, C.; Stoll, E.; Spring, C.; Lewis, R.; Nair, A.; Goutman, S.A.; Sakowski, S.A.; et al. Extracellular Vesicles in Serum and Central Nervous System Tissues Contain microRNA Signatures in Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2021, 14, 739016. [Google Scholar] [CrossRef]
- Sung, C.Y.; Huang, C.C.; Chen, Y.S.; Hsu, K.F.; Lee, G.B. Isolation and quantification of extracellular vesicle-encapsulated microRNA on an integrated microfluidic platform. Lab. Chip 2021, 21, 4660–4671. [Google Scholar] [CrossRef]
- Rima, X.Y.; Zhang, J.; Nguyen, L.T.; Rajasuriyar, A.; Yoon, M.J.; Chiang, C.L.; Walters, N.; Kwak, K.J.; Lee, L.J.; Reategui, E. Microfluidic harvesting of breast cancer tumor spheroid-derived extracellular vesicles from immobilized microgels for single-vesicle analysis. Lab. Chip 2022, 22, 2502–2518. [Google Scholar] [CrossRef]
- Niu, Q.; Gao, J.; Zhao, K.; Chen, X.; Lin, X.; Huang, C.; An, Y.; Xiao, X.; Wu, Q.; Cui, L.; et al. Fluid nanoporous microinterface enables multiscale-enhanced affinity interaction for tumor-derived extracellular vesicle detection. Proc. Natl. Acad. Sci. USA 2022, 119, e2213236119. [Google Scholar] [CrossRef]
- Guo, J.; Wu, C.; Lin, X.; Zhou, J.; Zhang, J.; Zheng, W.; Wang, T.; Cui, Y. Establishment of a simplified dichotomic size-exclusion chromatography for isolating extracellular vesicles toward clinical applications. J. Extracell. Vesicles 2021, 10, e12145. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Cheng, L.; Deng, C.; Huang, L.; Li, J.; Wang, Y.; Li, M.; Yang, Q.; Dong, X.; Su, J.; et al. The genetic source tracking of human urinary exosomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2108876118. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Huang, L.; Chen, J.; Ni, F.; Zhang, Y.; Liu, F. Isolation of Exosome Nanoparticles from Human Cerebrospinal Fluid for Proteomic Analysis. ACS Appl. Nano Mater. 2021, 4, 3351–3359. [Google Scholar] [CrossRef]
- Zhu, Q.; Huang, L.; Yang, Q.; Ao, Z.; Yang, R.; Krzesniak, J.; Lou, D.; Hu, L.; Dai, X.; Guo, F.; et al. Metabolomic analysis of exosomal-markers in esophageal squamous cell carcinoma. Nanoscale 2021, 13, 16457–16464. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, T.; Ma, H.; Pan, Y.; Wang, S.; Liu, X.; Dai, X.; Zheng, Y.; Lee, L.P.; Liu, F. Discovering the Secret of Diseases by Incorporated Tear Exosomes Analysis via Rapid-Isolation System: iTEARS. ACS Nano 2022, 16, 11720–11732. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, Y.H.; Ding, Y.; Zhang, G.; Liu, Y.; Sun, J.; Yang, Y.; Zhan, Z.; Iliuk, A.; Gu, Z.; et al. Proteomics, Phosphoproteomics and Mirna Analysis of Circulating Extracellular Vesicles through Automated and High-Throughput Isolation. Cells 2022, 11, 2070. [Google Scholar] [CrossRef]
- Buck, K.M.; Roberts, D.S.; Aballo, T.J.; Inman, D.R.; Jin, S.; Ponik, S.; Brown, K.A.; Ge, Y. One-Pot Exosome Proteomics Enabled by a Photocleavable Surfactant. Anal. Chem. 2022, 94, 7164–7168. [Google Scholar] [CrossRef]
- Wang, S.; He, Y.; Lu, J.; Wang, Y.; Wu, X.; Yan, G.; Fang, X.; Liu, B. All-in-One Strategy for Downstream Molecular Profiling of Tumor-Derived Exosomes. ACS Appl. Mater. Interfaces 2022, 14, 36341–36352. [Google Scholar] [CrossRef]
- Heo, Y.J.; Hwa, C.; Lee, G.H.; Park, J.M.; An, J.Y. Integrative Multi-Omics Approaches in Cancer Research: From Biological Networks to Clinical Subtypes. Mol. Cells 2021, 44, 433–443. [Google Scholar] [CrossRef]
- Aerqin, Q.; Wang, Z.T.; Wu, K.M.; He, X.Y.; Dong, Q.; Yu, J.T. Omics-based biomarkers discovery for Alzheimer’s disease. Cell. Mol. Life Sci. 2022, 79, 585. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; van der Ploeg, K.; Chakraborty, S.; Arunachalam, P.S.; Mori, D.A.; Jacobson, K.B.; Bonilla, H.; Parsonnet, J.; Andrews, J.R.; Holubar, M.; et al. Early immune markers of clinical, virological, and immunological outcomes in patients with COVID-19: A multi-omics study. Elife 2022, 11, e77943. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Lazaro-Ibanez, E.; Lasser, C.; Shelke, G.V.; Crescitelli, R.; Jang, S.C.; Cvjetkovic, A.; Garcia-Rodriguez, A.; Lotvall, J. DNA analysis of low- and high-density fractions defines heterogeneous subpopulations of small extracellular vesicles based on their DNA cargo and topology. J. Extracell. Vesicles 2019, 8, 1656993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.D.; Jiang, L.H.; Hou, J.C.; Zhong, S.L.; Zhu, L.P.; Wang, D.D.; Zhou, S.Y.; Yang, S.J.; Wang, J.Y.; Zhang, Q.; et al. Exosome: A novel mediator in drug resistance of cancer cells. Epigenomics 2018, 10, 1499–1509. [Google Scholar] [CrossRef]
- Carter, A.C.; Chang, H.Y.; Church, G.; Dombkowski, A.; Ecker, J.R.; Gil, E.; Giresi, P.G.; Greely, H.; Greenleaf, W.J.; Hacohen, N.; et al. Challenges and recommendations for epigenomics in precision health. Nat. Biotechnol. 2017, 35, 1128–1132. [Google Scholar] [CrossRef]
- Luo, T.; Chen, S.Y.; Qiu, Z.X.; Miao, Y.R.; Ding, Y.; Pan, X.Y.; Li, Y.; Lei, Q.; Guo, A.Y. Transcriptomic Features in a Single Extracellular Vesicle via Single-Cell RNA Sequencing. Small Methods 2022, 6, e2200881. [Google Scholar] [CrossRef]
- Lai, J.J.; Chau, Z.L.; Chen, S.Y.; Hill, J.J.; Korpany, K.V.; Liang, N.W.; Lin, L.H.; Lin, Y.H.; Liu, J.K.; Liu, Y.C.; et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022, 9, e2103222. [Google Scholar] [CrossRef]
- Williams, C.; Palviainen, M.; Reichardt, N.C.; Siljander, P.R.; Falcon-Perez, J.M. Metabolomics Applied to the Study of Extracellular Vesicles. Metabolites 2019, 9, 276. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, H.; Ao, Z.; Xu, H.; Luo, J.; Kaurich, C.; Yang, R.; Zhu, P.W.; Chen, S.D.; Wang, X.D.; et al. Lipidomic identification of urinary extracellular vesicles for non-alcoholic steatohepatitis diagnosis. J. Nanobiotechnol. 2022, 20, 349. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Yoshida, M.; Wagatsuma, T.; Sato, T.; Miyagishi, M.; Zhao, J.; Suematsu, M.; Kabe, Y.; Narimatsu, H. Comparative Glycomic Analysis of Exosome Subpopulations Derived from Pancreatic Cancer Cell Lines. J. Proteome Res. 2020, 19, 2516–2524. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, N.; Faghihkhorasani, F.; Fakhr, S.S.; Moghaddam, P.R.; Yazdani, E.; Kheradmand, Z.; Rezaei-Tazangi, F.; Adelian, S.; Mobarak, H.; Hamblin, M.R.; et al. Tumor-derived exosomal non-coding RNAs as diagnostic biomarkers in cancer. Cell. Mol. Life Sci. 2022, 79, 572. [Google Scholar] [CrossRef] [PubMed]
- Brunet, M.A.; Brunelle, M.; Lucier, J.F.; Delcourt, V.; Levesque, M.; Grenier, F.; Samandi, S.; Leblanc, S.; Aguilar, J.D.; Dufour, P.; et al. OpenProt: A more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Res. 2019, 47, D403–D410. [Google Scholar] [CrossRef]
- Jackson, R.; Kroehling, L.; Khitun, A.; Bailis, W.; Jarret, A.; York, A.G.; Khan, O.M.; Brewer, J.R.; Skadow, M.H.; Duizer, C.; et al. The translation of non-canonical open reading frames controls mucosal immunity. Nature 2018, 564, 434–438. [Google Scholar] [CrossRef]
- Yekula, A.; Muralidharan, K.; Kang, K.M.; Wang, L.; Balaj, L.; Carter, B.S. From laboratory to clinic: Translation of extracellular vesicle based cancer biomarkers. Methods 2020, 177, 58–66. [Google Scholar] [CrossRef]
- ClinicalTrials. Available online: https://clinicaltrials.gov/ct2/results?cond=exosome&term=biomarker&cntry=&state=&city=&dist= (accessed on 23 December 2022).
- ExoDxLung. Available online: https://exosome-rna.com/tag/exodx-lungalk/ (accessed on 17 December 2022).
- Sang, L.; Guo, X.; Fan, H.; Shi, J.; Hou, S.; Lv, Q. Mesenchymal Stem Cell-Derived Extracellular Vesicles as Idiopathic Pulmonary Fibrosis Microenvironment Targeted Delivery. Cells 2022, 11, 2322. [Google Scholar] [CrossRef]
- Exosome Diagnostics and Therapeutics: Global Markerts. Available online: https://www.researchandmarkets.com/reports/4538516/exosome-diagnostics-and-therapeutics-global (accessed on 17 December 2022).
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal. 2022, 20, 145. [Google Scholar] [CrossRef]
- Zipkin, M. Big pharma buys into exosomes for drug delivery. Nat. Biotechnol. 2020, 38, 1226–1228. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e1018. [Google Scholar] [CrossRef]
- Raimondo, F.; Morosi, L.; Chinello, C.; Magni, F.; Pitto, M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics 2011, 11, 709–720. [Google Scholar] [CrossRef]
- Carnino, J.M.; Ni, K.; Jin, Y. Post-translational Modification Regulates Formation and Cargo-Loading of Extracellular Vesicles. Front. Immunol. 2020, 11, 948. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Guan, S.; Wang, X.; Zhao, J.; Gao, M.; Zhang, X. Deconstruction of Heterogeneity of Size-Dependent Exosome Subpopulations from Human Urine by Profiling N-Glycoproteomics and Phosphoproteomics Simultaneously. Anal. Chem. 2020, 92, 9239–9246. [Google Scholar] [CrossRef] [PubMed]
- Nunez Lopez, Y.O.; Iliuk, A.; Petrilli, A.M.; Glass, C.; Casu, A.; Pratley, R.E. Proteomics and Phosphoproteomics of Circulating Extracellular Vesicles Provide New Insights into Diabetes Pathobiology. Int. J. Mol. Sci. 2022, 23, 5779. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Hoshina, T.; Matsuzaki, J.; Yoshioka, Y.; Kadota, T.; Hosaka, Y.; Fujimoto, S.; Kawamoto, H.; Watanabe, N.; Sawaki, K.; et al. Early prediction of COVID-19 severity using extracellular vesicle COPB2. J. Extracell. Vesicles 2021, 10, e12092. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhou, L.; Qiao, H.; Wang, Y.; Xiao, Y.; Fang, L.; Yang, B.; Wang, Z. Comparative proteomics identify HSP90A, STIP1 and TAGLN-2 in serum extracellular vesicles as potential circulating biomarkers for human adenomyosis. Exp. Ther. Med. 2022, 23, 374. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Im, E.J.; Moon, P.G.; Baek, M.C. Discovery of a diagnostic biomarker for colon cancer through proteomic profiling of small extracellular vesicles. BMC Cancer 2018, 18, 1058. [Google Scholar] [CrossRef]
- Kasahara, K.; Narumi, R.; Nagayama, S.; Masuda, K.; Esaki, T.; Obama, K.; Tomonaga, T.; Sakai, Y.; Shimizu, Y.; Adachi, J. A large-scale targeted proteomics of plasma extracellular vesicles shows utility for prognosis prediction subtyping in colorectal cancer. Cancer Med. 2022, 12, 7616–7626. [Google Scholar] [CrossRef]
- Shiromizu, T.; Kume, H.; Ishida, M.; Adachi, J.; Kano, M.; Matsubara, H.; Tomonaga, T. Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci. Rep. 2017, 7, 12782. [Google Scholar] [CrossRef]
- Fu, H.; Yang, H.; Zhang, X.; Wang, B.; Mao, J.; Li, X.; Wang, M.; Zhang, B.; Sun, Z.; Qian, H.; et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J. Exp. Clin. Cancer Res. 2018, 37, 162. [Google Scholar] [CrossRef]
- Huang, Q.; Hsueh, C.Y.; Shen, Y.J.; Guo, Y.; Huang, J.M.; Zhang, Y.F.; Li, J.Y.; Gong, H.L.; Zhou, L. Small extracellular vesicle-packaged TGFbeta1 promotes the reprogramming of normal fibroblasts into cancer-associated fibroblasts by regulating fibronectin in head and neck squamous cell carcinoma. Cancer Lett. 2021, 517, 1–13. [Google Scholar] [CrossRef]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.J.; Erice, O.; Gonzalez, E.; Jimenez-Aguero, R.; Lacasta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef] [PubMed]
- Vykoukal, J.; Sun, N.; Aguilar-Bonavides, C.; Katayama, H.; Tanaka, I.; Fahrmann, J.F.; Capello, M.; Fujimoto, J.; Aguilar, M.; Wistuba, I.I.; et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget 2017, 8, 95466–95480. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.S.; Faruque, H.A.; Kim, J.H.; Kim, K.J.; Choi, J.E.; Kim, B.A.; Kim, B.; Kim, Y.J.; Woo, M.H.; Park, J.Y.; et al. CD5L as an Extracellular Vesicle-Derived Biomarker for Liquid Biopsy of Lung Cancer. Diagnostics 2021, 11, 620. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, 6232. [Google Scholar] [CrossRef]
- Jiang, C.; Hopfner, F.; Katsikoudi, A.; Hein, R.; Catli, C.; Evetts, S.; Huang, Y.; Wang, H.; Ryder, J.W.; Kuhlenbaeumer, G.; et al. Serum neuronal exosomes predict and differentiate Parkinson’s disease from atypical parkinsonism. J. Neurol. Neurosurg. Psychiatry. 2020, 91, 720–729. [Google Scholar] [CrossRef]
- Tomlinson, P.R.; Zheng, Y.; Fischer, R.; Heidasch, R.; Gardiner, C.; Evetts, S.; Hu, M.; Wade-Martins, R.; Turner, M.R.; Morris, J.; et al. Identification of distinct circulating exosomes in Parkinson’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 353–361. [Google Scholar] [CrossRef]
- Nielsen, C.T.; Ostergaard, O.; Stener, L.; Iversen, L.V.; Truedsson, L.; Gullstrand, B.; Jacobsen, S.; Heegaard, N.H. Increased IgG on cell-derived plasma microparticles in systemic lupus erythematosus is associated with autoantibodies and complement activation. Arthritis Rheum. 2012, 64, 1227–1236. [Google Scholar] [CrossRef]
- Ostergaard, O.; Nielsen, C.T.; Iversen, L.V.; Tanassi, J.T.; Knudsen, S.; Jacobsen, S.; Heegaard, N.H. Unique protein signature of circulating microparticles in systemic lupus erythematosus. Arthritis Rheum. 2013, 65, 2680–2690. [Google Scholar] [CrossRef]
- Tomiyama, E.; Matsuzaki, K.; Fujita, K.; Shiromizu, T.; Narumi, R.; Jingushi, K.; Koh, Y.; Matsushita, M.; Nakano, K.; Hayashi, Y.; et al. Proteomic analysis of urinary and tissue-exudative extracellular vesicles to discover novel bladder cancer biomarkers. Cancer Sci. 2021, 112, 2033–2045. [Google Scholar] [CrossRef]
- Smalley, D.M.; Sheman, N.E.; Nelson, K.; Theodorescu, D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J. Proteome Res. 2008, 7, 2088–2096. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.Y.; Lee, C.H.; Choi, J.Y.; Cho, J.H.; Park, S.H.; Kim, Y.L.; Moon, P.G.; Baek, M.C.; Berm Park, J.; Hoon Kim, Y.; et al. Potential urinary extracellular vesicle protein biomarkers of chronic active antibody-mediated rejection in kidney transplant recipients. J. Chromatogr. B Analyt Technol. Biomed. Life Sci. 2020, 1138, 121958. [Google Scholar] [CrossRef]
- Wang, S.; Kojima, K.; Mobley, J.A.; West, A.B. Proteomic analysis of urinary extracellular vesicles reveal biomarkers for neurologic disease. EBioMedicine 2019, 45, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Barros, E.R.; Rigalli, J.P.; Tapia-Castillo, A.; Vecchiola, A.; Young, M.J.; Hoenderop, J.G.; Bindels, R.J.; Fardella, C.E.; Carvajal, C.A. Proteomic Profile of Urinary Extracellular Vesicles Identifies AGP1 as a Potential Biomarker of Primary Aldosteronism. Endocrinology 2021, 162, bqab032. [Google Scholar] [CrossRef]
- Raimondo, F.; Morosi, L.; Corbetta, S.; Chinello, C.; Brambilla, P.; Della Mina, P.; Villa, A.; Albo, G.; Battaglia, C.; Bosari, S.; et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. Biosyst. 2013, 9, 1220–1233. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, F.; Zhang, Q.; Liu, Y.; You, P.; Sun, S.; Lin, J.; Chen, N. Salivary exosomal PSMA7: A promising biomarker of inflammatory bowel disease. Protein Cell 2017, 8, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Aqrawi, L.A.; Galtung, H.K.; Guerreiro, E.M.; Ovstebo, R.; Thiede, B.; Utheim, T.P.; Chen, X.; Utheim, O.A.; Palm, O.; Skarstein, K.; et al. Proteomic and histopathological characterisation of sicca subjects and primary Sjogren’s syndrome patients reveals promising tear, saliva and extracellular vesicle disease biomarkers. Arthritis Res. Ther. 2019, 21, 181. [Google Scholar] [CrossRef]
- Xiao, K.; Li, S.; Ding, J.; Wang, Z.; Wang, D.; Cao, X.; Zhang, Y.; Dong, Z. Expression and clinical value of circRNAs in serum extracellular vesicles for gastric cancer. Front. Oncol. 2022, 12, 962831. [Google Scholar] [CrossRef]
- Iparraguirre, L.; Alberro, A.; Hansen, T.B.; Castillo-Trivino, T.; Munoz-Culla, M.; Otaegui, D. Profiling of Plasma Extracellular Vesicle Transcriptome Reveals That circRNAs Are Prevalent and Differ between Multiple Sclerosis Patients and Healthy Controls. Biomedicines 2021, 9, 1850. [Google Scholar] [CrossRef]
- Takahashi, K.; Ota, Y.; Kogure, T.; Suzuki, Y.; Iwamoto, H.; Yamakita, K.; Kitano, Y.; Fujii, S.; Haneda, M.; Patel, T.; et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020, 111, 98–111. [Google Scholar] [CrossRef]
- Kim, S.S.; Baek, G.O.; Ahn, H.R.; Sung, S.; Seo, C.W.; Cho, H.J.; Nam, S.W.; Cheong, J.Y.; Eun, J.W. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol. Oncol. 2020, 14, 2646–2659. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.; Yang, Y.; Jiang, J.; Yang, B.; Yang, Y.; Sun, F.; Zhang, J.; Lin, Y.; Xu, H. Extracellular vesicle-derived long non-coding RNA as circulating biomarkers for endometriosis. Reprod. Biomed. Online 2022, 44, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Huai, Q.; Liu, T.; Zhang, G.; Liang, N.; Ma, Q.; Liu, X.; Tan, F.; Xue, Q.; Gao, S.; et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for stage I lung adenocarcinoma. Transl. Lung Cancer Res. 2022, 11, 572–587. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, W.; Liu, T.; Liang, N.; Ma, Q.; Gao, Y.; Tan, F.; Xue, Q.; He, J. Plasma extracellular vesicle microRNA profiling and the identification of a diagnostic signature for stage I lung adenocarcinoma. Cancer Sci. 2022, 113, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, J.; Chen, Y.; Jiang, F.; Liao, H.; Liu, X.; Wang, Y.; Kong, G.; Zhang, X.; Li, J.; et al. miRNAs derived from plasma small extracellular vesicles predict organo-tropic metastasis of gastric cancer. Gastric Cancer 2022, 25, 360–374. [Google Scholar] [CrossRef]
- Perge, P.; Butz, H.; Pezzani, R.; Bancos, I.; Nagy, Z.; Paloczi, K.; Nyiro, G.; Decmann, A.; Pap, E.; Luconi, M.; et al. Evaluation and diagnostic potential of circulating extracellular vesicle-associated microRNAs in adrenocortical tumors. Sci. Rep. 2017, 7, 5474. [Google Scholar] [CrossRef]
- Chand, S.; Gowen, A.; Savine, M.; Moore, D.; Clark, A.; Huynh, W.; Wu, N.; Odegaard, K.; Weyrich, L.; Bevins, R.A.; et al. A comprehensive study to delineate the role of an extracellular vesicle-associated microRNA-29a in chronic methamphetamine use disorder. J. Extracell. Vesicles 2021, 10, e12177. [Google Scholar] [CrossRef]
- Yu, B.; Xiao, M.; Yang, F.; Xiao, J.; Zhang, H.; Su, L.; Zhang, X.; Li, X. MicroRNA-431–5p encapsulated in serum extracellular vesicles as a biomarker for proliferative diabetic retinopathy. Int. J. Biochem. Cell. Biol. 2021, 135, 105975. [Google Scholar] [CrossRef]
- Panvongsa, W.; Siripoon, T.; Worakitchanon, W.; Arsa, L.; Trachu, N.; Jinawath, N.; Ngamphaiboon, N.; Chairoungdua, A. Plasma extracellular vesicle microRNA-491–5p as diagnostic and prognostic marker for head and neck squamous cell carcinoma. Cancer Sci. 2021, 112, 4257–4269. [Google Scholar] [CrossRef]
- Zhang, K.; Dong, C.; Chen, M.; Yang, T.; Wang, X.; Gao, Y.; Wang, L.; Wen, Y.; Chen, G.; Wang, X.; et al. Extracellular vesicle-mediated delivery of miR-101 inhibits lung metastasis in osteosarcoma. Theranostics 2020, 10, 411–425. [Google Scholar] [CrossRef]
- Zheng, D.; Zhu, Y.; Zhang, J.; Zhang, W.; Wang, H.; Chen, H.; Wu, C.; Ni, J.; Xu, X.; Nian, B.; et al. Identification and evaluation of circulating small extracellular vesicle microRNAs as diagnostic biomarkers for patients with indeterminate pulmonary nodules. J. Nanobiotechnol. 2022, 20, 172. [Google Scholar] [CrossRef] [PubMed]
- Cavalleri, T.; Angelici, L.; Favero, C.; Dioni, L.; Mensi, C.; Bareggi, C.; Palleschi, A.; Rimessi, A.; Consonni, D.; Bordini, L.; et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS ONE 2017, 12, e0176680. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, A.; Kirchner, B.; Meidert, A.S.; Brandes, F.; Lindemann, A.; Doose, G.; Doege, A.; Weidenhagen, R.; Reithmair, M.; Schelling, G.; et al. Detection of Atherosclerosis by Small RNA-Sequencing Analysis of Extracellular Vesicle Enriched Serum Samples. Front. Cell. Dev. Biol. 2021, 9, 729061. [Google Scholar] [CrossRef] [PubMed]
- Ueta, E.; Tsutsumi, K.; Kato, H.; Matsushita, H.; Shiraha, H.; Fujii, M.; Matsumoto, K.; Horiguchi, S.; Okada, H. Extracellular vesicle-shuttled miRNAs as a diagnostic and prognostic biomarker and their potential roles in gallbladder cancer patients. Sci. Rep. 2021, 11, 12298. [Google Scholar] [CrossRef] [PubMed]
- Drees, E.E.; Roemer, M.G.; Groenewegen, N.J.; Perez-Boza, J.; van Eijndhoven, M.A.; Prins, L.I.; Verkuijlen, S.; Tran, X.M.; Driessen, J.; Zwezerijnen, G.J.; et al. Extracellular vesicle miRNA predict FDG-PET status in patients with classical Hodgkin Lymphoma. J. Extracell. Vesicles 2021, 10, e12121. [Google Scholar] [CrossRef]
- Jiang, L.; Zhang, Y.; Li, B.; Kang, M.; Yang, Z.; Lin, C.; Hu, K.; Wei, Z.; Xu, M.; Mi, J.; et al. miRNAs derived from circulating small extracellular vesicles as diagnostic biomarkers for nasopharyngeal carcinoma. Cancer Sci. 2021, 112, 2393–2404. [Google Scholar] [CrossRef]
- Kuhlmann, J.D.; Chebouti, I.; Kimmig, R.; Buderath, P.; Reuter, M.; Puppel, S.H.; Wimberger, P.; Kasimir-Bauer, S. Extracellular vesicle-associated miRNAs in ovarian cancer—Design of an integrated NGS-based workflow for the identification of blood-based biomarkers for platinum-resistance. Clin. Chem. Lab. Med. 2019, 57, 1053–1062. [Google Scholar] [CrossRef]
- Wang, Y.; Fang, Y.X.; Dong, B.; Du, X.; Wang, J.; Wang, X.; Gao, W.Q.; Xue, W. Discovery of extracellular vesicles derived miR-181a-5p in patient’s serum as an indicator for bone-metastatic prostate cancer. Theranostics 2021, 11, 878–892. [Google Scholar] [CrossRef]
- Koi, Y.; Tsutani, Y.; Nishiyama, Y.; Ueda, D.; Ibuki, Y.; Sasada, S.; Akita, T.; Masumoto, N.; Kadoya, T.; Yamamoto, Y.; et al. Predicting the presence of breast cancer using circulating small RNAs, including those in the extracellular vesicles. Cancer Sci. 2020, 111, 2104–2115. [Google Scholar] [CrossRef]
- Wang, W.; Li, W.; Cao, L.; Wang, B.; Liu, C.; Qin, Y.; Guo, B.; Huang, C. Serum extracellular vesicle MicroRNAs as candidate biomarkers for acute rejection in patients subjected to liver transplant. Front. Genet. 2022, 13, 1015049. [Google Scholar] [CrossRef]
- Sundar, I.K.; Li, D.; Rahman, I. Small RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J. Extracell. Vesicles 2019, 8, 1684816. [Google Scholar] [CrossRef]
- O’Farrell, H.E.; Bowman, R.V.; Fong, K.M.; Yang, I.A. Plasma Extracellular Vesicle miRNA Profiles Distinguish Chronic Obstructive Pulmonary Disease Exacerbations and Disease Severity. Int. J. Chron. Obs. Pulmon Dis. 2022, 17, 2821–2833. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.F.; Wei, S.N.; Geng, N.; Qin, W.W.; He, X.; Wang, X.H.; Qi, Y.P.; Song, S.; Wang, P. Evaluation of circulating small extracellular vesicle-derived miRNAs as diagnostic biomarkers for differentiating between different pathological types of early lung cancer. Sci. Rep. 2022, 12, 17201. [Google Scholar] [CrossRef] [PubMed]
- Go, H.; Maeda, H.; Miyazaki, K.; Maeda, R.; Kume, Y.; Namba, F.; Momoi, N.; Hashimoto, K.; Otsuru, S.; Kawasaki, Y.; et al. Extracellular vesicle miRNA-21 is a potential biomarker for predicting chronic lung disease in premature infants. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L845–L851. [Google Scholar] [CrossRef]
- Zhang, J.T.; Qin, H.; Man Cheung, F.K.; Su, J.; Zhang, D.D.; Liu, S.Y.; Li, X.F.; Qin, J.; Lin, J.T.; Jiang, B.Y.; et al. Plasma extracellular vesicle microRNAs for pulmonary ground-glass nodules. J. Extracell. Vesicles 2019, 8, 1663666. [Google Scholar] [CrossRef] [PubMed]
- Vadla, G.P.; Daghat, B.; Patterson, N.; Ahmad, V.; Perez, G.; Garcia, A.; Manjunath, Y.; Kaifi, J.T.; Li, G.; Chabu, C.Y. Combining plasma extracellular vesicle Let-7b-5p, miR-184 and circulating miR-22–3p levels for NSCLC diagnosis and drug resistance prediction. Sci. Rep. 2022, 12, 6693. [Google Scholar] [CrossRef] [PubMed]
- Durur, D.Y.; Tastan, B.; Ugur Tufekci, K.; Olcum, M.; Uzuner, H.; Karakulah, G.; Yener, G.; Genc, S. Alteration of miRNAs in Small Neuron-Derived Extracellular Vesicles of Alzheimer’s Disease Patients and the Effect of Extracellular Vesicles on Microglial Immune Responses. J. Mol. Neurosci. 2022, 72, 1182–1194. [Google Scholar] [CrossRef]
- Zheng, R.; Du, M.; Tian, M.; Zhu, Z.; Wei, C.; Chu, H.; Gan, C.; Liang, J.; Xue, R.; Gao, F.; et al. Fine Particulate Matter Induces Childhood Asthma Attacks via Extracellular Vesicle-Packaged Let-7i-5p-Mediated Modulation of the MAPK Signaling Pathway. Adv. Sci. 2022, 9, e2102460. [Google Scholar] [CrossRef]
- Lipps, C.; Northe, P.; Figueiredo, R.; Rohde, M.; Brahmer, A.; Kramer-Albers, E.M.; Liebetrau, C.; Wiedenroth, C.B.; Mayer, E.; Kriechbaum, S.D.; et al. Non-Invasive Approach for Evaluation of Pulmonary Hypertension Using Extracellular Vesicle-Associated Small Non-Coding RNA. Biomolecules 2019, 9, 666. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Seo, Y.; Im, J.H.; Rhim, J.; Baek, W.; Kim, S.; Kwon, J.W.; Yoo, B.C.; Shin, S.H.; Yoo, H.; et al. Molecular Signature of Extracellular Vesicular Small Non-Coding RNAs Derived from Cerebrospinal Fluid of Leptomeningeal Metastasis Patients: Functional Implication of miR-21 and Other Small RNAs in Cancer Malignancy. Cancers 2021, 13, 209. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Moon, H.Y. Urinary extracellular vesicle as a potential biomarker of exercise-induced fatigue in young adult males. Eur. J. Appl. Physiol. 2022, 122, 2175–2188. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Ebrahim Khani, S.; Wei, H.; Lee, M.Y.; Sim, H.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Alexander-Kaufman, K.L. Deep Sequencing of Small RNAs from Neurosurgical Extracellular Vesicles Substantiates miR-486–3p as a Circulating Biomarker that Distinguishes Glioblastoma from Lower-Grade Astrocytoma Patients. Int. J. Mol. Sci. 2020, 21, 4954. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Molins, L.; He, Y.; Vinolas, N.; Sanchez-Lorente, D.; Boada, M.; Guirao, A.; Diaz, T.; Martinez, D.; Ramirez, J.; et al. Characterization of the MicroRNA Cargo of Extracellular Vesicles Isolated from a Pulmonary Tumor-Draining Vein Identifies miR-203a-3p as a Relapse Biomarker for Resected Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 7138. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Simpson, R.J.; Greening, D.W. A Protocol for Isolation and Proteomic Characterization of Distinct Extracellular Vesicle Subtypes by Sequential Centrifugal Ultrafiltration. Methods Mol. Biol. 2017, 1545, 91–116. [Google Scholar] [PubMed]
- Hurwitz, S.N.; Meckes, D.G., Jr. An Adaptable Polyethylene Glycol-Based Workflow for Proteomic Analysis of Extracellular Vesicles. Methods Mol. Biol. 2017, 1660, 303–317. [Google Scholar] [PubMed]
- Kreimer, S.; Ivanov, A.R. Rapid Isolation of Extracellular Vesicles from Blood Plasma with Size-Exclusion Chromatography Followed by Mass Spectrometry-Based Proteomic Profiling. Methods Mol. Biol. 2017, 1660, 295–302. [Google Scholar]
- Andaluz Aguilar, H.; Iliuk, A.B.; Chen, I.H.; Tao, W.A. Sequential phosphoproteomics and N-glycoproteomics of plasma-derived extracellular vesicles. Nat. Protoc. 2019, 15, 161–180. [Google Scholar] [CrossRef]
- Rosa-Fernandes, L.; Rocha, V.B.; Carregari, V.C.; Urbani, A.; Palmisano, G. A Perspective on Extracellular Vesicles Proteomics. Front. Chem. 2017, 5, 102. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandao, B.B.; Zanotto, T.M.; Shah, S.; Kumar Patel, S.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef]
- Wu, D.; Yan, J.; Shen, X.; Sun, Y.; Thulin, M.; Cai, Y.; Wik, L.; Shen, Q.; Oelrich, J.; Qian, X.; et al. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nat. Commun. 2019, 10, 3854. [Google Scholar] [CrossRef]
- Ko, J.; Wang, Y.; Sheng, K.; Weitz, D.A.; Weissleder, R. Sequencing-Based Protein Analysis of Single Extracellular Vesicles. ACS Nano 2021, 15, 5631–5638. [Google Scholar] [CrossRef]
- Banijamali, M.; Hojer, P.; Nagy, A.; Haag, P.; Gomero, E.P.; Stiller, C.; Kaminskyy, V.O.; Ekman, S.; Lewensohn, R.; Karlstrom, A.E.; et al. Characterizing single extracellular vesicles by droplet barcode sequencing for protein analysis. J. Extracell. Vesicles 2022, 11, e12277. [Google Scholar] [CrossRef]
- Choi, D.; Montermini, L.; Jeong, H.; Sharma, S.; Meehan, B.; Rak, J. Mapping Subpopulations of Cancer Cell-Derived Extracellular Vesicles and Particles by Nano-Flow Cytometry. ACS Nano 2019, 13, 10499–10511. [Google Scholar] [CrossRef]
- Marassi, V.; Maggio, S.; Battistelli, M.; Stocchi, V.; Zattoni, A.; Reschiglian, P.; Guescini, M.; Roda, B. An ultracentrifugation—Hollow-fiber flow field-flow fractionation orthogonal approach for the purification and mapping of extracellular vesicle subtypes. J. Chromatogr. A 2021, 1638, 461861. [Google Scholar] [CrossRef]
- Bandu, R.; Oh, J.W.; Kim, K.P. Mass spectrometry-based proteome profiling of extracellular vesicles and their roles in cancer biology. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef]
- Karayel, O.; Virreira Winter, S.; Padmanabhan, S.; Kuras, Y.I.; Vu, D.T.; Tuncali, I.; Merchant, K.; Wills, A.M.; Scherzer, C.R.; Mann, M. Proteome profiling of cerebrospinal fluid reveals biomarker candidates for Parkinson’s disease. Cell. Rep. Med. 2022, 3, 100661. [Google Scholar] [CrossRef]


| Feature | Exosome | Microvesicle | Apoptotic Body |
|---|---|---|---|
| Size (nm) | 40–150 | 150–1000 | 1000–5000 |
| Density (g/mL) | 1.13–1.19 | 1.25–1.30 | 1.16–1.28 |
| Origin | Living cell | Living cell | Dying cell |
| Process | Releasing ILVs during plasma membrane fusion of MVBs | Budding from the plasma membrane directly | Blebbing from the plasma membrane during cell apoptosis |
| Contents | Nucleic acid, protein, lipid, etc. | Nucleic acid, protein, lipid, etc. | Fragments of the cellular components |
| Markers [28] | CD63, TSG101, Alix, HSP70, etc. | Integrins, selections, CD40 | Histones, TSP, C3b |
| Clinical application | Diagnosis, therapy [1,29] | Diagnosis, therapy [27,30] | Emerging [31] |
| Biomarker and therapeutic research ^ | High [32] | Medium [15] | Low [31,33] |
| Method | Yield | Purity | Time | Workload | Price | Practice |
|---|---|---|---|---|---|---|
| Conventional approaches | ||||||
| Differential ultracentrifugation | High | Medium | Long | >100 mL | Low | Easy |
| Ultrafiltration | High | Low | Medium | >100 mL | Medium | Easy |
| Tangential flow filtration | High | Medium | Medium | >100 mL | Medium | Medium |
| Size-exclusion chromatography | Low | Medium | Short | Up to a few mL | Medium | Easy |
| Density gradient ultracentrifugation | Low | High | Long | Up to 1 mL | Medium | Medium |
| Precipitation | High | Low | Medium | >100 µL | Low | Easy |
| Affinity | Low | High | Long | Up to 1 mL | High | Medium |
| Advanced approaches | ||||||
| AF4 | Low | High | Medium | 100 µL | / # | Medium |
| Microfluidic-based technologies | Low | High | - * | >10 µL | High | Medium |
| Dichotomic SEC | Medium | High | Short | 20 mL | Medium | Easy |
| EXODUS | High | High | Short | >100 mL | Medium | Easy |
| EVrich | - * | High | - * | / # | / # | Easy |
| Commercial EV isolation kits | High | High | Various | Various | High | Easy |
| Commercial Exosomes Isolation Kits | Separation Principle | Company | Cat. No. |
|---|---|---|---|
| Capturem | Affinity, lectin | Takara | 635741 |
| EasySep | Affinity, antibody | Stem Cell | 100-0812 |
| exoEasy | Affinity, membrane-based | Qiagen | 76064 |
| ExoPure | Precipitation | Abcam | ab287883 |
| ExoQuick | Precipitation | System Biosciences | EXOQ20A-1 |
| Exo-spin | Size-exclusion chromatography | Cell Guidance Systems | EX05 |
| ExoSure | Size-exclusion chromatography | Gene Copoeia | EP001 |
| Hieff | Precipitation | YEASEN | 41201ES25 |
| MagCapture | Affinity, phosphatidylserine | FUJIFILM Wako | 290-84103 |
| miRCURY | Precipitation | Qiagen | 76603 |
| PureExo | Precipitation | 101Bio | P101 |
| Total Exosome Isolation Reagent | Precipitation | Invitrogen | 4478359 |
| Omics | Subject | Current Challenges |
|---|---|---|
| Genomics | DNA [116] | The DNA of EVs is still difficult to preserve and isolate [117]. |
| Epigenomics | DNA [118] | The interpretation of the data from dynamic and specific tissue [119]. |
| Transcriptomics | RNA | A relatively robust method has been studied in high throughput even at a single EV level [120]. Now, analysis is the bottleneck. |
| Proteomics | Protein | Comprehensive, reproducible, and accurate data depends on the purity of EVs [121]; sensitivity and throughput. |
| Metabolomics/Lipidomics/Glycomics | Metabolite [122]/Lipid [123]/Glycan [124] | The sensitivity, reproducibility, robustness, speed, and accuracy; the investigation of EV subpopulation; comprehensive analysis. |
| Challenges | Future Directions |
|---|---|
| 1. The heterogeneity of EVs. | Applying integrated module approaches at a single EV level. |
| 2. The purity of EVs. | Selecting the most appropriate approach for each specific research scenario and sample type. |
| 3. The identification methods of EVs. | Considering reproducible and accurate approaches. |
| 4. The proteome coverage of EVs by LC/ESI-MS/MS and DDA/DIA. | Alternative data analysis strategies according to set parameters with robust bioinformatic tools. |
| 5. The PTM-proteome of EVs. | High capture efficiency of enrichment strategy. |
| 6. Tradeoffs between throughput and depth of LC-MS/MS. | Robust, automated, and high-throughput workflow. |
| 7. Systems biology overview of the EVs function. | Advanced computational and biological algorithms. |
| 8. Final biological effects validation in EVs. | A routine research workflow between labs and collaborative efforts from many different fields. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Poetsch, A. Proteomic Research of Extracellular Vesicles in Clinical Biofluid. Proteomes 2023, 11, 18. https://doi.org/10.3390/proteomes11020018
Fan S, Poetsch A. Proteomic Research of Extracellular Vesicles in Clinical Biofluid. Proteomes. 2023; 11(2):18. https://doi.org/10.3390/proteomes11020018
Chicago/Turabian StyleFan, Shipan, and Ansgar Poetsch. 2023. "Proteomic Research of Extracellular Vesicles in Clinical Biofluid" Proteomes 11, no. 2: 18. https://doi.org/10.3390/proteomes11020018
APA StyleFan, S., & Poetsch, A. (2023). Proteomic Research of Extracellular Vesicles in Clinical Biofluid. Proteomes, 11(2), 18. https://doi.org/10.3390/proteomes11020018