A Proteomic Perspective on the Bacterial Adaptation to Cold: Integrating OMICs Data of the Psychrotrophic Bacterium Exiguobacterium antarcticum B7
Abstract
:1. Introduction
2. Mechanisms of Bacterial Adaptation to Cold
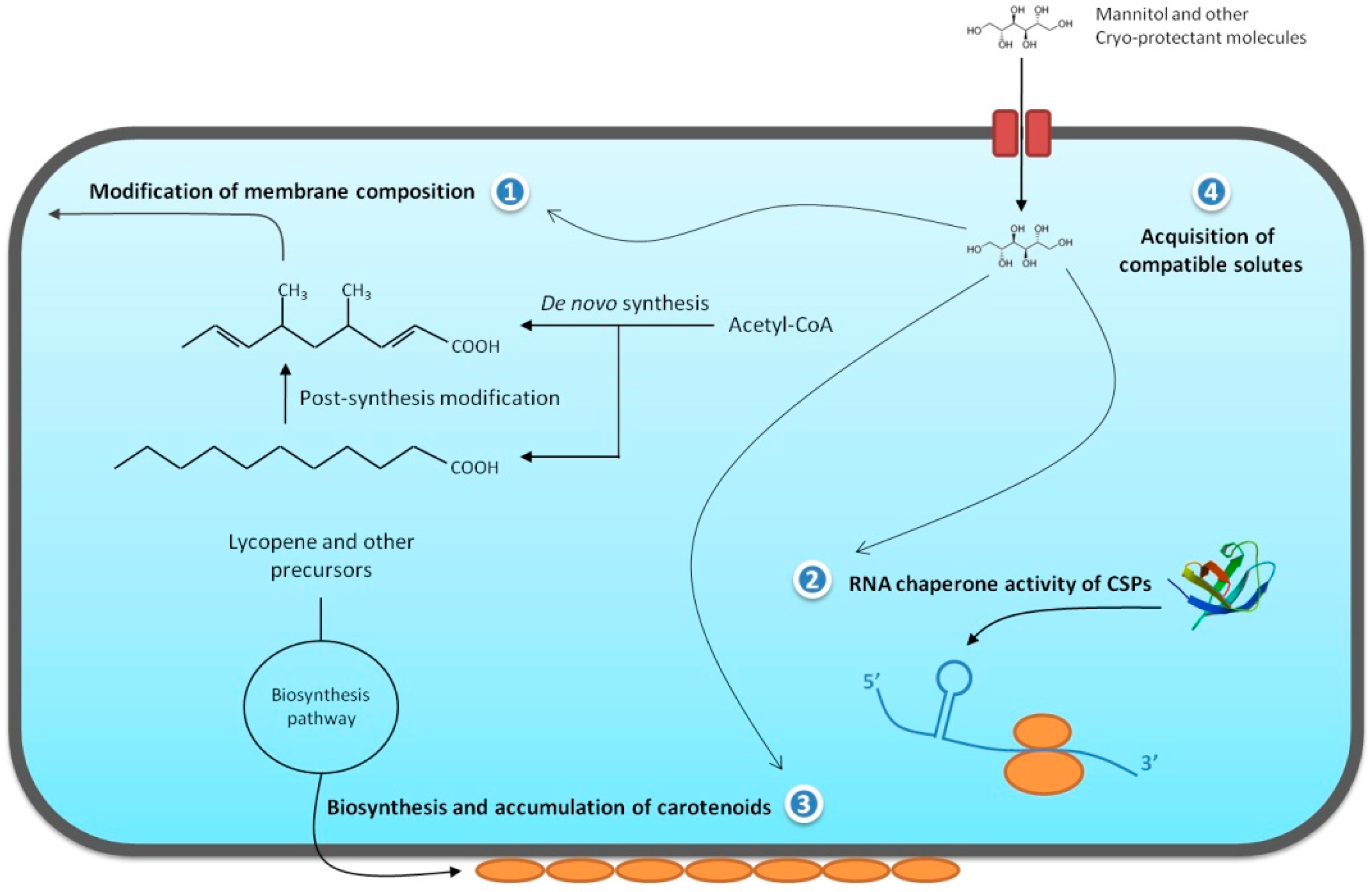
2.1. Chemical Modification of the Cellular Membrane
2.2. Cold-Adapted Enzymes
2.3. Cold Shock and Cold Acclimation Proteins
2.4. Other Important Aspects
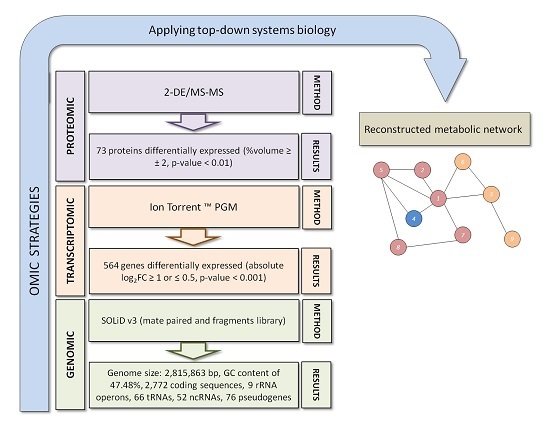
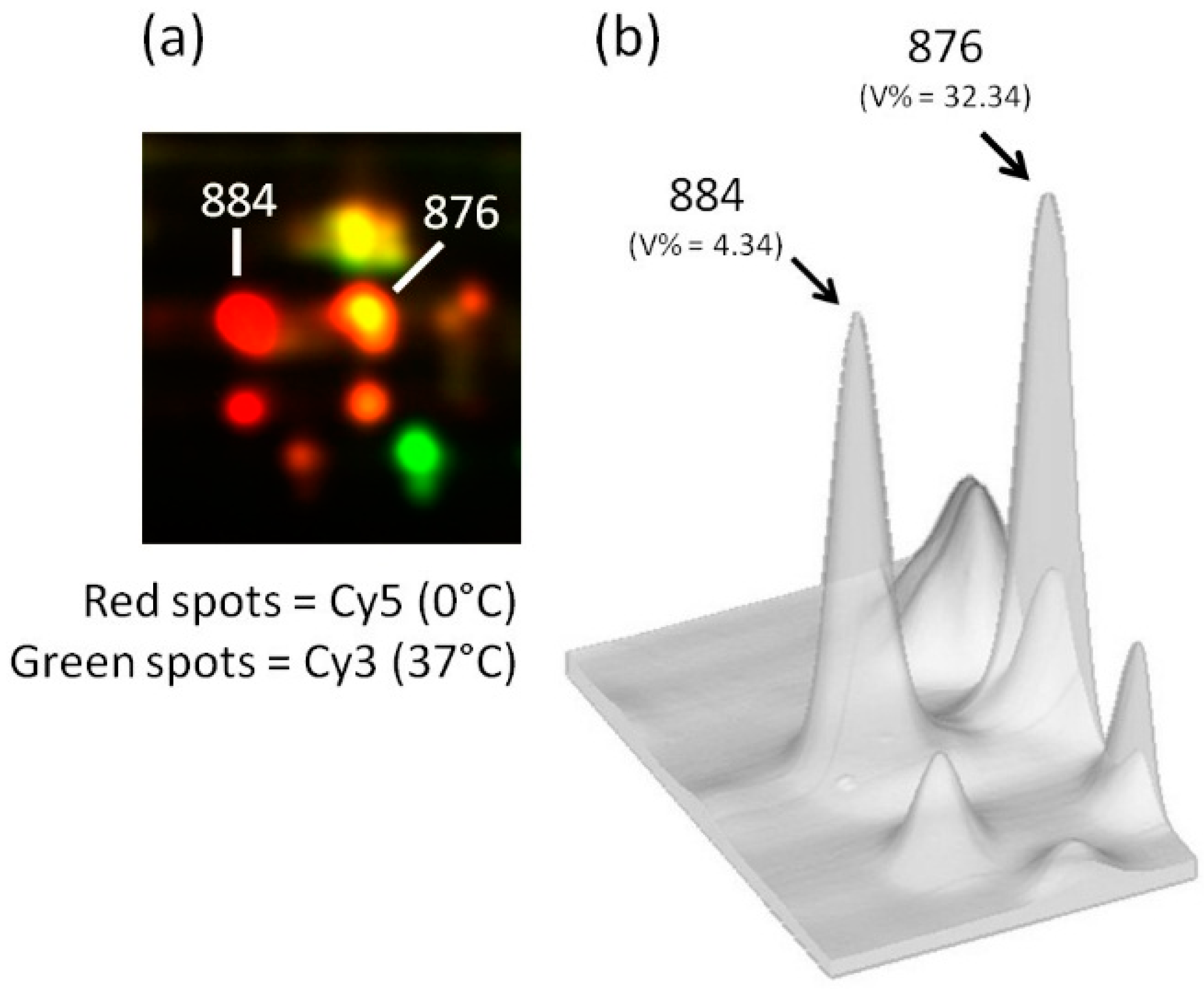
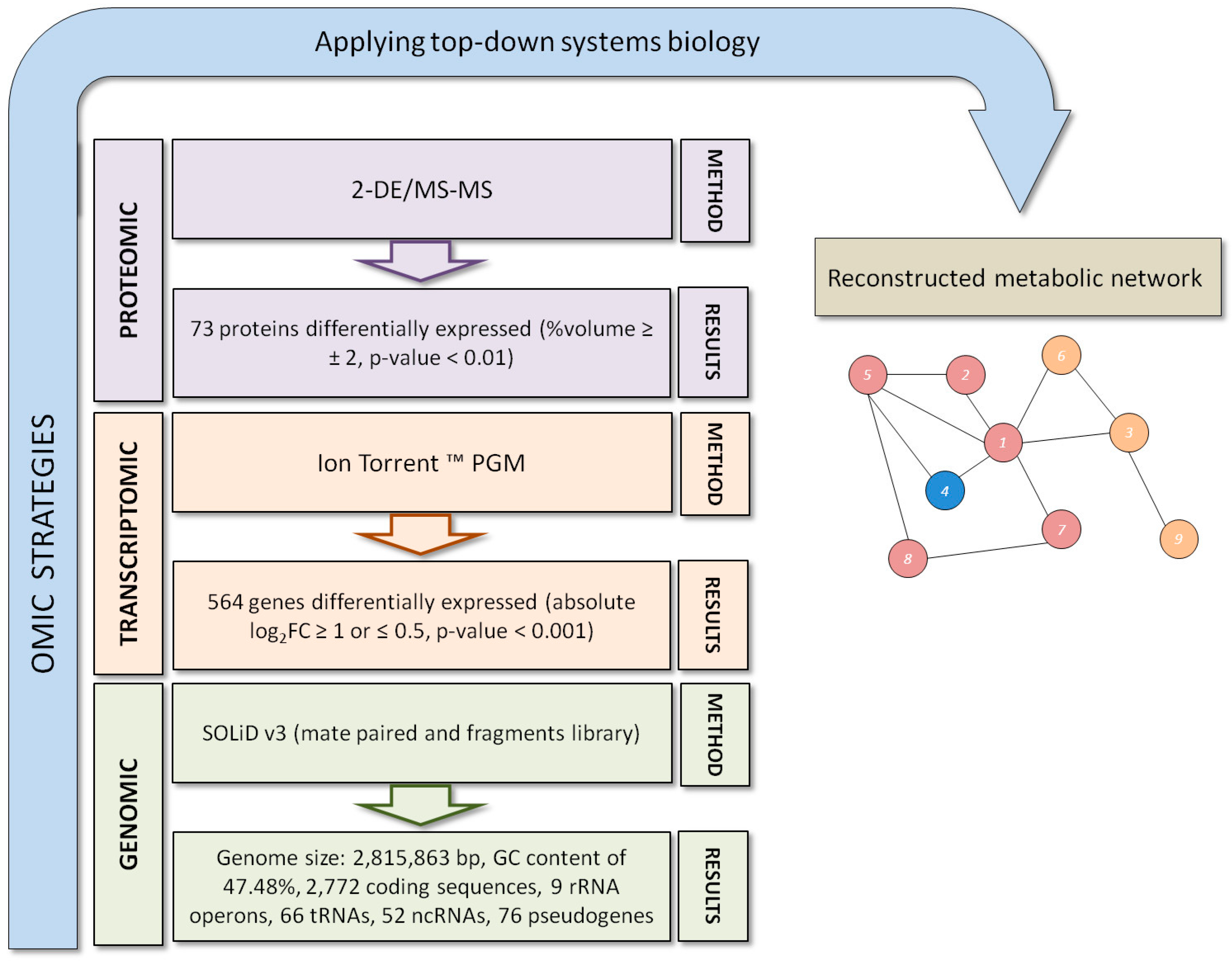
3. Exiguobacterium antarcticum B7 as a Model Organism for Studies of Cold Adaptation
4. Conclusions and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
References
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of protein. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [PubMed]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshikazu, Y.; Yoshida, Y.; Matsuo, T. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Fenn, J.B.; Mann, M.; Meng, C.K.; Wong, S.F. Electrospray ionization for mass spectrometry of large biomolecules. Science 1989, 246, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Rampelotto, P.H. Resistance of microorganisms to extreme environmental conditions and its contribution to astrobiology. Sustainability 2010, 2, 1602–1623. [Google Scholar] [CrossRef]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of “omic” technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Tiedje, J.M. Multi-locus real-time PCR for quantitation of bacteria in the environment reveals Exiguobacterium to be prevalent in permafrost. FEMS Microbiol. Ecol. 2007, 59, 489–499. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskaya, T.A.; Kathariou, S.; Tiedje, J.M. The Exiguobacterium genus: Biodiversity and biogeography. Extremophiles 2009, 13, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, A.R.; Ramos, R.T.J.; Dall’Agnol, H.; Pinto, A.C.; Soares, S.C.; Santos, A.R.; Guimarães, L.C.; Almeida, S.S.; Baraúna, R.A.; Graças, D.A.; et al. Genome sequence of Exiguobacterium antarcticum B7, isolated from a biofilm in Ginger Lake, King George Island, Antarctica. J. Bacteriol. 2012, 194, 6689–6690. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Ivanova, N.; He, Z.; Huebner, M.; Zhou, J.; Tiedje, J.M. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: A genome and transcriptome approach. BMC Genom. 2008, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskaya, T.A.; Lucas, S.; Copeland, A.; Lapidus, A.; del Rio, T.G.; Dalin, E.; Tice, H.; Bruce, D.C.; Goodwin, L.A.; Pitluk, S.; et al. Complete genome sequence of the thermophilic Exiguobacterium sp. AT1b. J. Bacteriol. 2011, 193, 2880–2881. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhang, Y.; Meng, H.; Xue, Z.; Ma, J. Complete genome sequence of Exiguobacterium sp. strain MH3, isolated from rhizosphere of Lemma minor. Genome Announc. 2013, 1, 2012–2013. [Google Scholar] [CrossRef] [PubMed]
- Baraúna, R.A.; das Graças, D.A.; Nunes, C.I.P.; Schneider, M.P.C.; Silva, A.; Carepo, M.S.P. De novo synthesis of fatty acids is regulated by FapR protein in Exiguobacterium antarcticum B7, a psychrotrophic bacterium isolated from Antarctica. BMC Res. Notes 2016, 9, 447. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, R.; Baraúna, R.A.; Silva, A.; Carepo, M.S.P.; Oliveira, R.; Marques, R.; Ramos, R.T.J.; Schneider, M.P.C. Reconstruction of the fatty acid biosynthetic pathway of Exiguobacterium antarcticum B7 based on genomic and bibliomic data. Biomed. Res. Int. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dall’Agnol, H.P.M.B.; Baraúna, R.A.; de Sá, P.H.C.G.; Ramos, R.T.J.; Nóbrega, F.; Nunes, C.I.P.; Graças, D.A.; Carneiro, A.R.; Santos, D.M.; Pimenta, A.M.C.; et al. Omics profiles used to evaluate the gene expression of Exiguobacterium antarcticum B7 during cold adaptation. BMC Genom. 2014, 15, 986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, N.J. Mechanisms of thermal adaptation in bacteria: Blueprints for survival. Trends Biochem. Sci. 1984, 9, 108–112. [Google Scholar] [CrossRef]
- Sinensky, M. Homeoviscous adaptation a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Nat. Acad. Sci. USA 1974, 71, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Rock, C.O. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008, 6, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, P.S.; Cronan, J.E.; de Mendoza, D. A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipid desaturase. J. Bacteriol. 1998, 180, 2194–2200. [Google Scholar] [PubMed]
- Albnesi, D.; Mansilla, M.C.; de Mendoza, D. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J. Bacteriol. 2004, 186, 2655–2663. [Google Scholar] [CrossRef]
- Aguilar, P.S.; Hernandez-Arriaga, A.M.; Cybulski, L.E.; Erazo, A.C.; de Mendoza, D. Molecular basis of thermosensing: A two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001, 20, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- De Mendoza, D. Temperature sensing by membranes. Annu. Rev. Microbiol. 2014, 68, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Methé, B.A.; Nelson, K.E.; Deming, J.W.; Momen, B.; Melamud, E.; Zhang, X.; Moult, J.; Madupu, R.; Nelson, W.C.; Dodson, R.J.; et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Nat. Acad. Sci. USA 2005, 102, 10913–10918. [Google Scholar] [CrossRef] [PubMed]
- Nunn, B.L.; Slattery, K.V.; Cameron, K.A.; Schiffman, E.T.; Junge, K. Proteomics of Colwellia psychrerythraea at subzero temperatures—A life with limited movement, flexible membranes and vital DNA repair. Environ. Microbiol. 2015, 17, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hashimoto, M.; Hori, R.; Adachi, T.; Okuyama, H.; Orikasa, Y.; Nagamine, T.; Shimizu, S.; Ueno, A.; Morita, N. Bacterial long-chain polyunsaturated fatty acids: Their biosynthetic genes, functions and practical use. Mar. Drugs 2016, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Ting, L.; Williams, T.J.; Cowley, M.J.; Lauro, F.M.; Guilhaus, M.; Raftery, M.J.; Cavicchioli, R. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environ. Microbiol. 2010, 12, 2658–2676. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, P.W.; Bakermans, C.; Tiedje, J.M. Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J. Bacteriol. 2009, 191, 2340–2352. [Google Scholar] [CrossRef] [PubMed]
- Mykytczuk, N.C.S.; Foote, S.J.; Omelon, C.R.; Southam, G.; Greer, C.W.; Whyte, L.G. Bacterial growth at −15 °C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 2013, 7, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Ross, T.; Bowman, J.P. Characterisation of the transcriptomes of genetically diverse Listeria monocytogenes exposed to hyperosmotic and low temperature conditions reveal global stress-adaptation mechanisms. PLoS ONE 2013, 8, e73603. [Google Scholar] [CrossRef] [PubMed]
- Sandermann, H., Jr. Regulation of membrane enzymes by lipids. Biochim. Biophys. Acta 1978, 515, 209–237. [Google Scholar] [CrossRef]
- Kovacic, F.; Mandrysch, A.; Poojari, C.; Strodel, B.; Jaeger, K.E. Structural features determining thermal adaptation of esterases. Protein Eng. Des. Sel. 2016, 29, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Feller, G. Psychrophilic enzymes: From folding to function and biotechnology. Scientifica 2013. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Marx, J.C.; Collins, T.; D’Amico, S.; Feller, G.; Gerday, C. Cold-adapted enzymes from marine Antarctic microorganisms. Mar. Biotechnol. 2007, 9, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein disorder prediction: Implications for structural proteomics. Structure 2003, 11, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Ayala-del-Río, H.L.; Chain, P.S.; Grzymski, J.J.; Ponder, M.A.; Ivanova, N.; Bergholz, P.W.; Di Bartolo, G.; Hauser, L.; Land, M.; Bakermans, C.; et al. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl. Environ. Microbiol. 2010, 76, 2304–2312. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yang, G.; Aprile, L.; Turturo, V.; Pucciarelli, S.; Pucciarelli, S.; Miceli, C. Characterization and comparative analysis of psychrophilic and mesophilic alpha-amilases from Euplotes species: A contribution to the understanding of enzyme thermal adaptation. Biochem. Biophys. Res. Commun. 2013, 4, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsen, S.; Hjerde, E.; Fenton, C.; Willassen, N.P. Molecular characterization of cold adaptation based on ortholog protein sequences from Vibrionaceae species. Extremophiles 2007, 11, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Fields, P.A. Review: Protein function at thermal extremes: Balancing stability and flexibility. Comp. Biochem. Phys. A 2001, 129, 417–431. [Google Scholar] [CrossRef]
- Adekoya, O.A.; Helland, R.; Willassen, N.P.; Sylte, I. Comparative sequence and structure analysis reveal features of cold adaptation of an enzyme in the thermolysin family. Proteins Struct. Funct. Bioinform. 2006, 62, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Panoff, J.M.; Corroler, D.; Thammavongs, B.; Boutibonnes, P. Differentiation between cold shock and cold acclimation proteins in a mesophilic gram-positive bacterium, Enterococcus faecalis JH2-2. J. Bacteriol. 1997, 179, 4451–4454. [Google Scholar] [CrossRef] [PubMed]
- Phadtare, S. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 2004, 6, 125–136. [Google Scholar] [PubMed]
- Graumann, P.L.; Marahiel, M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998, 23, 286–290. [Google Scholar] [CrossRef]
- Pierce, A.; Gillette, D.; Jones, P.G. Escherichia coli cold shock proteins CsdA effects an increase in septation and the resultant formation of coccobacilli at low temperatures. Arch. Microbiol. 2011, 193, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Ke, H.; Shinde, U.; Inouye, M. The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J. Mol. Biol. 2003, 332, 575–584. [Google Scholar] [CrossRef]
- Yamanaka, K.; Inouye, M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 2001, 183, 2808–2816. [Google Scholar] [CrossRef] [PubMed]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold shock proteins: A minireview with special emphasis on Csp-family of Enteropathogenic Yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, K.; Saxena, J.; Tripathi, B.K.; Agarwal, M.K. Detection of carotenoids in psychrotrophic bacteria by spectroscopic approach. J. BioSci. Biotechnol. 2014, 3, 253–260. [Google Scholar]
- Takano, H.; Asker, D.; Beppu, T.; Ueda, K. Genetic control for light-induced carotenoid production in non-phototrophic bacteria. J. Ind. Microbiol. Biotechnol. 2006, 33, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Dsouza, M.; Taylor, M.W.; Turner, S.J.; Aislabie, J. Genomic and phenotypic insights into the ecology of Arthrobacter from Antarctic soils. BMC Genom. 2015, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Jagannadham, M.V.; Chattopadhyay, M.K.; Subbalakshmi, C.; Vairamani, M.; Narayanan, K.; Rao, C.M.; Shivaji, S. Carotenoids of an Antarctic psychrotolerant bacterium, Sphingobacterium antarcticus, and a mesophilic bacterium, Sphingobacterium multivorum. Arch. Microbiol. 2000, 173, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid pigmentation in Antarctic heterotrophic bacteria as a strategy to withstand environmental stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Fong, N.J.C.; Burgess, M.L.; Barrow, K.D.; Glenn, D.R. Carotenoid accumulation in the psychrotrophic bacterium Arthrobacter agilis in response to thermal and salt stress. Appl. Microbiol. Biotechnol. 2001, 56, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K.; Jagannadham, M.V.; Vairamani, M.; Shivaji, S. Carotenoid pigments of an Antarctic psychrotrophic bacterium Micrococcus roseus: Temperature dependent biosynthesis, structure, and interaction with synthetic membranes. Biochem. Biophys. Res. Commun. 1997, 239, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Nupur, L.N.U.; Vats, A.; Dhanda, S.K.; Raghava, G.P.S.; Pinnaka, A.K.; Kumar, A. ProCarDB: A database of bacterial carotenoids. BMC Microbiol. 2016, 16, 96. [Google Scholar] [CrossRef] [PubMed]
- Ghobakhlou, A.F.; Johnston, A.; Harris, L.; Antoun, H.; Laberge, S. Microarray transcriptional profiling of Arctic Mesorhizobium strain N33 at low temperature provides insights into cold adaption strategies. BMC Genom. 2015, 16, 383. [Google Scholar] [CrossRef] [PubMed]
- Palsson, B. In silico biology through “omics”. Nat. Biotechnol. 2002, 20, 649–650. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; de Hoon, M.J.L.; Grimmond, S.M.; Daub, C.O.; Hayashizaki, Y.; Faulkner, G.J. Probabilistic resolution of multi-mapping reads in massively parallel sequencing data using MuMRescueLite. Bioinformatics 2009, 25, 2613–2614. [Google Scholar] [CrossRef] [PubMed]
- Petrak, J.; Ivanek, R.; Toman, O.; Cmejla, R.; Cmejlova, J.; Vyoral, D.; Zivny, J.; Vulpe, C.D. Déjà vu in proteomics: A hit parade of repeatedly identified differentially expressed proteins. Proteomics 2008, 8, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.J.; Wilkins, D.; Long, E.; Evans, F.; DeMaere, M.Z.; Raftery, M.J.; Cavicchioli, R. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ. Microbiol. 2013, 15, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Karlson, C.; Malmström, L.; Aebersold, R.; Malmström, J. Proteome-wide selected reaction monitoring assays for the human pathogen Streptococcus pyogenes. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Olaya-Abril, A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Rodríguez-Ortega, M.J. Surfomics: Shaving live organisms for a fast proteomic identification of surface proteins. J. Proteom. 2014, 97, 164–176. [Google Scholar] [CrossRef] [PubMed]
| Genes | Log2FC | p-Value | Genes | Log2FC | p-Value |
|---|---|---|---|---|---|
| Cold shock proteins | De novo synthesis of fatty acids | ||||
| csp1 | 1.94 | 0 | accA | 0.45 | 0.01 |
| csp2 | 2.16 | 5.43 × 10−194 | accB | −0.05 | 0.01 |
| csp3 | 2.30 | 0 | accC | −0.56 | 5.68 × 10−27 |
| csp4 | 2.46 | 0 | accD | −0.28 | 7.55 × 10−4 |
| csp5 | −1.06 | 3.73 × 10−35 | fapR | 0.58 | 4.80 × 10−21 |
| csp6 | −1.28 | 4.41 × 10−194 | plsX | 0.77 | 2.31 × 10−46 |
| Desaturation of membrane fatty acids | fabD | 0.94 | 2.92 × 10−85 | ||
| desK | 7.03 | 8.15 × 10−16 | fabG | 0.85 | 8.42 × 10−63 |
| desR | −0.48 | 9.37 × 10−8 | fabH1 | 0.85 | 2.67 × 10−26 |
| Transport of compatible solutes | fabF | 0.69 | 3.46 × 10−16 | ||
| opuCA | 3.17 | 1.99 × 10−62 | fabI | −1.74 | 0 |
| opuCC | 1.61 | 3.06 × 10−29 | plsC | −0.82 | 1.28 × 10−6 |
| opuE | 3.70 | 4.26 × 10−16 | Carotenoid biosynthesis | ||
| opuCD | −2.72 | 8.67 × 10−24 | crtI (pseudo) | 3.94 | 4.65 × 10−43 |
| opuBA | −0.52 | 6.11 × 10−4 | yisP1 (pseudo) | −1.33 | 3.09 × 10−41 |
| yisP2 | 0.75 | 2.16 × 10−7 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baraúna, R.A.; Freitas, D.Y.; Pinheiro, J.C.; Folador, A.R.C.; Silva, A. A Proteomic Perspective on the Bacterial Adaptation to Cold: Integrating OMICs Data of the Psychrotrophic Bacterium Exiguobacterium antarcticum B7. Proteomes 2017, 5, 9. https://doi.org/10.3390/proteomes5010009
Baraúna RA, Freitas DY, Pinheiro JC, Folador ARC, Silva A. A Proteomic Perspective on the Bacterial Adaptation to Cold: Integrating OMICs Data of the Psychrotrophic Bacterium Exiguobacterium antarcticum B7. Proteomes. 2017; 5(1):9. https://doi.org/10.3390/proteomes5010009
Chicago/Turabian StyleBaraúna, Rafael A., Dhara Y. Freitas, Juliana C. Pinheiro, Adriana R. C. Folador, and Artur Silva. 2017. "A Proteomic Perspective on the Bacterial Adaptation to Cold: Integrating OMICs Data of the Psychrotrophic Bacterium Exiguobacterium antarcticum B7" Proteomes 5, no. 1: 9. https://doi.org/10.3390/proteomes5010009
APA StyleBaraúna, R. A., Freitas, D. Y., Pinheiro, J. C., Folador, A. R. C., & Silva, A. (2017). A Proteomic Perspective on the Bacterial Adaptation to Cold: Integrating OMICs Data of the Psychrotrophic Bacterium Exiguobacterium antarcticum B7. Proteomes, 5(1), 9. https://doi.org/10.3390/proteomes5010009