Detection of Functional Overreaching in Endurance Athletes Using Proteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Baseline Testing
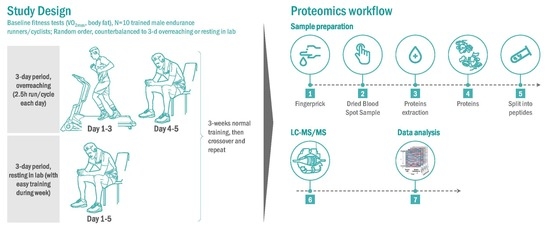
2.3. Research Design for the Randomized Trials
2.4. Proteomics Procedures
2.5. Data Processing
2.6. Statistics
2.7. Protein–Protein Interaction Network Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CV | Coefficient of variation |
| DIA | Data independent acquisition |
| DBS | Dried blood spot |
| FDR | False discovery rate |
| FOR | Functional overreaching |
| GEE | Generalized estimating equation |
| GLMM | Generalized linear mixed models |
| NFOR | Nonfunctional overreaching |
| OTS | Overtraining syndrome |
| RPE | Rating of perceived exertion |
| SAA | Serum amyloid A |
| STRING | Search tool for the retrieval of interacting genes/proteins |
| TDS | Training distress scale |
| VO2max | Maximal volume of oxygen consumption |
References
- Meeusen, R.; Duclos, M.; Foster, C.; Fry, A.; Gleeson, M.; Nieman, D.; Raglin, J.; Rietjens, G.; Steinacker, J.; Urhausen, A. European College of Sport Science; American College of Sports Medicine. Prevention, diagnosis, and treatment of the overtraining syndrome: Joint consensus statement of the European College of Sport Science and the American College of Sports Medicine. Med. Sci. Sports Exerc. 2013, 45, 186–205. [Google Scholar] [PubMed]
- Aubry, A.; Hausswirth, C.; Louis, J.; Coutts, A.J.; Le Meur, Y. Functional overreaching: The key to peak performance during the taper? Med. Sci. Sports Exerc. 2014, 46, 1769–1777. [Google Scholar] [CrossRef] [PubMed]
- Soligard, T.; Schwellnus, M.; Alonso, J.M.; Bahr, R.; Clarsen, B.; Dijkstra, H.P.; Gabbett, T.; Gleeson, M.; Hägglund, M.; Hutchinson, M.R.; et al. How much is too much? (Part 1) International Olympic Committee consensus statement on load in sport and risk of injury. Br. J. Sports Med. 2016, 50, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Saw, A.E.; Main, L.C.; Gastin, P.B. Monitoring the athlete training response: Subjective self-reported measures trump commonly used objective measures: A systematic review. Br. J. Sports Med. 2016, 50, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Joro, R.; Uusitalo, A.; DeRuisseau, K.C.; Atalay, M. Changes in cytokines, leptin, and IGF-1 levels in overtrained athletes during a prolonged recovery phase: A case-control study. J. Sports Sci. 2017, 35, 2342–2349. [Google Scholar] [CrossRef] [PubMed]
- Jürimäe, J.; Mäestu, J.; Jürimäe, T.; Mangus, B.; von Duvillard, S.P. Peripheral signals of energy homeostasis as possible markers of training stress in athletes: A review. Metabolism 2011, 60, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Halson, S.L.; Jeukendrup, A.E. Does overtraining exist? An analysis of overreaching and overtraining research. Sports Med. 2004, 34, 967–981. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Suárez, E.; Whetton, A.D. The application of quantification techniques in proteomics for biomedical research. Mass. Spectrom. Rev. 2013, 32, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Balfoussia, E.; Skenderi, K.; Tsironi, M.; Anagnostopoulos, A.K.; Parthimos, N.; Vougas, K.; Papassotiriou, I.; Tsangaris, G.T.; Chrousos, G.P. A proteomic study of plasma protein changes under extreme physical stress. J. Proteom. 2014, 98, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cywinska, A.; Gorecka, R.; Szarska, E.; Witkowski, L.; Dziekan, P.; Schollenberger, A. Serum amyloid A level as a potential indicator of the status of endurance horses. Equine Vet. J. Suppl. 2010, 38, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Cywinska, A.; Witkowski, L.; Szarska, E.; Schollenberger, A.; Winnicka, A. Serum amyloid A (SAA) concentration after training sessions in Arabian race and endurance horses. BMC Vet. Res. 2013, 9, 91. [Google Scholar] [CrossRef] [PubMed]
- Valle, E.; Zanatta, R.; Odetti, P.; Traverso, N.; Furfaro, A.; Bergero, D.; Badino, P.; Girardi, C.; Miniscalco, B.; Bergagna, S.; et al. Effects of competition on acute phase proteins and lymphocyte subpopulations—Oxidative stress markers in eventing horses. J. Anim. Physiol. Anim. Nutr. 2015, 99, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Casella, S.; Fazio, F.; Russo, C.; Giudice, E.; Piccione, G. Acute phase proteins response in hunting dogs. J. Vet. Diagn. Investig. 2013, 25, 577–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakshlag, J.J.; Stokol, T.; Geske, S.M.; Greger, C.E.; Angle, C.T.; Gillette, R.L. Evaluation of exercise-induced changes in concentrations of C-reactive protein and serum biochemical values in sled dogs completing a long-distance endurance race. Am. J. Vet. Res. 2010, 71, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Art, T.; Franck, T.; Gangl, M.; Votion, D.; Kohnen, S.; Deby-Dupont, G.; Serteyn, D. Plasma concentrations of myeloperoxidase in endurance and 3-day event horses after a competition. Equine Vet. J. Suppl. 2006, 36, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Serteyn, D.; Sandersen, C.; Lejeune, J.P.; de la Rebière de Pouyade, G.; Ceusters, J.; Mouithys-Mickalad, A.; Niesten, A.; Fraipont, A.; van Erck, E.; Goachet, A.G.; et al. Effect of a 120 km endurance race on plasma and muscular neutrophil elastase and myeloperoxidase concentrations in horses. Equine Vet. J. Suppl. 2010, 38, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, J.P.; Sandersen, C.; Votion, D.; Caudron, I.; Vander Heyden, L.; Franck, T.; Ceusters, J.; Mouithys-Mickalad, A.; Niesten, A.; De La Rebière de Pouyade, G.; et al. Effect of intensive exercise on plasmatic neutrophil elastase level in eventing and endurance horses. Equine Vet. J. Suppl. 2010, 38, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Petibois, C.; Cazorla, G.; Poortmans, J.R.; Déléris, G. Biochemical aspects of overtraining in endurance sports: A review. Sports Med. 2002, 32, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.R.; Main, L.C.; Partridge, K.; Bishop, D.J.; Russell, S.; Shepherdson, A.; Ferguson, L. Training distress and performance readiness: Laboratory and field validation of a brief self-report measure. Scand. J. Med. Sci. Sports 2014, 24, e483–e490. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Röst, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteom. 2012, 11, O111-016717. [Google Scholar] [CrossRef] [PubMed]
- Bilbao, A.; Zhang, Y.; Varesio, E.; Luban, J.; Strambio-De-Castillia, C.; Lisacek, F.; Hopfgartner, G. Ranking fragment ions based on outlier detection for improved label-free quantification in data-independent acquisition LC-MS/MS. J. Proteome Res. 2015, 14, 4581–4593. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.J.; Bunch, J.; Cooper, H.J. Dried blood spot proteomics: Surface extraction of endogenous proteins coupled with automated sample preparation and mass spectrometry analysis. J. Am. Soc. Mass Spectrom. 2013, 24, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, O.H.; Andersen, K.; Fischer, C.; Nielsen, A.R.; Nielsen, S.; Akerström, T.; Aastrøm, M.B.; Borup, R.; Pedersen, B.K. Calprotectin is released from human skeletal muscle tissue during exercise. J. Physiol. 2008, 586, 3551–3562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neubauer, O.; König, D.; Wagner, K.H. Recovery after an Ironman triathlon: Sustained inflammatory responses and muscular stress. Eur. J. Appl. Physiol. 2008, 104, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Schiopu, A.; Cotoi, O.S. S100A8 and S100A9: DAMPs at the crossroads between innate immunity, traditional risk factors, and cardiovascular disease. Mediat. Inflamm. 2013, 2013, 828354. [Google Scholar] [CrossRef] [PubMed]
- Oesterle, A.; Bowman, M.A. S100A12 and the S100/calgranulins: Emerging biomarkers for atherosclerosis and possibly therapeutic targets. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2496–2507. [Google Scholar] [CrossRef] [PubMed]
- Peake, J.M.; Neubauer, O.; Della Gatta, P.A.; Nosaka, K. Muscle damage and inflammation during recovery from exercise. J. Appl. Physiol. 2017, 122, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, B.; Horwitz, M.S.; Jenne, D.E.; Gauthier, F. Neutrophil elastase, proteinase 3, and cathepsin G as therapeutic targets in human diseases. Pharmacol. Rev. 2010, 62, 726–759. [Google Scholar] [CrossRef] [PubMed]
- Schleicher, M.; André, B.; Andréoli, C.; Eichinger, L.; Haugwitz, M.; Hofmann, A.; Karakesisoglou, J.; Stöckelhuber, M.; Noegel, A.A. Structure/function studies on cytoskeletal proteins in Dictyostelium amoebae as a paradigm. FEBS Lett. 1995, 369, 38–42. [Google Scholar] [CrossRef] [Green Version]
- Weiner, O.D.; Servant, G.; Welch, M.D.; Mitchison, T.J.; Sedat, J.W.; Bourne, H.R. Spatial control of actin polymerization during neutrophil chemotaxis. Nat. Cell Biol. 1999, 1, 75–81. [Google Scholar] [PubMed]
- Semple, S.J.; Smith, L.L.; McKune, A.J.; Hoyos, J.; Mokgethwa, B.; San Juan, A.F.; Lucia, A.; Wadee, A.A. Serum concentrations of C reactive protein, α1 antitrypsin, and complement (C3, C4, C1 esterase inhibitor) before and during the Vuelta a Espańa. Br. J. Sports Med. 2006, 40, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Margeli, A.; Skenderi, K.; Tsironi, M.; Hantzi, E.; Matalas, A.L.; Vrettou, C.; Kanavakis, E.; Chrousos, G.; Papassotiriou, I. Dramatic elevations of interleukin-6 and acute-phase reactants in athletes participating in the ultradistance foot race spartathlon: Severe systemic inflammation and lipid and lipoprotein changes in protracted exercise. J. Clin. Endocrinol. Metab. 2005, 90, 3914–3918. [Google Scholar] [CrossRef] [PubMed]
- Liesen, H.; Dufaux, B.; Hollmann, W. Modifications of serum glycoproteins the days following a prolonged physical exercise and the influence of physical training. Eur. J. Appl. Physiol. Occup. Physiol. 1977, 37, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Papassotiriou, I.; Alexiou, V.G.; Tsironi, M.; Skenderi, K.; Spanos, A.; Falagas, M.E. Severe aseptic inflammation caused by long distance running (246 km) does not increase procalcitonin. Eur. J. Clin. Investig. 2008, 38, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Fallon, K.E. The acute phase response and exercise: The ultramarathon as prototype exercise. Clin. J. Sport Med. 2001, 11, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Weight, L.M.; Alexander, D.; Jacobs, P. Strenuous exercise: Analogous to the acute-phase response? Clin. Sci. 1991, 81, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Peake, J.; Nosaka, K.; Okutsu, M.; Abbiss, C.R.; Surriano, R.; Bishop, D.; Quod, M.J.; Lee, H.; Martin, D.T.; et al. Changes in markers of muscle damage, inflammation and HSP70 after an Ironman Triathlon race. Eur. J. Appl. Physiol. 2006, 98, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Ye, R.D.; Sun, L. Emerging functions of serum amyloid A in inflammation. J. Leukoc. Biol. 2015, 98, 923–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabay, C.; Kushner, I. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 1999, 340, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Melanson, S.E.; Green, S.M.; Wood, M.J.; Neilan, T.G.; Lewandrowski, E.L. Elevation of myeloperoxidase in conjunction with cardiac-specific markers after marathon running. Am. J. Clin. Pathol. 2006, 126, 888–893. [Google Scholar] [CrossRef] [PubMed]
- Nieman, D.C.; Luo, B.; Dréau, D.; Henson, D.A.; Shanely, R.A.; Dew, D.; Meaney, M.P. Immune and inflammation responses to a 3-day period of intensified running versus cycling. Brain Behav. Immun. 2014, 39, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.A.; Davis, J.M.; Smith, J.A.; Nieman, D.C. Exercise and cellular innate immune function. Med. Sci. Sports Exerc. 1999, 31, 57–66. [Google Scholar] [CrossRef] [PubMed]










| Variable | Mean ± SE |
|---|---|
| Age (years) | 38.3 ± 3.4 |
| Height (m) | 1.81 ± 0.02 |
| Weight (kg) | 85.6 ± 1.3 |
| Body fat (%) | 20.9 ± 2.1 |
| VO2max (mL kg−1 min−1) | 42.0 ± 1.3 |
| Maximal heart rate (beats/min) | 176 ± 3.7 |
| UniProt Protein | Protein Name | Basic Function |
|---|---|---|
| P61626 | Lysozyme C | Monocyte/macrophage bacterilytic function |
| P08246 | Neutrophil elastase | Modifies the functions of natural killer cells, monocytes and granulocytes |
| P59665; P59666 | Neutrophil defensin 1 | Antibacterial, fungicide, antiviral activity; kills by permeabilizing membrane |
| P80511 | Protein S100-A12 | Ca, Zn, Cu binding protein; prominent role, regulation inflammation/immune |
| P05109 | Protein S100-A8 | Ca, Zn binding protein; regulate inflammation/immune; chemotaxis |
| P49913 | Cathelicidin antimicrobial peptide | Binds to bacterial lipopolysaccharides (LPS), has antibacterial activity |
| P12814 | Alpha-actinin-1 | F-actin cross-linking protein to anchor actin to intracellular structures |
| P60709 | Actin, cytoplasmic 1 | Cell motility; granulocytes |
| P07737 | Profilin-1 | Binds to actin; granulocyte motility/chemotaxis |
| P02776 | Platelet factor 4 | Released during platelet aggregation; chemokine activity; chemotaxis |
| P60660 | Myosin light polypeptide 6 | Regulatory light chain myosin; muscle development |
| Q96QV6; Q93077 | Histone H2A types | Component of nucleosome; transcription regulation, DNA repair |
| P62805 | Histone H4 | Component of nucleosome; transcription regulation, DNA repair |
| P05204 | Non-histone chromosomal protein HMG-17 | Binds nucleosomal DNA |
| O95810 | Serum deprivation-response protein | Targets protein kinase C-alpha on lipid rafts |
| UniProt Protein | Protein Name | Function |
|---|---|---|
| P35542 | Serum amyloid A-4 protein | Major acute phase reactant; cell chemotaxis |
| P05164 | Myeloperoxidase | Granulocyte microbicidal activity against wide range of pathogens; production of hypochlorous acid |
| P07360 | Complement component C8 gamma chain | Part of membrane attack complex that plays key role in immune response; forms pores in target cells |
| P0C0L5 | Complement C4B | Non-enzymatic component C3, C5 convertases and thus essential for complement activation |
| P05155 | Plasma protease C1 inhibitor | Crucial role in regulation of complement activation |
| Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | Acute-phase protein involved in trauma inflammatory response |
| P19652 | Alpha-1-acid glycoprotein 2 | Transport protein; modulates immune function during the acute-phase reaction; inflammation |
| P10643 | Complement component C7 | Part of membrane attack complex that plays key role in immune response; forms pores in target cells |
| P02765 | Alpha-2-HS-glycoprotein | Promotes endocytosis; part of acute-phase response; phagocytosis; bone mineral influence |
| P01834 | Immunoglobulin kappa constant | Constant region of immunoglobulin heavy chains; complement activation; defense immune response; phagocytosis recognition and engulfment |
| P01871; P04220 | Immunoglobulin heavy constant mu | Constant region of immunoglobulin heavy chains; C region; antigen binding; immune response |
| P08185 | Corticosteroid-binding globulin | Major transport protein for glucocorticoids and progestins |
| P35754 | Glutaredoxin-1 | Glutathione activity; cell redox homeostasis |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nieman, D.C.; Groen, A.J.; Pugachev, A.; Vacca, G. Detection of Functional Overreaching in Endurance Athletes Using Proteomics. Proteomes 2018, 6, 33. https://doi.org/10.3390/proteomes6030033
Nieman DC, Groen AJ, Pugachev A, Vacca G. Detection of Functional Overreaching in Endurance Athletes Using Proteomics. Proteomes. 2018; 6(3):33. https://doi.org/10.3390/proteomes6030033
Chicago/Turabian StyleNieman, David C., Arnoud J. Groen, Artyom Pugachev, and Gianmarco Vacca. 2018. "Detection of Functional Overreaching in Endurance Athletes Using Proteomics" Proteomes 6, no. 3: 33. https://doi.org/10.3390/proteomes6030033
APA StyleNieman, D. C., Groen, A. J., Pugachev, A., & Vacca, G. (2018). Detection of Functional Overreaching in Endurance Athletes Using Proteomics. Proteomes, 6(3), 33. https://doi.org/10.3390/proteomes6030033