P40 and P90 from Mpn142 are Targets of Multiple Processing Events on the Surface of Mycoplasma pneumoniae
Abstract
:1. Introduction
2. Experimental Section
2.1. Strains and Cultures and Reagents
2.2. Enrichment of M. pneumoniae Surface Proteins
2.2.1. Biotinylation
2.2.2. Trypsin Shaving
2.3. Preparation of M. pneumoniae Whole Cell Lysates for One- and Two-Dimensional Gel Electrophoresis
2.4. One- and Two-dimensional Polyacrylamide Gel Electrophoresis (PAGE)
2.4.1. 1D SDS-PAGE
2.4.2. 2D SDS-PAGE
2.4.3. Trypsin Digest
2.5. Heparin Affinity Chromatography
2.6. Identification of Host Binding Proteins (Fibronectin, Plasminogen, Actin and Fetuin)
2.7. Identification of A549 Binding Proteins
2.8. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS)
2.9. MS/MS Data Analysis
2.10. Dimethyl Labelling of M. pneumoniae Proteins
2.11. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS): Sciex 5600
2.12. Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS): Thermo Scientific Q Exactive™
2.13. Bioinformatic Analysis of Mpn142
3. Results
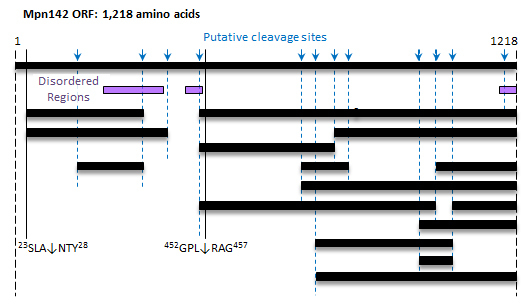
3.1. Defining P40 and P90 on the Surface of M. pneumoniae

| # | Peptide Sequence | Score | E-Value | 5600 | QE |
|---|---|---|---|---|---|
| N-terminal Dimethyl Labelled | |||||
| N1 | -.1MKSKLKLKR9.Y | 11 * | 0.72 | 1 | 1 |
| N2 | L.25ANTYLLQDHNTLTPYTPFTTPLNGGLDVVR54.A | 36 | 0.0024 | 1 | 3 |
| N3 | A.26NTYLLQDHNTLTPYTPFTTPLNGGLDVVR54.A | 142 | 3.5 × 10−13 | 4 | 4 |
| N4 | R.153KIKLALPYVKQESQGSGDQGSNGKGSLYKTLQDLLVEQPVTPYTPNAGLARVNGV207.A | 66 | 2.3 × 10−5 | - | 1 |
| N5 | R.368TASDTATFSK377.Y | 50 | 0.00068 | - | 1 |
| N6 | R.368TASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVAS409.R | 55 | 1.2 × 10−5 | 2 | - |
| N7 | R.368TASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVASR410.Q | 146 | 1.1 × 10−13 | 2 | 3 |
| N8 | R.368TASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVASRQ411.D | 148 | 5.2 × 10−14 | 1 | 4 |
| N9 | R.368TASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVASRQD412.A | 163 | 1.2 × 10−15 | - | 2 |
| N10 | R.368TASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVASRQDA413.T | 80 | 9.8 × 10−8 | - | 2 |
| N11 | R.368TASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVASRQDAT414.S | 62 | 3.9 × 10−6 | - | 2 |
| N12 | T.369ASDTATFSKYLNTAQALHQMGVIVPGLEKWGGNNGTGVVASR410.Q | 138 | 2.7 × 10−13 | 2 | 4 |
| N13 | R.446AVTVVAGPLR455.A | 72 | 1.2 × 10−5 | 3 | 4 |
| N14 | A.447VTVVAGPLR455.A | 48 | 0.0024 | 3 | 4 |
| N15 | L.696GLNFNFKLNEER707.L | 13 * | 1.3 | 2 | 2 |
| N16 | R.731ENAQSTSDDNSNTKVKWTNTASHYLPVPYYYSANFPEAGNRR772.R | 21 * | 0.015 | - | 1 |
| N17 | A.775EQRNGVKISTLESQATDGFANSLLNFGTGLKAGVDPAPVAR815.G | 41 | 0.00025 | - | 1 |
| N18 | A.1022NSVLQARNLTDKTVDEVINNPDILQSFFKFTPAFDNQR1059.A | 84 | 8.8 × 10−7 | 3 | 1 |
| N19 | K.1190AANNAAPKAPVKPAAPTAPRPPVQPPKKA1218.- | 59 | 4.7 × 10−6 | - | 1 |
| N20 | A.1191ANNAAPKAPVKPAAPTAPRPPVQPPKKA1218.- | 99 | 1.7 × 10−9 | 1 | 3 |
| N21 | A.1192NNAAPKAPVKPAAPTAPRPPVQPPKKA1218.- | 41 | 0.00014 | - | 1 |
| N22 | N.1194AAPKAPVKPAAPTAPRPPVQPPKKA1218.- | 44 | 0.00019 | - | 2 |
| N23 | A.1195APKAPVKPAAPTAPRPPVQPPKKA1218.- | 18 * | 0.051 | - | 1 |
| N24 | P.1197KAPVKPAAPTAPRPPVQPPKKA1218.- | 21 * | 0.029 | - | 1 |
| Semi-tryptic Truncated C-terminal Peptides | |||||
| S1 | R.253KLNESWPVYEPLDSTKEGKGKDESSWKNSEKTTAENDAPLVGMVGSGAA301.G | 73 | 5.4 × 10−7 | 1 | - |
| S2 | R.253KLNESWPVYEPLDSTKEGKGKDESSWKNSEKTTAENDAPLVGMVGSGAAGS303.A | 87 | 1.4 × 10−8 | 1 | - |
| S3 | R.253KLNESWPVYEPLDSTKEGKGKDESSWKNSEKTTAENDAPLVGMVGSGAAGSA304.S | 30 * | 0.0071 | 1 | - |
| S4 | R.253KLNESWPVYEPLDSTKEGKGKDESSWKNSEKTTAENDAPLVGMVGSGAAGSAS305.S | 40 | 0.00072 | 1 | - |
| S5 | R.253KLNESWPVYEPLDSTKEGKGKDESSWKNSEKTTAENDAPLVGMVGSGAAGSASS306.L | 39 | 0.00077 | - | 1 |
| S6 | R.253KLNESWPVYEPLDSTKEGKGKDESSWKNSEKTTAENDAPLVGMVGSGAAGSASSLQ308.G | 86 | 1.4 × 10−7 | 1 | - |
| S7 | R.1029NLTDKTVDEVINNPDILQSFFKFTPAFDNQRAMLV1063.G | 45 | 0.0023 | - | 1 |
| Semi-tryptic Truncated N-terminal Peptides | |||||
| S8 | D.810PAPVARGHKPNYSAVLLVR828.G | 65 | 2.3 × 10−6 | - | 1 |
| S9 | N.983GLFEKDDQLSENVKRR998.D | 26 * | 0.033 | - | 1 |
| S10 | N.1193NAAPKAPVKPAAPTAPRPPVQPPKKA1218.- | 25 * | 0.015 | - | 1 |
| P1 adhesin Cleaved Peptides | |||||
| C1 | R.1594LKQTSAAKPG1603.A (Semi-tryptic) | 24 * | 0.045 | - | 1 |
| C2 | T.1598SAAKPGAPRPPVPPKPGAPKPPVQPPKKPA1627.- (Dimethyl labeled) | 58 | 4.5 × 10−6 | - | 2 |
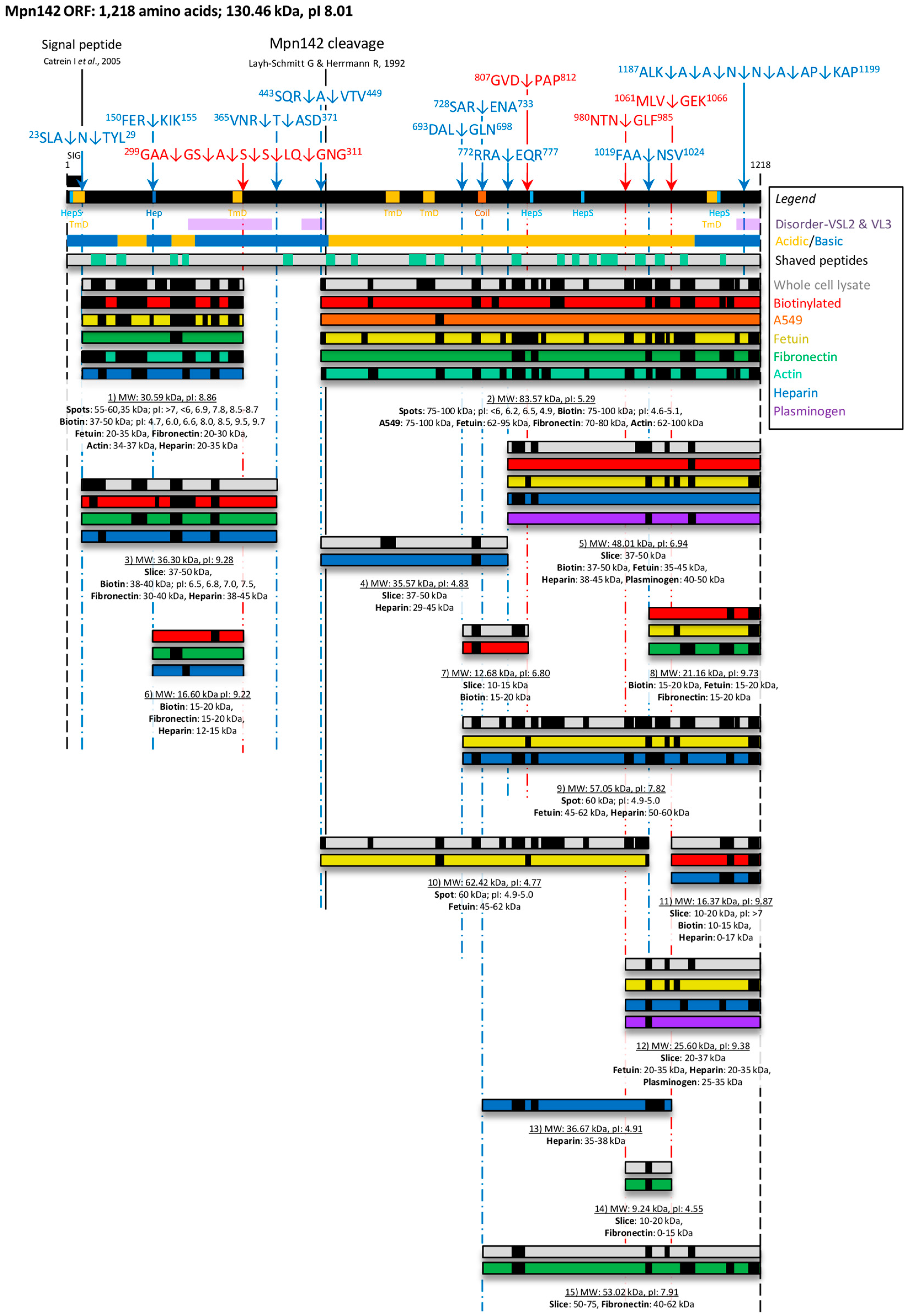
3.2. Bioinformatic Analysis of Mpn142
3.3. Cleavage Fragments in the N-terminus of Mpn142

3.4. Cleavage Fragments Residing in the C-terminus of Mpn142
4. Discussion
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Waites, K.B.; Talkington, D.F. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 2004, 17, 697–728. [Google Scholar] [CrossRef] [PubMed]
- Waites, K.B.; Atkinson, T.P. The role of mycoplasma in upper respiratory infections. Curr. Infect. Dis. Rep. 2009, 11, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Youn, Y.S.; Lee, K.Y. Mycoplasma pneumoniae pneumonia in children. Korean J. Pediatr. 2012, 55, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Robinson, D.; Bebear, C. Antibiotic susceptibilities of mycoplasmas and treatment of mycoplasmal infections. J. Antimicrob. Chemother. 1997, 40, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, G.D.; Gadsby, N.J.; Henderson, S.S.; Hardie, A.; Kalima, P.; Morris, A.C.; Hill, A.T.; Cunningham, S.; Templeton, K.E. Clinical outcomes and macrolide resistance in Mycoplasma pneumoniae infection in Scotland, UK. J. Med. Microbiol. 2013, 62, 1876–1882. [Google Scholar] [CrossRef] [PubMed]
- Peuchant, O.; Menard, A.; Renaudin, H.; Morozumi, M.; Ubukata, K.; Bebear, C.M.; Pereyre, S. Increased macrolide resistance of Mycoplasma pneumoniae in france directly detected in clinical specimens by real-time pcr and melting curve analysis. J. Antimicrob. Chemother. 2009, 64, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Dumke, R.; von Baum, H.; Luck, P.C.; Jacobs, E. Occurrence of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Clin. Microbiol. Infect. 2010, 16, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Chironna, M.; Sallustio, A.; Esposito, S.; Perulli, M.; Chinellato, I.; Di Bari, C.; Quarto, M.; Cardinale, F. Emergence of macrolide-resistant strains during an outbreak of Mycoplasma pneumoniae infections in children. J. Antimicrob. Chemother. 2011, 66, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Meyer Sauteur, P.M.; Bleisch, B.; Voit, A.; Maurer, F.P.; Relly, C.; Berger, C.; Nadal, D.; Bloemberg, G.V. Survey of macrolide-resistant Mycoplasma pneumoniae in children with community-acquired pneumonia in Switzerland. Swiss Med. Wkly. 2014, 144, w14041. [Google Scholar] [CrossRef] [PubMed]
- Averbuch, D.; Hidalgo-Grass, C.; Moses, A.E.; Engelhard, D.; Nir-Paz, R. Macrolide resistance in Mycoplasma pneumoniae, Israel, 2010. Emerg. Infect. Dis. 2011, 17, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Morozumi, M.; Tajima, T.; Hasegawa, M.; Sakata, H.; Ohnari, S.; Chiba, N.; Iwata, S.; Ubukata, K. Rapid effectiveness of minocycline or doxycycline against macrolide-resistant Mycoplasma pneumoniae infection in a 2011 outbreak among Japanese children. Clin. Infect. Dis. 2012, 55, 1642–1649. [Google Scholar] [CrossRef] [PubMed]
- Wolff, B.J.; Thacker, W.L.; Schwartz, S.B.; Winchell, J.M. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time pcr and high-resolution melt analysis. Antimicrob. Agents Chemother. 2008, 52, 3542–3549. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Atkinson, T.P.; Hagood, J.; Makris, C.; Duffy, L.B.; Waites, K.B. Emerging macrolide resistance in Mycoplasma pneumoniae in children: Detection and characterization of resistant isolates. Pediatr. Infect. Dis. J. 2009, 28, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Buller, R.; Bledsoe, S.; Storch, G.A. Rising rates of macrolide-resistant Mycoplasma pneumoniae in the central United States. Pediatr. Infect. Dis. J. 2012, 31, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Liu, G.; Wu, J.; Cao, B.; Tao, X.; He, L.; Meng, F.; Zhu, L.; Lv, M.; Yin, Y.; et al. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob. Agents Chemother. 2013, 57, 1521–1523. [Google Scholar] [CrossRef] [PubMed]
- Kawai, Y.; Miyashita, N.; Kubo, M.; Akaike, H.; Kato, A.; Nishizawa, Y.; Saito, A.; Kondo, E.; Teranishi, H.; Wakabayashi, T.; et al. Nationwide surveillance of macrolide-resistant Mycoplasma pneumoniae infection in pediatric patients. Antimicrob. Agents Chemother. 2013, 57, 4046–4049. [Google Scholar] [CrossRef] [PubMed]
- Eshaghi, A.; Memari, N.; Tang, P.; Olsha, R.; Farrell, D.J.; Low, D.E.; Gubbay, J.B.; Patel, S.N. Macrolide-resistant Mycoplasma pneumoniae in humans, Ontario, Canada, 2010–2011. Emerg. Infect. Dis. 2013, 19, 1525–1527. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.H.; Benitez, A.J.; Winchell, J.M. Investigations of Mycoplasma pneumoniae infections in the United States: Trends in molecular typing and macrolide resistance from 2006 to 2013. J. Clin. Microbiol. 2015, 53, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Lee, S.; Selvarangan, R.; Qin, X.; Tang, Y.W.; Stiles, J.; Hong, T.; Todd, K.; Ratliff, A.E.; Crabb, D.M.; et al. Macrolide-resistant Mycoplasma pneumoniae, United States. Emerg. Infect. Dis. 2015, 21, 1470–1472. [Google Scholar] [CrossRef] [PubMed]
- Szczepanek, S.M.; Majumder, S.; Sheppard, E.S.; Liao, X.; Rood, D.; Tulman, E.R.; Wyand, S.; Krause, D.C.; Silbart, L.K.; Geary, S.J. Vaccination of balb/c mice with an avirulent Mycoplasma pneumoniae p30 mutant results in disease exacerbation upon challenge with a virulent strain. Infect. Immun. 2012, 80, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.; Yogev, D.; Naot, Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998, 62, 1094–1156. [Google Scholar] [PubMed]
- Meseguer, M.A.; Alvarez, A.; Rejas, M.T.; Sanchez, C.; Perez-Diaz, J.C.; Baquero, F. Mycoplasma pneumoniae: A reduced-genome intracellular bacterial pathogen. Infect. Genet. Evol. 2003, 3, 47–55. [Google Scholar] [CrossRef]
- Krause, D.C.; Balish, M.F. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 2001, 198, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Layh-Schmitt, G.; Podtelejnikov, A.; Mann, M. Proteins complexed to the p1 adhesin of Mycoplasma pneumoniae. Microbiology 2000, 146, (Pt 3). 741–747. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.C.; Balish, M.F. Cellular engineering in a minimal microbe: Structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 2004, 51, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.P.; Jensen, G.J. Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 2006, 60, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Seybert, A.; Herrmann, R.; Frangakis, A.S. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 2006, 156, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Hasselbring, B.M.; Jordan, J.L.; Krause, R.W.; Krause, D.C. Terminal organelle development in the cell wall-less bacterium Mycoplasma pneumoniae. Proc. Natl. Acad. Sci. USA 2006, 103, 16478–16483. [Google Scholar] [CrossRef] [PubMed]
- Hasselbring, B.M.; Krause, D.C. Proteins p24 and p41 function in the regulation of terminal-organelle development and gliding motility in Mycoplasma pneumoniae. J. Bacteriol. 2007, 189, 7442–7449. [Google Scholar] [CrossRef] [PubMed]
- Su, C.J.; Tryon, V.V.; Baseman, J.B. Cloning and sequence analysis of cytadhesin p1 gene from Mycoplasma pneumoniae. Infect. Immun. 1987, 55, 3023–3029. [Google Scholar] [PubMed]
- Inamine, J.M.; Denny, T.P.; Loechel, S.; Schaper, U.; Huang, C.H.; Bott, K.F.; Hu, P.C. Nucleotide sequence of the P1 attachment-protein gene of Mycoplasma pneumoniae. Gene 1988, 64, 217–229. [Google Scholar] [CrossRef]
- Chang, H.Y.; Prince, O.A.; Sheppard, E.S.; Krause, D.C. Processing is required for a fully functional protein p30 in Mycoplasma pneumoniae gliding and cytadherence. J. Bacteriol. 2011, 193, 5841–5846. [Google Scholar] [CrossRef] [PubMed]
- Layh-Schmitt, G.; Herrmann, R. Localization and biochemical characterization of the orf6 gene product of the Mycoplasma pneumoniae p1 operon. Infect. Immun. 1992, 60, 2906–2913. [Google Scholar] [PubMed]
- Catrein, I.; Herrmann, R.; Bosserhoff, A.; Ruppert, T. Experimental proof for a signal peptidase i like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type i spase. FEBS J. 2005, 272, 2892–2900. [Google Scholar] [CrossRef] [PubMed]
- Balish, M.F.; Krause, D.C. Mycoplasmas: A distinct cytoskeleton for wall-less bacteria. J. Mol. Microbiol. Biotechnol. 2006, 11, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Layh-Schmitt, G.; Herrmann, R. Spatial arrangement of gene products of the p1 operon in the membrane of Mycoplasma pneumoniae. Infect. Immun. 1994, 62, 974–979. [Google Scholar] [PubMed]
- Nakane, D.; Adan-Kubo, J.; Kenri, T.; Miyata, M. Isolation and characterization of p1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J. Bacteriol. 2011, 193, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Baseman, J.B.; Cole, R.M.; Krause, D.C.; Leith, D.K. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J. Bacteriol. 1982, 151, 1514–1522. [Google Scholar] [PubMed]
- Layh-Schmitt, G.; Harkenthal, M. The 40- and 90-kda membrane proteins (orf6 gene product) of Mycoplasma pneumoniae are responsible for the tip structure formation and P1 (adhesin) association with the triton shell. FEMS Microbiol. Lett. 1999, 174, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Waldo, R.H., 3rd; Jordan, J.L.; Krause, D.C. Identification and complementation of a mutation associated with loss of Mycoplasma pneumoniae virulence-specific proteins b and c. J. Bacteriol. 2005, 187, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Biberfeld, G.; Biberfeld, P. Ultrastructural features of Mycoplasma pneumoniae. J. Bacteriol. 1970, 102, 855–861. [Google Scholar] [PubMed]
- Hu, P.C.; Collier, A.M.; Baseman, J.B. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J. Exp. Med. 1977, 145, 1328–1343. [Google Scholar] [CrossRef] [PubMed]
- Feldner, J.; Gobel, U.; Bredt, W. Mycoplasma pneumoniae adhesin localized to tip structure by monoclonal antibody. Nature 1982, 298, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.C.; Cole, R.M.; Huang, Y.S.; Graham, J.A.; Gardner, D.E.; Collier, A.M.; Clyde, W.A., Jr. Mycoplasma pneumoniae infection: Role of a surface protein in the attachment organelle. Science 1982, 216, 313–315. [Google Scholar] [CrossRef] [PubMed]
- Baseman, J.B.; Morrison-Plummer, J.; Drouillard, D.; Puleo-Scheppke, B.; Tryon, V.V.; Holt, S.C. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr. J. Med. Sci. 1987, 23, 474–479. [Google Scholar] [PubMed]
- Dallo, S.F.; Chavoya, A.; Baseman, J.B. Characterization of the gene for a 30-kilodalton adhesion-related protein of Mycoplasma pneumoniae. Infect. Immun. 1990, 58, 4163–4165. [Google Scholar] [PubMed]
- Krause, D.C.; Leith, D.K.; Wilson, R.M.; Baseman, J.B. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect. Immun. 1982, 35, 809–817. [Google Scholar] [PubMed]
- Krause, D.C.; Leith, D.K.; Baseman, J.B. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect. Immun. 1983, 39, 830–836. [Google Scholar] [PubMed]
- Hu, P.C.; Collier, A.M.; Clyde, W.A., Jr. Serological comparison of virulent and avirulent Mycoplasma pneumoniae by monoclonal antibodies. Isr. J. Med. Sci. 1984, 20, 870–873. [Google Scholar] [PubMed]
- Inamine, J.M.; Loechel, S.; Hu, P.C. Analysis of the nucleotide sequence of the p1 operon of Mycoplasma pneumoniae. Gene 1988, 73, 175–183. [Google Scholar] [PubMed]
- Waldo, R.H., 3rd; Krause, D.C. Synthesis, stability, and function of cytadhesin p1 and accessory protein b/c complex of Mycoplasma pneumoniae. J. Bacteriol. 2006, 188, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Sperker, B.; Hu, P.; Herrmann, R. Identification of gene products of the p1 operon of Mycoplasma pneumoniae. Mol. Microbiol. 1991, 5, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. A novel family of predicted phosphoesterases includes drosophila prune protein and bacterial recj exonuclease. Trends Biochem. Sci. 1998, 23, 17–19. [Google Scholar] [CrossRef]
- Jaffe, J.D.; Berg, H.C.; Church, G.M. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 2004, 4, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Hansen, E.J.; Wilson, R.M.; Baseman, J.B. Two-dimensional gel electrophoretic comparison of proteins from virulent and avirulent strains of Mycoplasma pneumoniae. Infect. Immun. 1979, 24, 468–475. [Google Scholar] [PubMed]
- Lipman, R.P.; Clyde, W.A., Jr.; Denny, F.W. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J. Bacteriol. 1969, 100, 1037–1043. [Google Scholar] [PubMed]
- Jacobs, E.; Fuchte, K.; Bredt, W. Amino acid sequence and antigenicity of the amino-terminus of the 168 kDa adherence protein of Mycoplasma pneumoniae. J. Gen. Microbiol. 1987, 133, 2233–2236. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, S.P.; Cordwell, S.J.; Djordjevic, M.A.; Wilton, J.; Minion, F.C. Proteolytic processing of the mycoplasma hyopneumoniae cilium adhesin. Infect. Immun. 2004, 72, 2791–2802. [Google Scholar] [CrossRef] [PubMed]
- Burnett, T.A.; Dinkla, K.; Rohde, M.; Chhatwal, G.S.; Uphoff, C.; Srivastava, M.; Cordwell, S.J.; Geary, S.; Liao, X.; Minion, F.C.; et al. P159 is a proteolytically processed, surface adhesin of mycoplasma hyopneumoniae: Defined domains of p159 bind heparin and promote adherence to eukaryote cells. Mol. Microbiol. 2006, 60, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Wilton, J.; Jenkins, C.; Cordwell, S.J.; Falconer, L.; Minion, F.C.; Oneal, D.C.; Djordjevic, M.A.; Connolly, A.; Barchia, I.; Walker, M.J.; et al. Mhp493 (p216) is a proteolytically processed, cilium and heparin binding protein of mycoplasma hyopneumoniae. Mol. Microbiol. 2009, 71, 566–582. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, A.T.; Jenkins, C.; Minion, F.C.; Seymour, L.M.; Padula, M.P.; Dixon, N.E.; Walker, M.J.; Djordjevic, S.P. Repeat regions r1 and r2 in the p97 paralogue mhp271 of mycoplasma hyopneumoniae bind heparin, fibronectin and porcine cilia. Mol. Microbiol. 2010, 78, 444–458. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.M.; Deutscher, A.T.; Jenkins, C.; Kuit, T.A.; Falconer, L.; Minion, F.C.; Crossett, B.; Padula, M.; Dixon, N.E.; Djordjevic, S.P.; et al. A processed multidomain mycoplasma hyopneumoniae adhesin binds fibronectin, plasminogen, and swine respiratory cilia. J. Biol. Chem. 2010, 285, 33971–33978. [Google Scholar] [CrossRef] [PubMed]
- Bogema, D.R.; Scott, N.E.; Padula, M.P.; Tacchi, J.L.; Raymond, B.B.; Jenkins, C.; Cordwell, S.J.; Minion, F.C.; Walker, M.J.; Djordjevic, S.P. Sequence TTKF↓QE defines the site of proteolytic cleavage in mhp683 protein, a novel glycosaminoglycan and cilium adhesin of mycoplasma hyopneumoniae. J. Biol. Chem. 2011, 286, 41217–41229. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.M.; Falconer, L.; Deutscher, A.T.; Minion, F.C.; Padula, M.P.; Dixon, N.E.; Djordjevic, S.P.; Walker, M.J. Mhp107 is a member of the multifunctional adhesin family of mycoplasma hyopneumoniae. J. Biol. Chem. 2011, 286, 10097–10104. [Google Scholar] [CrossRef] [PubMed]
- Bogema, D.R.; Deutscher, A.T.; Woolley, L.K.; Seymour, L.M.; Raymond, B.B.; Tacchi, J.L.; Padula, M.P.; Dixon, N.E.; Minion, F.C.; Jenkins, C.; et al. Characterization of cleavage events in the multifunctional cilium adhesin mhp684 (p146) reveals a mechanism by which mycoplasma hyopneumoniae regulates surface topography. MBio 2012, 3, e00282-11. [Google Scholar] [CrossRef] [PubMed]
- Deutscher, A.T.; Tacchi, J.L.; Minion, F.C.; Padula, M.P.; Crossett, B.; Bogema, D.R.; Jenkins, C.; Kuit, T.A.; Walker, M.J.; Djordjevic, S.P. Mycoplasma hyopneumoniae surface proteins mhp385 and mhp384 bind host cilia and glycosaminoglycans and are endoproteolytically processed by proteases that recognize different cleavage motifs. J. Proteome Res. 2012, 11, 1924–1936. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.M.; Jenkins, C.; Deutscher, A.T.; Raymond, B.B.; Padula, M.P.; Tacchi, J.L.; Bogema, D.R.; Eamens, G.J.; Woolley, L.K.; Dixon, N.E.; et al. Mhp182 (p102) binds fibronectin and contributes to the recruitment of plasmin(ogen) to the mycoplasma hyopneumoniae cell surface. Cell Microbiol. 2012, 14, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.B.; Tacchi, J.L.; Jarocki, V.M.; Minion, F.C.; Padula, M.P.; Djordjevic, S.P. P159 from mycoplasma hyopneumoniae binds porcine cilia and heparin and is cleaved in a manner akin to ectodomain shedding. J. Proteome Res. 2013, 12, 5891–5903. [Google Scholar] [CrossRef] [PubMed]
- Tacchi, J.L.; Raymond, B.B.; Jarocki, V.M.; Berry, I.J.; Padula, M.P.; Djordjevic, S.P. Cilium adhesin p216 (mhj_0493) is a target of ectodomain shedding and aminopeptidase activity on the surface of mycoplasma hyopneumoniae. J. Proteome Res. 2014, 13, 2920–2930. [Google Scholar] [CrossRef] [PubMed]
- Raymond, B.B.; Jenkins, C.; Seymour, L.M.; Tacchi, J.L.; Widjaja, M.; Jarocki, V.M.; Deutscher, A.T.; Turnbull, L.; Whitchurch, C.B.; Padula, M.P.; et al. Proteolytic processing of the cilium adhesin mhj_0194 (p123j) in mycoplasma hyopneumoniae generates a functionally diverse array of cleavage fragments that bind multiple host molecules. Cell Microbiol. 2015, 17, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Calcutt, M.J.; Kim, M.F.; Karpas, A.B.; Muhlradt, P.F.; Wise, K.S. Differential posttranslational processing confers intraspecies variation of a major surface lipoprotein and a macrophage-activating lipopeptide of mycoplasma fermentans. Infect. Immun. 1999, 67, 760–771. [Google Scholar] [PubMed]
- Szczepanek, S.M.; Frasca, S., Jr.; Schumacher, V.L.; Liao, X.; Padula, M.; Djordjevic, S.P.; Geary, S.J. Identification of lipoprotein msla as a neoteric virulence factor of mycoplasma gallisepticum. Infect. Immun. 2010, 78, 3475–3483. [Google Scholar] [CrossRef] [PubMed]
- Suh, M.J.; Clark, D.J.; Parmer, P.P.; Fleischmann, R.D.; Peterson, S.N.; Pieper, R. Using chemical derivatization and mass spectrometric analysis to characterize the post-translationally modified staphylococcus aureus surface protein g. Biochim. Biophys. Acta 2010, 1804, 1394–1404. [Google Scholar] [CrossRef] [PubMed]
- Scott, N.E.; Marzook, N.B.; Deutscher, A.; Falconer, L.; Crossett, B.; Djordjevic, S.P.; Cordwell, S.J. Mass spectrometric characterization of the campylobacter jejuni adherence factor cadf reveals post-translational processing that removes immunogenicity while retaining fibronectin binding. Proteomics 2010, 10, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Veith, P.D.; Nor Muhammad, N.A.; Dashper, S.G.; Likic, V.A.; Gorasia, D.G.; Chen, D.; Byrne, S.J.; Catmull, D.V.; Reynolds, E.C. Protein substrates of a novel secretion system are numerous in the bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J. Proteome Res. 2013, 12, 4449–4461. [Google Scholar] [CrossRef] [PubMed]
- Dallo, S.F.; Kannan, T.R.; Blaylock, M.W.; Baseman, J.B. Elongation factor tu and e1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 2002, 46, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Dumke, R.; Hausner, M.; Jacobs, E. Role of Mycoplasma pneumoniae glyceraldehyde-3-phosphate dehydrogenase (gapdh) in mediating interactions with the human extracellular matrix. Microbiology 2011, 157, 2328–2338. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Jacobs, E.; Dumke, R. Characterization of pyruvate dehydrogenase subunit b and enolase as plasminogen-binding proteins in Mycoplasma pneumoniae. Microbiology 2013, 159, 352–365. [Google Scholar] [CrossRef] [PubMed]
- Grundel, A.; Friedrich, K.; Pfeiffer, M.; Jacobs, E.; Dumke, R. Subunits of the pyruvate dehydrogenase cluster of Mycoplasma pneumoniae are surface-displayed proteins that bind and activate human plasminogen. PLoS ONE 2015, 10, e0126600. [Google Scholar] [CrossRef] [PubMed]
- Loomes, L.M.; Uemura, K.; Childs, R.A.; Paulson, J.C.; Rogers, G.N.; Scudder, P.R.; Michalski, J.C.; Hounsell, E.F.; Taylor-Robinson, D.; Feizi, T. Erythrocyte receptors for Mycoplasma pneumoniae are sialylated oligosaccharides of ii antigen type. Nature 1984, 307, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Loomes, L.M.; Uemura, K.; Feizi, T. Interaction of Mycoplasma pneumoniae with erythrocyte glycolipids of i and i antigen types. Infect. Immun. 1985, 47, 15–20. [Google Scholar] [PubMed]
- Krivan, H.C.; Olson, L.D.; Barile, M.F.; Ginsburg, V.; Roberts, D.D. Adhesion of Mycoplasma pneumoniae to sulfated glycolipids and inhibition by dextran sulfate. J. Biol. Chem. 1989, 264, 9283–9288. [Google Scholar] [PubMed]
- Roberts, D.D.; Olson, L.D.; Barile, M.F.; Ginsburg, V.; Krivan, H.C. Sialic acid-dependent adhesion of Mycoplasma pneumoniae to purified glycoproteins. J. Biol. Chem. 1989, 264, 9289–9293. [Google Scholar] [PubMed]
- Hayflick, L. Tissue cultures and mycoplasmas. Tex. Rep. Biol. Med. 1965, 23 (Suppl. S1), 285–303. [Google Scholar] [PubMed]
- Webb, A. Systems Biology Mascot Server: Databases (mspnr100). Available online: http://www.wehi.edu.au/people/andrew-webb/1295/andrew-webb-resources (accessed on 1 September 2015).
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the expasy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the embl-ebi. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Hofmann K, S.W. Tmbase—A database of membrane spanning proteins segments. Biol. Chem. 1993, 374, 166. [Google Scholar]
- Lupas, A.; van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Radivojac, P.; Vucetic, S.; Dunker, A.K.; Obradovic, Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006, 7, 208. [Google Scholar] [CrossRef] [PubMed]
- Radivojac, P.; Obradovic, Z.; Brown, C.J.; Dunker, A.K. Prediction of boundaries between intrinsically ordered and disordered protein regions. Pac. Symp. Biocomput. 2003, 216–227. [Google Scholar]
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. Scanprosite: Detection of prosite signature matches and prorule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34, W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Cardin, A.D.; Weintraub, H.J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 1989, 9, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Klimstra, W.B.; Heidner, H.W.; Johnston, R.E. The furin protease cleavage recognition sequence of sindbis virus pe2 can mediate virion attachment to cell surface heparan sulfate. J. Virol. 1999, 73, 6299–6306. [Google Scholar] [PubMed]
- Regula, J.T.; Ueberle, B.; Boguth, G.; Gorg, A.; Schnolzer, M.; Herrmann, R.; Frank, R. Towards a two-dimensional proteome map of Mycoplasma pneumoniae. Electrophoresis 2000, 21, 3765–3780. [Google Scholar] [CrossRef]
- Seto, S.; Layh-Schmitt, G.; Kenri, T.; Miyata, M. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 2001, 183, 1621–1630. [Google Scholar] [CrossRef] [PubMed]
- Seto, S.; Miyata, M. Attachment organelle formation represented by localization of cytadherence proteins and formation of the electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 2003, 185, 1082–1091. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Widjaja, M.; Berry, I.J.; Pont, E.J.; Padula, M.P.; Djordjevic, S.P. P40 and P90 from Mpn142 are Targets of Multiple Processing Events on the Surface of Mycoplasma pneumoniae. Proteomes 2015, 3, 512-537. https://doi.org/10.3390/proteomes3040512
Widjaja M, Berry IJ, Pont EJ, Padula MP, Djordjevic SP. P40 and P90 from Mpn142 are Targets of Multiple Processing Events on the Surface of Mycoplasma pneumoniae. Proteomes. 2015; 3(4):512-537. https://doi.org/10.3390/proteomes3040512
Chicago/Turabian StyleWidjaja, Michael, Iain J. Berry, Elsa J. Pont, Matthew P. Padula, and Steven P. Djordjevic. 2015. "P40 and P90 from Mpn142 are Targets of Multiple Processing Events on the Surface of Mycoplasma pneumoniae" Proteomes 3, no. 4: 512-537. https://doi.org/10.3390/proteomes3040512
APA StyleWidjaja, M., Berry, I. J., Pont, E. J., Padula, M. P., & Djordjevic, S. P. (2015). P40 and P90 from Mpn142 are Targets of Multiple Processing Events on the Surface of Mycoplasma pneumoniae. Proteomes, 3(4), 512-537. https://doi.org/10.3390/proteomes3040512