Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. WGA-Bound Glycoproteins Enrichment
2.3. Assessment of Total Plasma Protein Levels
2.4. Sample Preparation for LC-MS/MS Analysis
2.5. LC-MS/MS Analysis
2.6. Label-Free Relative Quantification and Identification of Proteins
2.7. Protein-Protein Interaction (PPI) Network Analysis
2.8. Immunoblot Analysis
2.9. CEA ELISA Assay
2.10. Statistical Analysis
3. Results
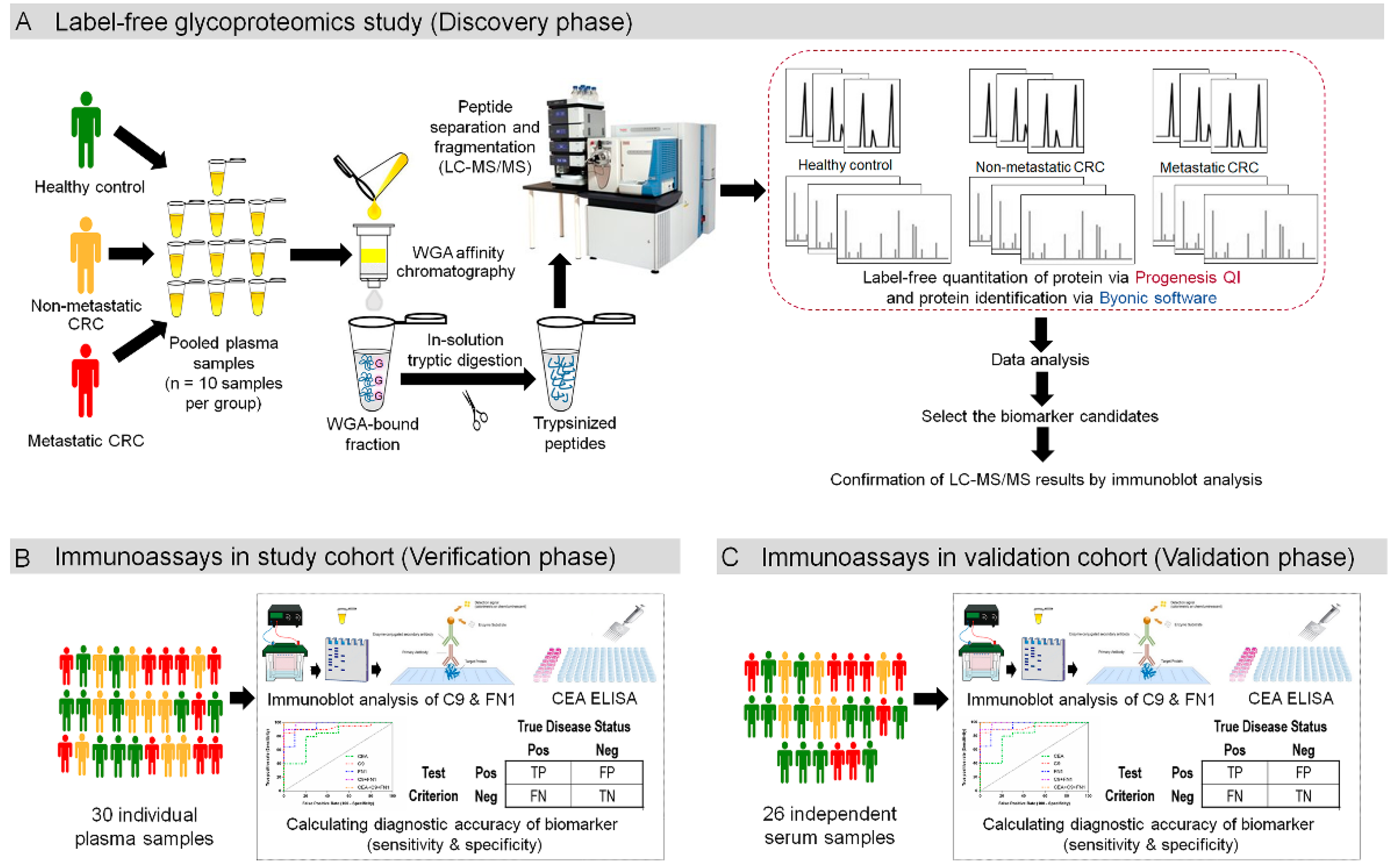
3.1. Experimental Overview to Study Plasma Glycoprotein Biomarkers for CRC
3.2. Enrichment of Plasma Glycoproteins Using WGA
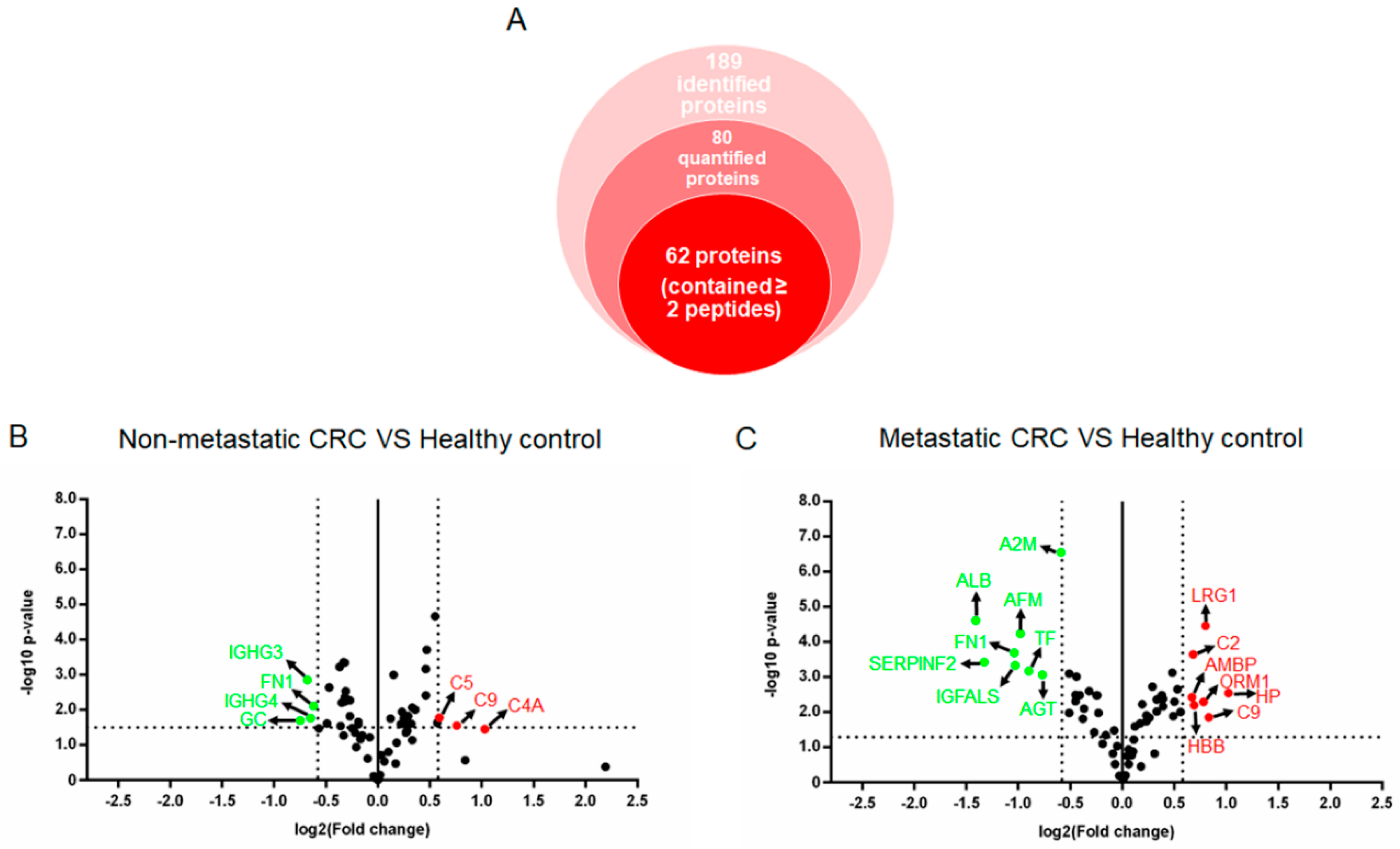
3.3. Label-free Relative Quantification of Plasma Glycoproteins
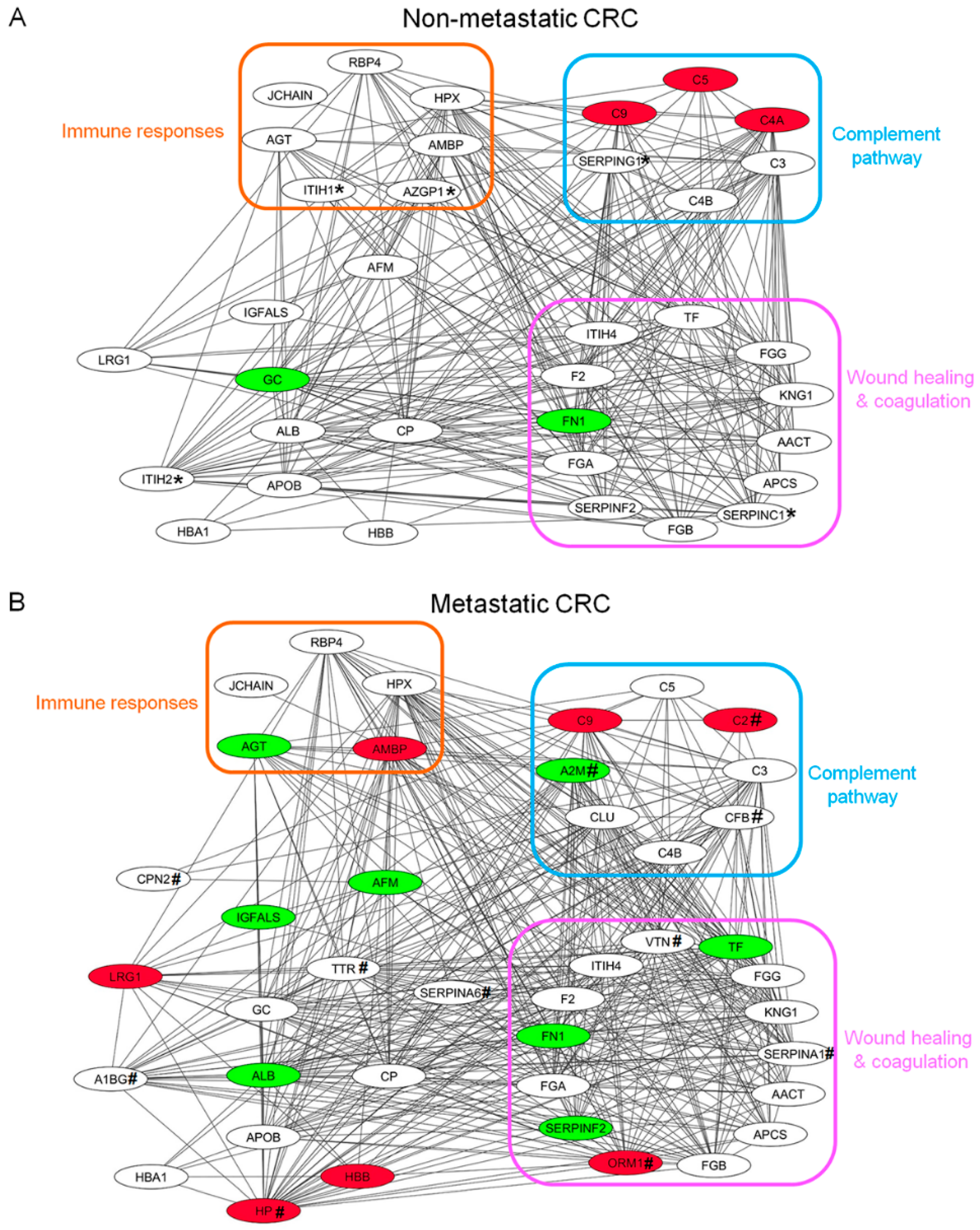
3.4. Protein-Protein Interaction (PPI) Network Analysis of Plasma Glycoproteins in CRC Patients
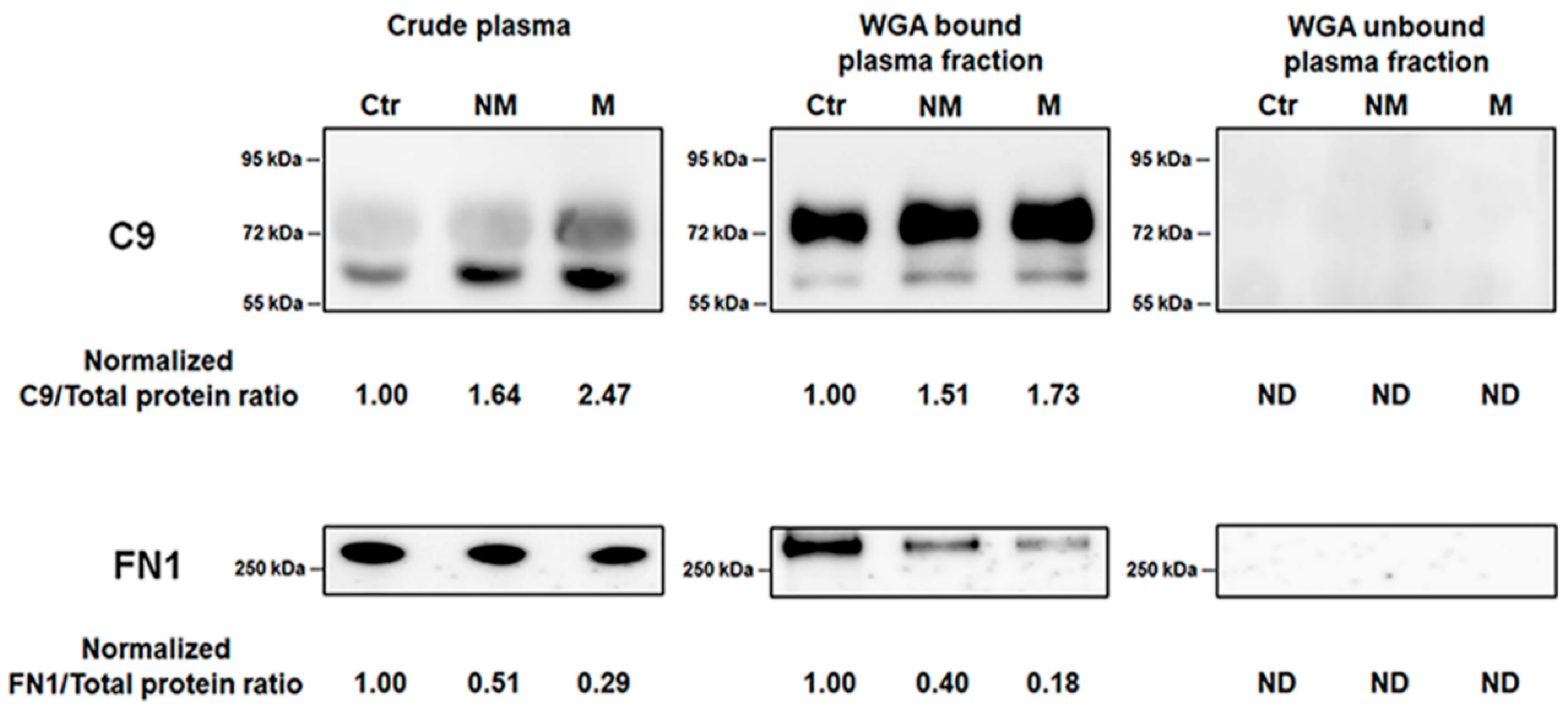
3.5. Verification of Biomaker Cadidates Identified by Quantitative Proteomics
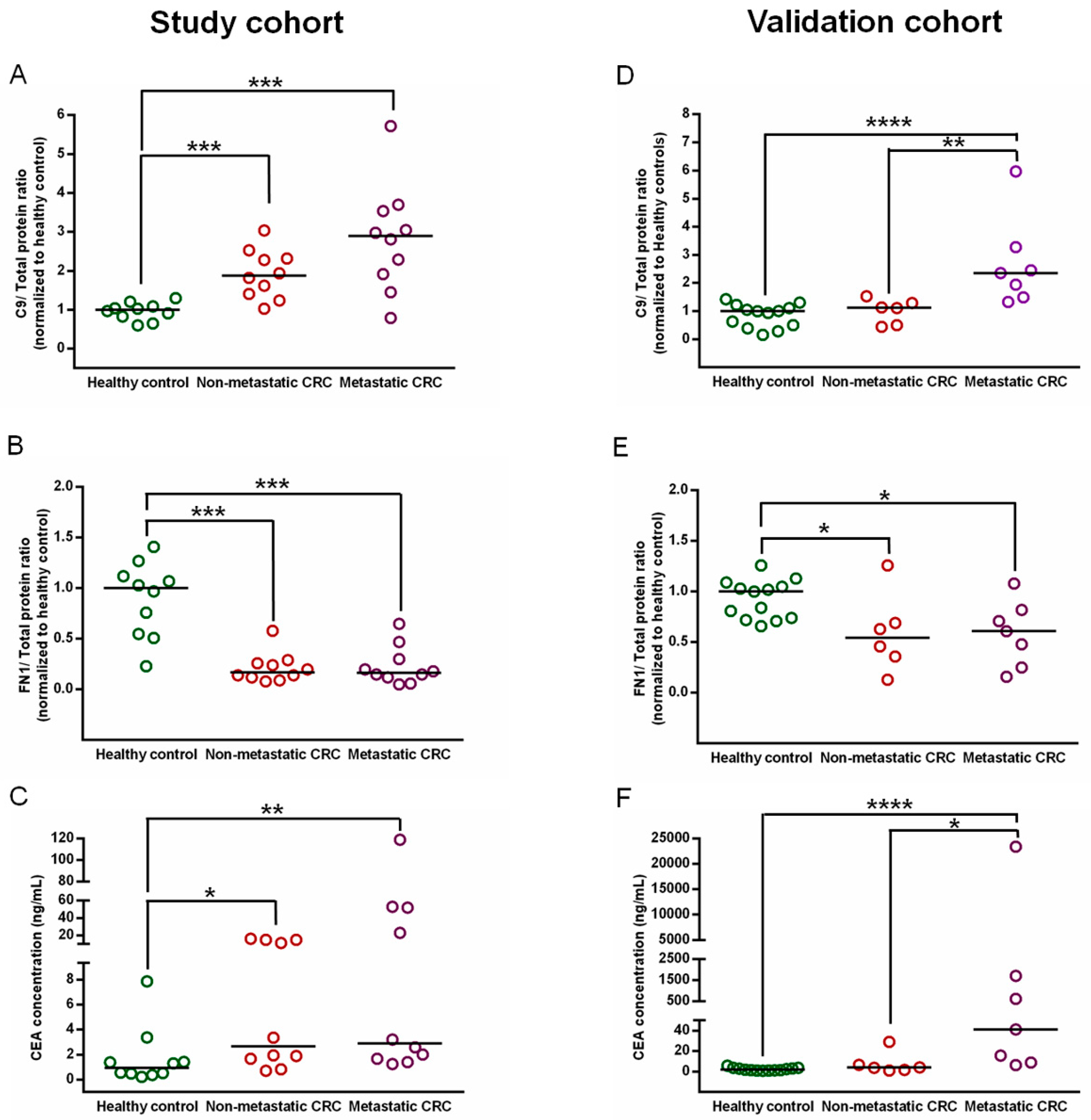
3.6. Confirmation of C9 and FN1 Levels in the Study Cohort
3.7. Validation of C9 and FN1 Expression in an Independent Cohort
3.8. Evaluation of Biomarker Candidate Performance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbo, C.; Cevenini, A.; Salvatore, F. Biomarker discovery by proteomics-based approaches for early detection and personalized medicine in colorectal cancer. Proteom. Clin. Appl. 2017, 11, 1–19. [Google Scholar] [CrossRef]
- Gonzalez-Pons, M.; Cruz-Correa, M. Colorectal cancer biomarkers: Where are we now? Biomed. Res. Int. 2015, 2015, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirwan, A.; Utratna, M.; O’Dwyer, M.E.; Joshi, L.; Kilcoyne, M. Glycosylation-based serum biomarkers for cancer diagnostics and prognostics. Biomed. Res. Int. 2015, 2015, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Simeone, D.M.; Heidt, D.; Anderson, M.A.; Lubman, D.M. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. J. Proteome Res. 2006, 5, 1792–1802. [Google Scholar] [CrossRef]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef]
- Kailemia, M.J.; Park, D.; Lebrilla, C.B. Glycans and glycoproteins as specific biomarkers for cancer. Anal. Bioanal. Chem. 2017, 409, 395–410. [Google Scholar] [CrossRef]
- Ho, W.L.; Hsu, W.M.; Huang, M.C.; Kadomatsu, K.; Nakagawara, A. Protein glycosylation in cancers and its potential therapeutic applications in neuroblastoma. J. Hematol. Oncol. 2016, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ahn, Y.H.; Kim, J.Y.; Yoo, J.S. Quantitative mass spectrometric analysis of glycoproteins combined with enrichment methods. Mass Spectrom. Rev. 2015, 34, 148–165. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.; Mao, L. Cancer biomarker discovery: Lectin-based strategies targeting glycoproteins. Dis. Markers 2012, 33, 1–10. [Google Scholar] [CrossRef]
- Hashim, O.H.; Jayapalan, J.J.; Lee, C.S. Lectins: An effective tool for screening of potential cancer biomarkers. Peer J. 2017, 5, e3784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagerbaumer, P.; Vieth, M.; Anders, M.; Schumacher, U. Lectin histochemistry shows WGA, PHA-l and HPA binding increases during progression of human colorectal cancer. Anticancer Res. 2015, 35, 5333–5339. [Google Scholar] [PubMed]
- Lee, A.; Nakano, M.; Hincapie, M.; Kolarich, D.; Baker, M.S.; Hancock, W.S.; Packer, N.H. The lectin riddle: Glycoproteins fractionated from complex mixtures have similar glycomic profiles. OMICS 2010, 14, 487–499. [Google Scholar] [CrossRef]
- Fanayan, S.; Hincapie, M.; Hancock, W.S. Using lectins to harvest the plasma/serum glycoproteome. Electrophoresis 2012, 33, 1746–1754. [Google Scholar] [CrossRef]
- Qiu, Y.; Patwa, T.H.; Xu, L.; Shedden, K.; Misek, D.E.; Tuck, M.; Jin, G.; Ruffin, M.T.; Turgeon, D.K.; Synal, S.; et al. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J. Proteome Res. 2008, 7, 1693–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.S.; Son, O.L.; Lee, J.Y.; Kim, S.H.; Oh, S.; Lee, Y.S.; Kim, C.H.; Yoo, J.S.; Lee, J.H.; Miyoshi, E.; et al. Lectin precipitation using phytohemagglutinin-l4 coupled to avidin–agarose for serological biomarker discovery in colorectal cancer. Proteomics 2008, 8, 3229–3235. [Google Scholar] [CrossRef] [PubMed]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Liu, Q.; Li, T.; Liao, Q.; Zhao, Y. Role of the complement system in the tumor microenvironment. Cancer Cell Int. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Chong, P.K.; Lee, H.; Loh, M.C.; Choong, L.Y.; Lin, Q.; So, J.B.; Lim, K.H.; Soo, R.A.; Yong, W.P.; Chan, S.P.; et al. Upregulation of plasma C9 protein in gastric cancer patients. Proteomics 2010, 10, 3210–3221. [Google Scholar] [CrossRef]
- Cheng, P.; Gong, J.; Wang, T.; Chen, J.; Liu, G.S.; Zhang, R. Gene expression in rats with barrett’s esophagus and esophageal adenocarcinoma induced by gastroduodenoesophageal reflux. World J. Gastroenterol. 2005, 11, 5117–5122. [Google Scholar] [CrossRef]
- Wang, J.P.; Hielscher, A. Fibronectin: How its aberrant expression in tumors may improve therapeutic targeting. J. Cancer 2017, 8, 674–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, W.; Xiao, E.; Ding, R.; Luo, P.; Yang, Y. High expression of fibronectin is associated with poor prognosis, cell proliferation and malignancy via the NF-ΚB/P53-apoptosis signaling pathway in colorectal cancer. Oncol. Rep. 2016, 36, 3145–3153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruhn, H.D.; Heimburger, N. Factor-VIII-related antigen and cold-insoluble globulin in leukemias and carcinomas. Haemostasis 1976, 5, 189–192. [Google Scholar] [CrossRef] [PubMed]
- Ruelland, A.; Kerbrat, P.; Clerc, C.; Legras, B.; Cloarec, L. Level of plasma fibronectin in patients with breast cancer. Clin. Chim. Acta 1988, 178, 283–287. [Google Scholar] [CrossRef]
- Tas, F.; Bilgin, E.; Karabulut, S.; Tastekin, D.; Duranyildiz, D. Levels of serum fibronectin as a biomarker in gastric cancer patients: Correlation with clinical diagnosis and outcome. Mol. Clin. Oncol. 2016, 4, 655–659. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.C.; Yang, C.H.; Cheng, L.H.; Chang, W.T.; Lin, Y.R.; Cheng, H.C. Fibronectin in cancer: Friend or foe. Cells 2019, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Taylor, G.A.; Jeffers, M.; Webb, C.P.; Koo, H.M.; Anver, M.; Sekiguchi, K.; Vande Woude, G.F. Decreased fibronectin expression in MET/HGF-mediated tumorigenesis. Oncogene 1998, 17, 1179–1183. [Google Scholar] [CrossRef]
- Cai, X.; Liu, C.; Zhang, T.N.; Zhu, Y.W.; Dong, X.; Xue, P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J. Cell Biochem. 2018, 119, 4717–4728. [Google Scholar] [CrossRef]
- Topalovski, M.; Brekken, R.A. Matrix control of pancreatic cancer: New insights into fibronectin signaling. Cancer Lett. 2016, 381, 252–258. [Google Scholar] [CrossRef] [Green Version]
- Kuonen, F.; Surbeck, I.; Sarin, K.Y.; Dontenwill, M.; Rüegg, C.; Gilliet, M.; Oro, A.E.; Gaide, O. TGFβ, fibronectin and integrin α5β1 promote invasion in basal cell carcinoma. J. Investig. Dermatol. 2018, 138, 2432–2442. [Google Scholar] [CrossRef] [Green Version]
- Peters, T.J. All about Albumins: Biochemistry, Genetics and Medical Applications; Academic Press: San Diego, CA, USA, 1996; p. 256. [Google Scholar]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Lammersfeld, C.A.; Vashi, P.G.; Dahlk, S.; Grutsch, J.F.; Lis, C.G. Is serum albumin an independent predictor of survival in ovarian cancer? Clin. Ovarian Cancer 2009, 2, 52–56. [Google Scholar] [CrossRef]
- Ohtani, H.; Saito, M.; Koshiba, K. Alpha-2-Macroglobulin deficiency in patients with advanced prostate cancer. Oncology 1985, 42, 341–344. [Google Scholar] [CrossRef] [PubMed]
- Vasishta, A.; Baker, P.R.; Preece, P.E.; Wood, R.A.; Cuschieri, A. Serum proteinase-like peptidase activities and proteinase inhibitors in women with breast disease. Eur. J. Cancer Clin. Oncol. 1984, 20, 197–202. [Google Scholar] [CrossRef]
- Gressner, A.M.; Peltzer, B. Amidolytic and immuno-nephelometric determination of alpha 1-proteinase inhibitor and alpha 2-macroglobulin in serum with calculation of specific inhibitor activities in health and disease. J. Clin. Chem. Clin. Biochem. 1984, 22, 633–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Hu, S.; Yu, L.; Guo, C.; Sun, L.; Yang, Z.; Qi, J.; Ran, Y. Serum haptoglobin as a novel molecular biomarker predicting colorectal cancer hepatic metastasis. Int. J. Cancer 2016, 138, 2724–2731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Lee, S.H.; Choi, S.; Kim, U.; Yeo, I.S.; Kim, S.H.; Oh, M.J.; Moon, H.; Lee, J.; Jeong, S.; et al. Direct analysis of aberrant glycosylation on haptoglobin in patients with gastric cancer. Oncotarget 2017, 8, 11094–11104. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, R.B.; Erasmo, M.-C.; Santiago, J. The Relevance of Alpha-1-Acid glycoprotein in human cancer: A minireview. Adv. Cancer Res. Clin. Imaging 2019, 2, 1–2. [Google Scholar] [CrossRef]


| Description | Study Cohort (n = 30) | Validation Cohort (n = 26) | ||||
|---|---|---|---|---|---|---|
| Healthy Control | Non-Metastatic CRC | Metastatic CRC | Healthy Control | Non-Metastatic CRC | Metastatic CRC | |
| N | 10 | 10 | 10 | 13 | 6 | 7 |
| Gender | ||||||
| Female | 4 | 4 | 4 | 8 | 5 | 6 |
| Male | 6 | 6 | 6 | 5 | 1 | 2 |
| Median age (range) | 51 (40–66) | 58 (41–78) | 59 (41–72) | 56 (50–63) | 59 (33–66) | 60 (56–64) |
| Histopathology | ||||||
| Well-differentiated adenocarcinoma | 4 | 2 | 1 | 3 | ||
| Moderately differentiated adenocarcinoma | 6 | 4 | 5 | 4 | ||
| Poorly differentiated adenocarcinoma | 0 | 4 | 0 | 0 | ||
| No. | UniProt | Gene | Description | Peptides Counts * | Non-Metastatic CRC vs. Healthy Control | Metastatic CRC vs. Healthy Control | ||
|---|---|---|---|---|---|---|---|---|
| p-Value | Fold Change (NM/Ctr) | p-Value | Fold Change (M/Ctr) | |||||
| 1 | sp|P00738 | HP | Haptoglobin ** | 60 | >0.05 | 1.07 | 0.0028 | 2.02 |
| 2 | sp|P02748 | C9 | Complement component C9 | 4 | 0.0278 | 1.69 | 0.0138 | 1.78 |
| 3 | sp|P02750 | LRG1 | Leucine-rich alpha-2-glycoprotein | 14 | 0.0098 | 1.28 | <0.0001 | 1.74 |
| 4 | sp|P02763 | ORM1 | Alpha-1-acid glycoprotein 1 ** | 14 | >0.05 | 1.26 | 0.0050 | 1.71 |
| 5 | sp|P68871 | HBB | Hemoglobin subunit beta | 15 | 0.0257 | 1.25 | 0.0063 | 1.61 |
| 6 | sp|P06681 | C2 | Complement C2 | 2 | >0.05 | −1.03 | 0.0002 | 1.60 |
| 7 | sp|P02760 | AMBP | Protein AMBP | 10 | 0.0007 | 1.38 | 0.0037 | 1.60 |
| 8 | sp|P02774 | GC | Vitamin D-binding protein | 2 | 0.0198 | −1.69 | 0.0097 | 1.47 |
| 9 | sp|P01031 | C5 | Complement C5 | 6 | 0.0165 | 1.51 | 0.0208 | 1.12 |
| 10 | sp|P0C0L4 | C4A | Complement C4-A ** | 5 | 0.0350 | 2.05 | >0.05 | −1.06 |
| 11 | sp|P01861 | IGHG4 | Immunoglobulin heavy constant gamma 4 | 5 | 0.0169 | −1.57 | >0.05 | −1.14 |
| 12 | sp|P01860 | IGHG3 | Immunoglobulin heavy constant gamma 3 | 3 | 0.0014 | −1.61 | 0.0370 | −1.21 |
| 13 | sp|P01023 | A2M | Alpha-2-macroglobulin **,*** | 168 | >0.05 | 1.00 | <0.0001 | −1.51 |
| 14 | sp|P01019 | AGT | Angiotensinogen | 7 | 0.0288 | −1.29 | 0.0008 | −1.71 |
| 15 | sp|P02787 | TF | Serotransferrin | 2 | 0.0061 | −1.28 | 0.0007 | −1.87 |
| 16 | sp|P43652 | AFM | Afamin | 19 | 0.0029 | −1.24 | 0.0001 | −1.98 |
| 17 | sp|P35858 | IGFALS | Insulin-like growth factor-binding protein complex acid labile subunit | 6 | 0.0049 | −1.23 | 0.0005 | −2.05 |
| 18 | sp|P02751 | FN1 | Fibronectin | 5 | 0.0077 | −1.54 | 0.0002 | −2.05 |
| 19 | sp|P08697 | SERPINF2 | Alpha-2-antiplasmin | 4 | 0.0151 | −1.21 | 0.0004 | −2.52 |
| 20 | sp|P02768 | ALB | Serum albumin | 63 | 0.0333 | −1.49 | <0.0001 | −2.66 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chantaraamporn, J.; Champattanachai, V.; Khongmanee, A.; Verathamjamras, C.; Prasongsook, N.; Mingkwan, K.; Luevisadpibul, V.; Chutipongtanate, S.; Svasti, J. Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer. Proteomes 2020, 8, 26. https://doi.org/10.3390/proteomes8030026
Chantaraamporn J, Champattanachai V, Khongmanee A, Verathamjamras C, Prasongsook N, Mingkwan K, Luevisadpibul V, Chutipongtanate S, Svasti J. Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer. Proteomes. 2020; 8(3):26. https://doi.org/10.3390/proteomes8030026
Chicago/Turabian StyleChantaraamporn, Juthamard, Voraratt Champattanachai, Amnart Khongmanee, Chris Verathamjamras, Naiyarat Prasongsook, Kanokwan Mingkwan, Virat Luevisadpibul, Somchai Chutipongtanate, and Jisnuson Svasti. 2020. "Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer" Proteomes 8, no. 3: 26. https://doi.org/10.3390/proteomes8030026
APA StyleChantaraamporn, J., Champattanachai, V., Khongmanee, A., Verathamjamras, C., Prasongsook, N., Mingkwan, K., Luevisadpibul, V., Chutipongtanate, S., & Svasti, J. (2020). Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer. Proteomes, 8(3), 26. https://doi.org/10.3390/proteomes8030026





