SIP-UNet: Sequential Inputs Parallel UNet Architecture for Segmentation of Brain Tissues from Magnetic Resonance Images
Abstract
:1. Introduction
- We propose to use multiple slices as input that include neighboring slices, to extract correlated information from them.
- We introduce a novel parallel UNet to preserve individual spatial information of each input slice.
- We propose integration of the outputs of parallel Unets using a residual network with late fusion to improve the performance.
- We experiment with resizing images from OASIS data. Apart from resizing the 2D images, the proposed method does not use any augmentation, patch-wise method, pre- or post-processing of skull-stripped images.
- We also experiment with the latest state-of-the-art methods, typical UNet, and modified Unet that takes three slices. The proposed method outperforms rest of these methods.
2. Materials and Methods
2.1. Data
2.2. Method
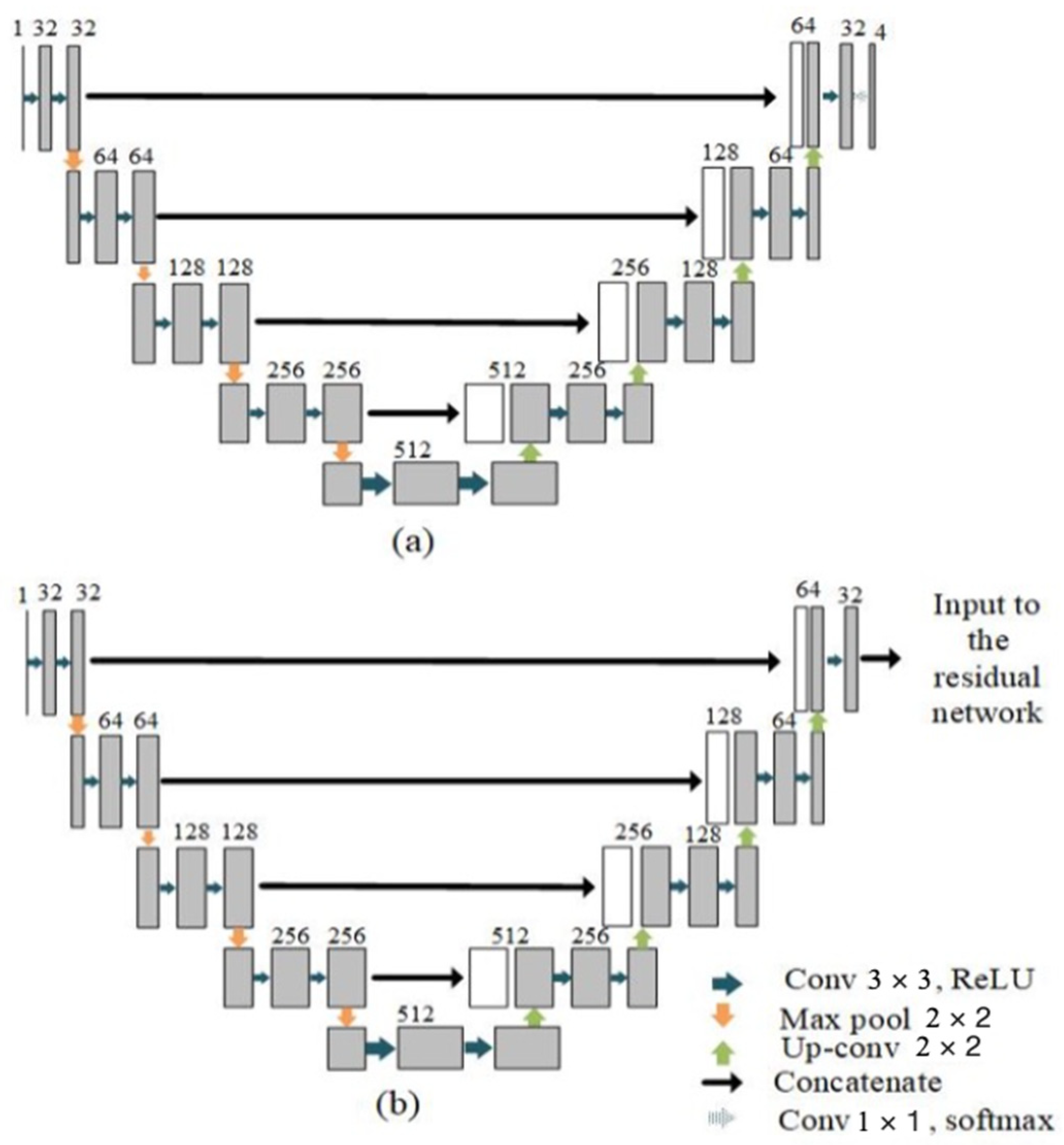
2.2.1. Parallel UNet
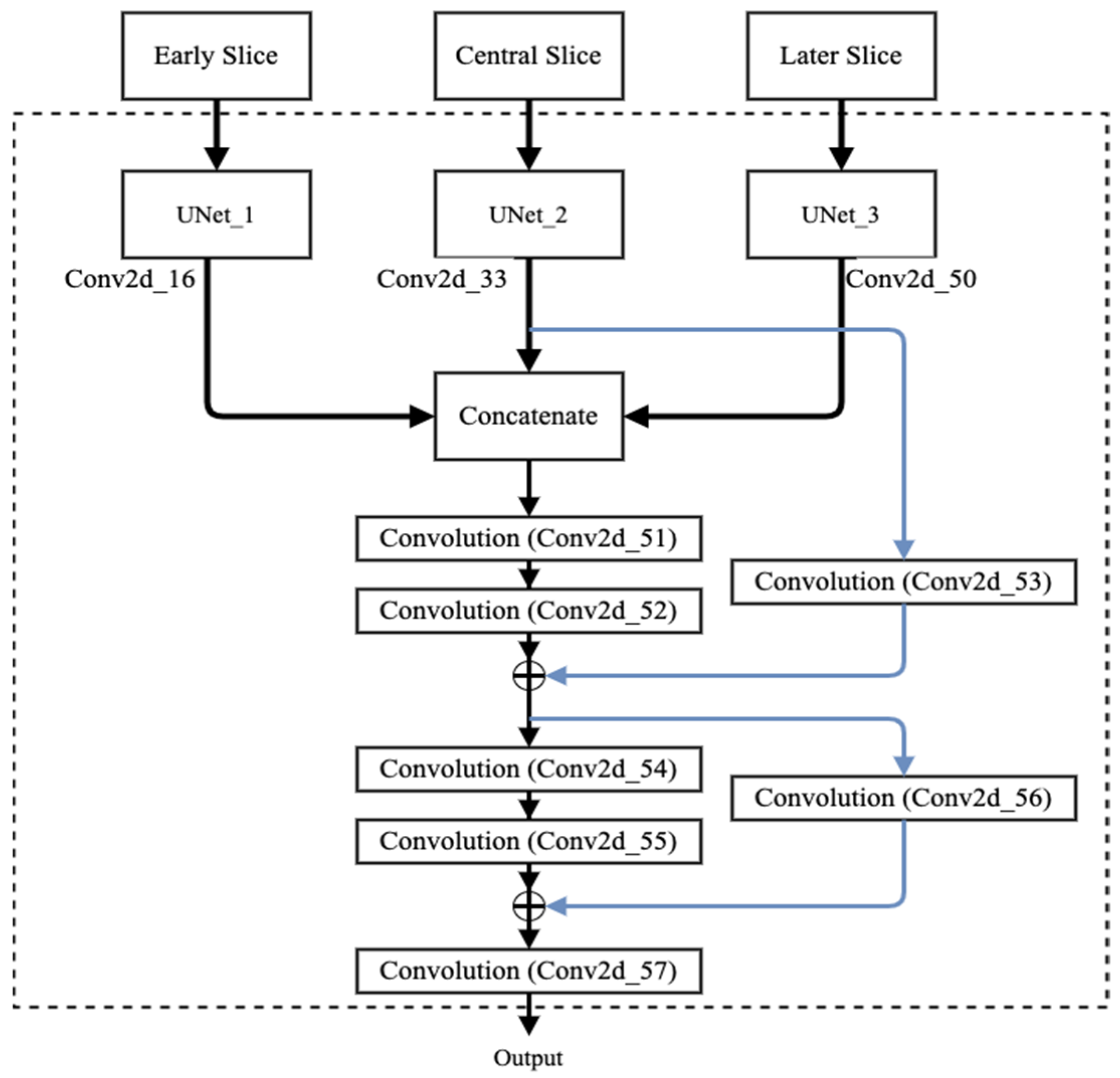
2.2.2. Proposed Fusion Using Residual Network
| Layer Name | Output Shape | Connected to |
|---|---|---|
| Input_1 | 256 × 256 × 1 | |
| Conv2d | 256 × 256 × 32 | Input_1 |
| Conv2d_1 | 256 × 256 × 32 | Conv2d |
| Max_pooling2d | 128 × 128 × 32 | Conv2d_1 |
| Conv2d_2 | 128 × 128 × 64 | Max_pooling2d |
| Conv2d_3 | 128 × 128 × 64 | Conv2d_2 |
| Max_pooling2d_1 | 64 × 64 × 64 | Conv2d_3 |
| Conv2d_4 | 64 × 64 × 128 | Max_pooling2d_1 |
| Conv2d_5 | 64 × 64 × 128 | Conv2d_4 |
| Max_pooling2d_2 | 32 × 32 × 128 | Conv2d_5 |
| Conv2d_6 | 32 × 32 × 256 | Max_pooling2d_2 |
| Conv2d_7 | 32 × 32 × 256 | Conv2d_6 |
| Max_pooling2d_3 | 16 × 16 × 256 | Conv2d_7 |
| Conv2d_8 | 16 × 16 × 512 | Max_pooling2d_3 |
| Conv2d_9 | 16 × 16 × 512 | Conv2d_8 |
| Conv2d_transpose | 32 × 32 × 256 | Conv2d_9 |
| Concatenate | 32 × 32 × 512 | Conv2d_transpose, Conv2d_7 |
| Conv2d_10 | 32 × 32 × 256 | Concatenate |
| Conv2d_11 | 32 × 32 × 256 | Conv2d_10 |
| Conv2d_transpose_1 | 64 × 64 × 128 | Conv2d_11 |
| Concatenate_1 | 64 × 64 × 256 | Conv2d_transpose_1, Conv2d_5 |
| Conv2d_12 | 64 × 64 × 128 | Concatenate_1 |
| Conv2d_13 | 64 × 64 × 128 | Conv2d_12 |
| Conv2d_transpose_2 | 128 × 128 × 64 | Conv2d_13 |
| Concatenate_2 | 128 × 128 × 128 | Conv2d_transpose_2, Conv2d_3 |
| Conv2d_14 | 128 × 128 × 64 | Concatenate_2 |
| Conv2d_15 | 128 × 128 × 64 | Conv2d_14 |
| Conv2d_transpose_3 | 256 × 256 × 32 | Conv2d_15 |
| Concatenate_3 | 256 × 256 × 64 | Conv2d_transpose_3, Conv2d_1 |
| Conv2d_16 | 256 × 256 × 32 | Concatenate_3 |
| Conv2d_17 | 256 × 256 × 32 | Conv2d_16 |
| Conv2d_18 | 256 × 256 × 4 | Conv2d_17 |
| Layer Name | Output Shape | Connected to |
|---|---|---|
| Concatenate_12 | 256 × 256 × 96 | Conv2d_16, Conv2d_33, Conv2d_50 |
| Conv2d_51 | 256 × 256 × 64 | Concatenate_12 |
| Conv2d_52 | 256 × 256 × 64 | Conv2d_51 |
| Conv2d_53 | 256 × 256 × 64 | Conv2d_33 |
| Add | 256 × 256 × 64 | Conv2d_52, Conv2d_53 |
| Conv2d_54 | 256 × 256 × 32 | Add |
| Conv2d_55 | 256 × 256 × 32 | Conv2d_54 |
| Conv2d_56 | 256 × 256 × 32 | Add |
| Add_1 | 256 × 256 × 32 | Conv2d_55, Conv2d_56 |
| Conv2d_57 | 256 × 256 × 4 | Add_1 |
2.3. Loss Function
2.4. Training and Testing Schemes
2.5. Evaluation Metrices
3. Results
3.1. Analysis and Comparison with Single-Slice and Multiple-Slice Input UNet
3.2. Comparisons with Other Methods
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Wiest, R.; Nolte, L.; Reyes, M. A survey of MRI-based medical image analysis for brain tumor studies. Phys. Med. Biol. 2013, 58, R97–R129. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C.J.; Hing, E.; Ashman, J. Trends in Electronic Health Record System Use Among Office-based Physicians: United States, 2007–2012. Natl. Health Stat. Rep. 2014, 1, 1–18. [Google Scholar]
- Despotović, I.; Goossens, B.; Philips, W. MRI segmentation of the human brain: Challenges, methods, and applications. Comput. Math. Methods Med. 2015, 2015, 450341. [Google Scholar] [CrossRef] [Green Version]
- Ulku, I.; Akagunduz, E. A Survey on Deep Learning-based Architectures for Semantic Segmentation on 2D images. arXiv 2019, arXiv:1912.10230. [Google Scholar] [CrossRef]
- Lafferty, J.; McCalium, A.; Pereira, F.C. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. In Proceedings of the Eighteenth International Conference on Machine Learning, Williamstown, MA, USA, 28 June–1 July 2001; pp. 282–289. [Google Scholar]
- Ganin, Y.; Lempitsky, V. N4-Fields: Neural Network Nearest Neighbor Fields for Image Transforms. arXiv 2014, arXiv:1406.6558. [Google Scholar]
- Ning, F.; Delhomme, D.; LeCun, Y.; Piano, F.; Bottou, L.; Barbano, P.E. Toward automatic phenotyping of developing embryos from videos. IEEE Trans. Image Process. 2005, 14, 1360–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibtehaz, N.; Sohel Rahman, M. MultiResUNet: Rethinking the U-Net Architecture for Multimodal Biomedical Image Segmentation. arXiv 2019, arXiv:1902.04049. [Google Scholar] [CrossRef]
- Khagi, B.; Kwon, G.R. Pixel-Label-Based Segmentation of Cross-Sectional Brain MRI Using Simplified SegNet Architecture-Based CNN. J. Healthc. Eng. 2018, 2018, 3640705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Cheng, J.; Fu, H.; Zhou, K.; Hao, H.; Zhao, Y.; Zhang, T.; Gao, S.; Liu, J. CE-Net: Context Encoder Network for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 2019, 38, 2281–2292. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. arXiv 2014, arXiv:1411.4038. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Yamanakkanavar, N.; Choi, J.; Lee, B. MRI Segmentation and Classification of Human Brain Using Deep Learning for Diagnosis of Alzheimer’s Disease: A Survey. Sensors 2020, 20, 3243. [Google Scholar] [CrossRef] [PubMed]
- Punn, S.N.; Agarwal, S. Modality specific U-Net variants for biomedical image segmentation: A survey. arXiv 2021, arXiv:2107.04537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Mu, H.; Gao, M.; Ni, H.; Chen, J.; Yang, H.; Qi, D. A Novel Multi-Scale Attention PFE-UNet for Forest Image Segmentation. Forests 2021, 12, 937. [Google Scholar] [CrossRef]
- Rehman, M.U.; Cho, S.; Kim, J.H.; Chong, K.T. BU-Net: Brain Tumor Segmentation Using Modified U-Net Architecture. Electronics 2020, 9, 2203. [Google Scholar] [CrossRef]
- Comelli, A.; Dahiya, N.; Stefano, A.; Vernuccio, F.; Portoghese, M.; Cutaia, G.; Bruno, A.; Salvaggio, G.; Yezzi, A. Deep Learning-Based Methods for Prostate Segmentation in Magnetic Resonance Imaging. Appl. Sci. 2021, 11, 782. [Google Scholar] [CrossRef] [PubMed]
- Gadosey, P.K.; Li, Y.; Agyekum, E.A.; Zhang, T.; Liu, Z.; Yamak, P.T.; Essaf, F. SD-UNet: Stripping down U-Net for Segmentation of Biomedical Images on Platforms with Low Computational Budgets. Diagnostics 2020, 10, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Blanc-Durand, P.; Gucht, A.V.D.; Schaefer, N.; Itti, E.; Prior, J.O. Automatic lesion detection and segmentation of 18F-FET PET in gliomas: A full 3D U-Net convolutional neural network study. PLoS ONE 2018, 13, e0195798. [Google Scholar] [CrossRef] [PubMed]
- Tong, G.; Li, Y.; Chen, H.; Zhang, Q.; Jiang, H. Improved U-NET network for pulmonary nodules segmentation. Optik 2018, 174, 460–469. [Google Scholar] [CrossRef]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic Brain Tumor Detection and Segmentation Using U-Net Based Fully Convolutional Networks. arXiv 2017, arXiv:1705.03820. [Google Scholar]
- Que, Q.; Tang, Z.; Wang, R.; Zeng, Z.; Wang, J.; Chua, M.; Sin Gee, T.; Yang, X.; Veeravalli, B. CardioXNet: Automated Detection for Cardiomegaly Based on Deep Learning. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 612–615. [Google Scholar]
- Zhang, Y.; Wu, J.; Liu, Y.; Chen, Y.; Wu, E.X.; Tang, X. MI-UNet: Multi-Inputs UNet Incorporating Brain Parcellation for Stroke Lesion Segmentation From T1-Weighted Magnetic Resonance Images. IEEE J. Biomed. Health Inform. 2021, 25, 526–535. [Google Scholar] [CrossRef]
- Kong, Y.; Li, H.; Ren, Y.; Genchev, G.Z.; Wang, X.; Zhao, H.; Xie, Z.; Lu, H. Automated yeast cells segmentation and counting using a parallel U-Net based two-stage framework. OSA Continuum 2020, 3, 982–992. [Google Scholar] [CrossRef]
- Dolz, J.; Ben Ayed, I.; Desrosiers, C. Dense Multi-path U-Net for Ischemic Stroke Lesion Segmentation in Multiple Image Modalities. arXiv 2018, arXiv:1810.07003. [Google Scholar]
- Tran, S.T.; Cheng, C.H.; Nguyen, T.T.; Le, M.H.; Liu, D.G. TMD-Unet: Triple-Unet with Multi-Scale Input Features and Dense Skip Connection for Medical Image Segmentation. Healthcare 2021, 9, 54. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Diakogiannis, F.I.; Waldner, F.; Caccetta, P.; Wu, C. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote Sens. 2020, 162, 94–114. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Jiang, J. U-Net Based Multi-instance Video Object Segmentation. arXiv 2019, arXiv:1905.07826. [Google Scholar]
- Perazzi, F.; Khoreva, A.; Benenson, R.; Schiele, B.; Sorkine-Hornung, A. Learning Video Object Segmentation from Static Images. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3491–3500. [Google Scholar]
- Vu, M.; Grimbergen, G.; Nyholm, T.; Löfstedt, T. Evaluation of multislice inputs to convolutional neural networks for medical image segmentation. Med. Phys. 2020, 47, 6216–6231. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Wang, L.; Gao, Y.; Shen, D. Fully convolutional networks for multi-modality isointense infant brain image segmentation. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1342–1345. [Google Scholar]
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef] [Green Version]
- Nair, V.; Hinton, G. Rectified Linear Units Improve Restricted Boltzmann Machines. ICML 2010, 27, 807–814. [Google Scholar]
- Rohlfing, T. Image Similarity and Tissue Overlaps as Surrogates for Image Registration Accuracy: Widely Used but Unreliable. IEEE Trans. Med. Imaging 2021, 31, 153–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powers, D.M. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar]
- Yeghiazaryan, V.; Voiculescu, I.D. Family of boundary overlap metrics for the evaluation of medical image segmentation. J. Med. Imaging 2018, 5, 015006. [Google Scholar] [CrossRef] [PubMed]
- Yamanakkanavar, N.; Lee, B. A novel M-SegNet with global attention CNN architecture for automatic segmentation of brain MRI. Comput. Biol. Med. 2021, 136, 104761. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Yamanakkanavar, N.; Choi, J. Automatic segmentation of brain MRI using a novel patch-wise U-net deep architecture. PLoS ONE 2020, 15, e0236493. [Google Scholar] [CrossRef] [PubMed]
- Yamanakkanavar, N.; Lee, B. Brain Tissue Segmentation using Patch-wise M-net Convolutional Neural Network. In Proceedings of the 2020 IEEE International Conference on Consumer Electronics—Asia (ICCE-Asia), Seoul, Korea, 1–3 November 2020; pp. 1–4. [Google Scholar]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Ji, S.; Shen, D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef] [Green Version]

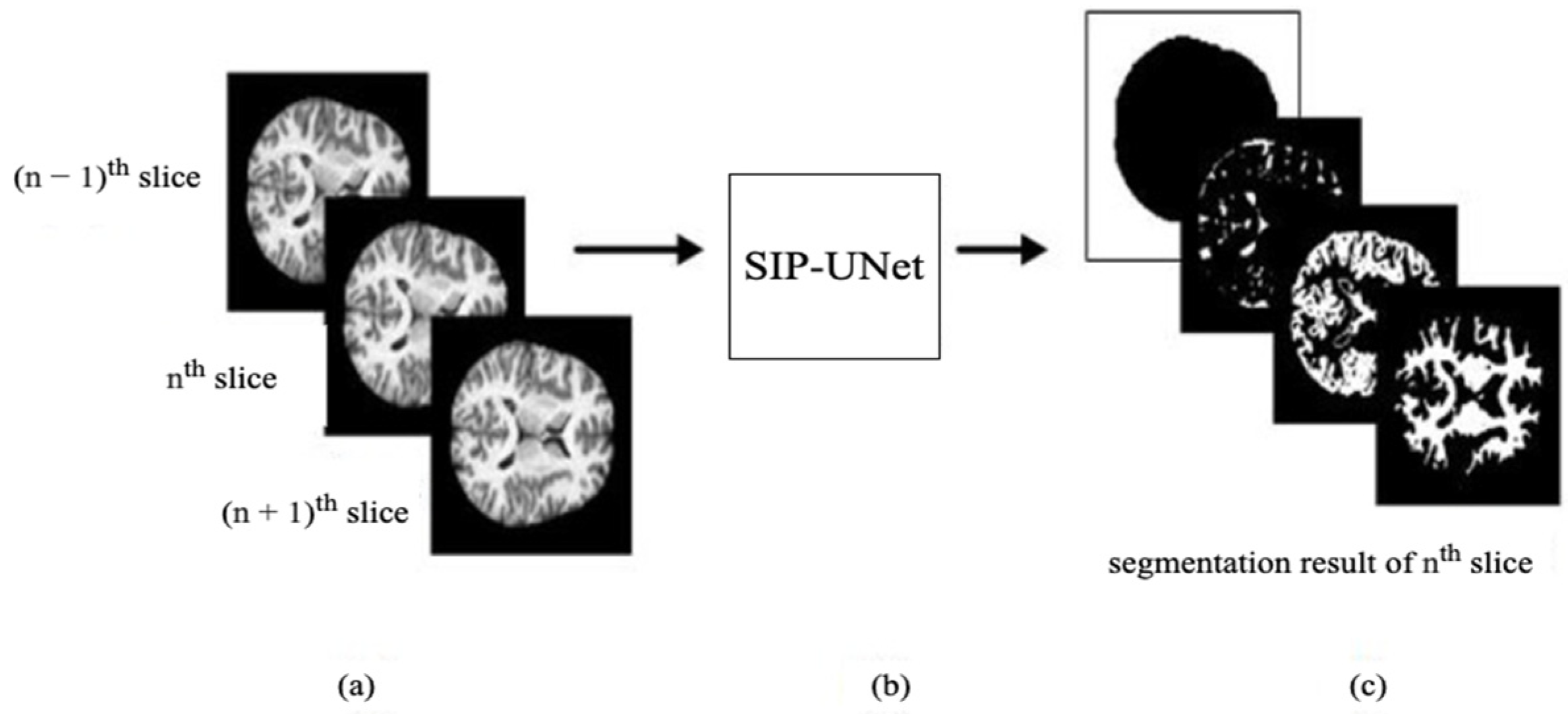
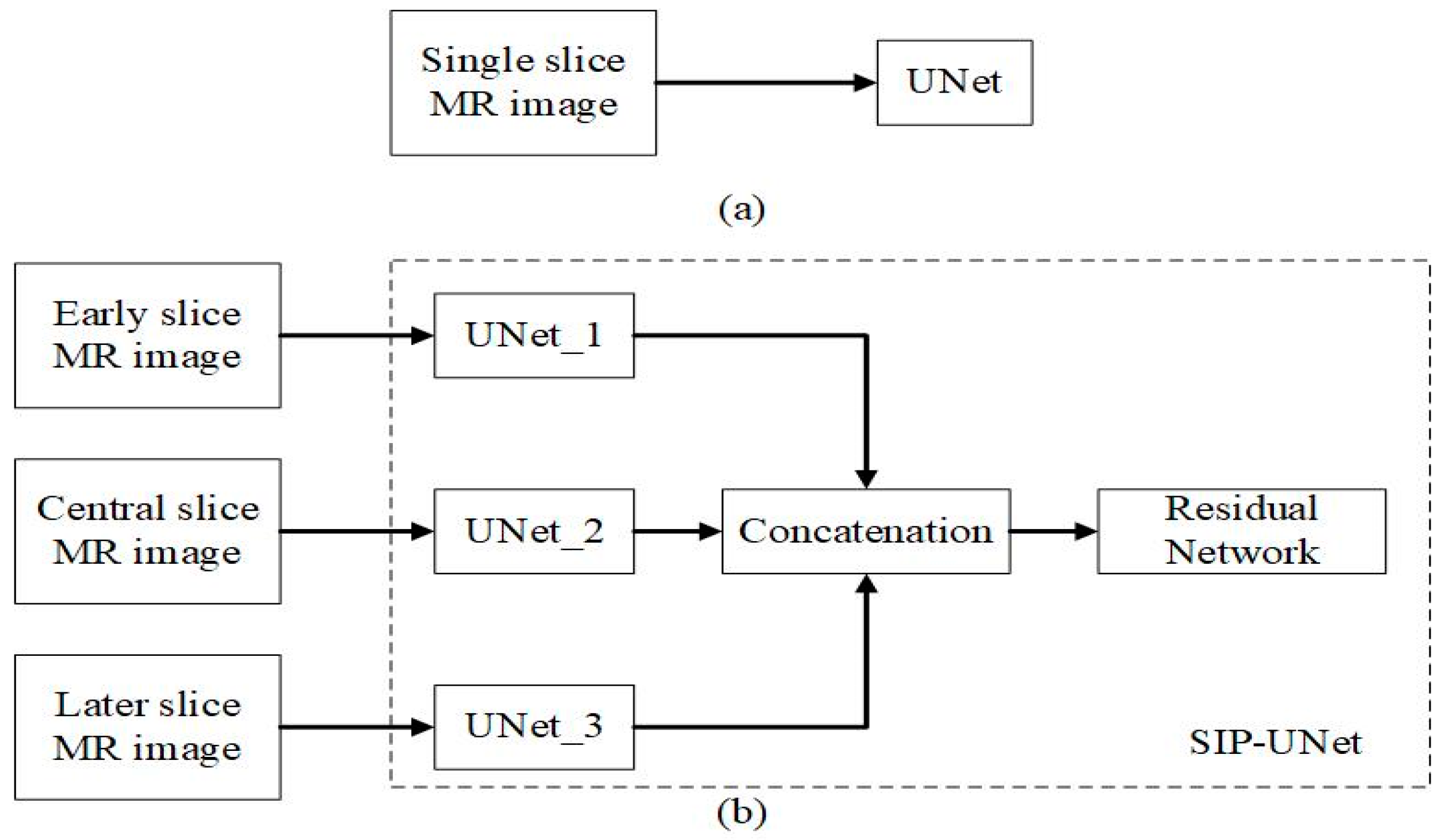

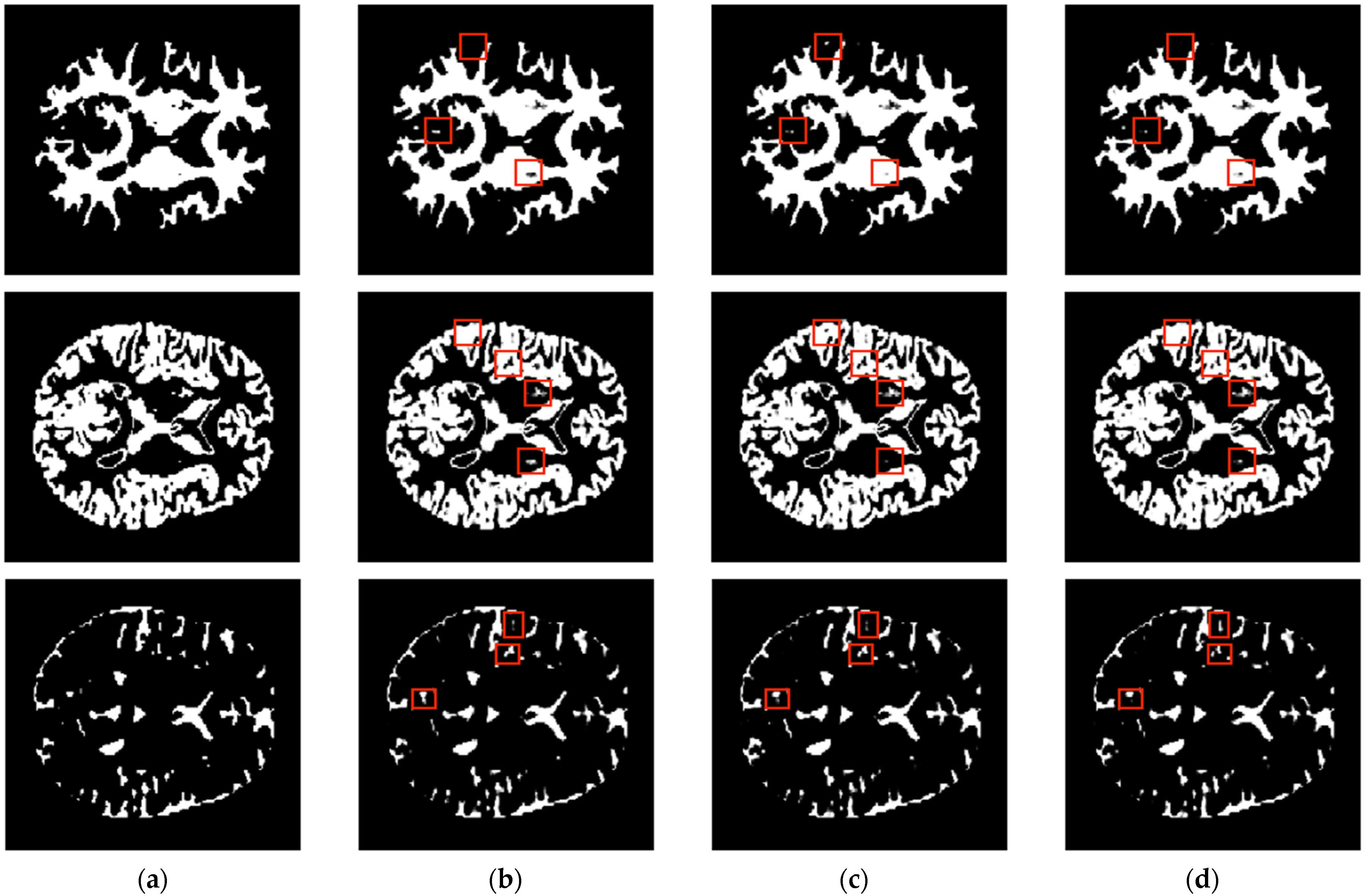

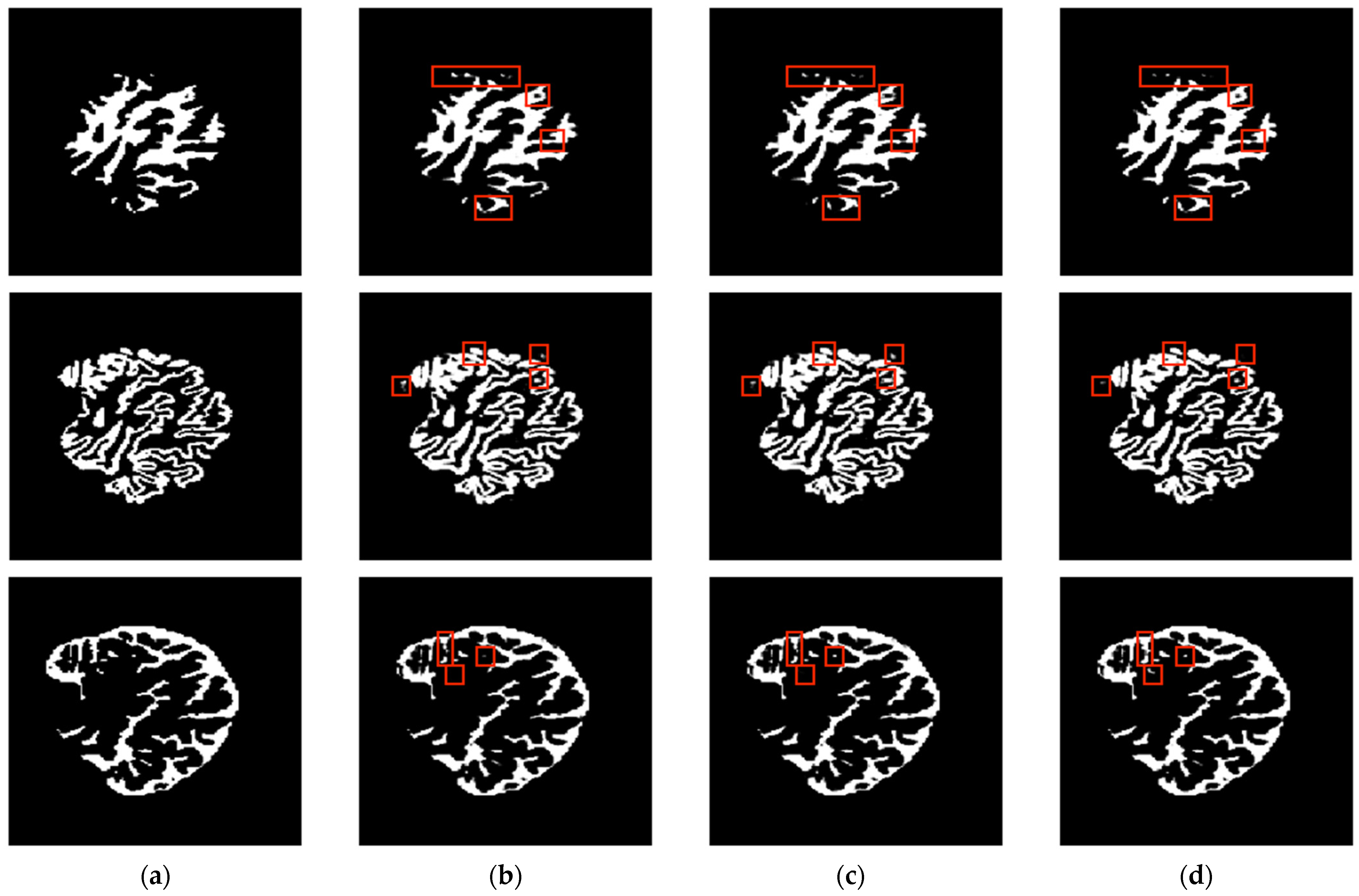
| Axial Plane | ||||||||
|---|---|---|---|---|---|---|---|---|
| Methods | Input Slices | Epochs | WM | GM | CSF | |||
| DSC | JI | DSC | JI | DSC | JI | |||
| Multiresnet [21] | 1 | 38 | 0.679 ± 0.180 | 0.538 ± 0.172 | 0.750 ± 0.073 | 0.605 ± 0.084 | 0.725 ± 0.079 | 0.574 ± 0.090 |
| SegNet [20] | 1 | 72 | 0.857 ± 0.087 | 0.758 ± 0.110 | 0.873 ± 0.050 | 0.778 ± 0.076 | 0.848 ± 0.041 | 0.738 ± 0.060 |
| Unet | 1 | 82 | 0.948 ± 0.075 | 0.908 ± 0.090 | 0.954 ± 0.027 | 0.914 ± 0.068 | 0.942 ± 0.032 | 0.893 ± 0.052 |
| Unet (modified) | 3 | 69 | 0.948 ± 0.075 | 0.908 ± 0.091 | 0.956 ± 0.027 | 0.917 ± 0.042 | 0.947 ± 0.030 | 0.900 ± 0.050 |
| Proposed method | 3 | 67 | 0.951 ± 0.074 | 0.912 ± 0.089 | 0.954 ± 0.026 | 0.923 ± 0.041 | 0.951 ± 0.074 | 0.912 ± 0.089 |
| Coronal plane | ||||||||
| Multiresnet [21] | 1 | 50 | 0.737 ± 0.090 | 0.590 ± 0.101 | 0.762 ± 0.050 | 0.617 ± 0.063 | 0.736 ± 0.056 | 0.585 ± 0.068 |
| SegNet [20] | 1 | 70 | 0.889 ± 0.048 | 0.803 ± 0.073 | 0.886 ± 0.032 | 0.796 ± 0.049 | 0.861 ± 0.039 | 0.758 ± 0.058 |
| Unet | 1 | 64 | 0.959 ± 0.027 | 0.924 ± 0.044 | 0.954 ± 0.022 | 0.912 ± 0.035 | 0.941 ± 0.031 | 0.881 ± 0.049 |
| Unet (modified) | 3 | 101 | 0.962 ± 0.028 | 0.928 ± 0.046 | 0.958 ± 0.022 | 0.919 ± 0.036 | 0.948 ± 0.030 | 0.902 ± 0.048 |
| Proposed method | 3 | 82 | 0.962 ± 0.027 | 0.928 ± 0.044 | 0.959 ± 0.022 | 0.921 ± 0.035 | 0.951 ± 0.028 | 0.907 ± 0.046 |
| Sagittal plane | ||||||||
| Multiresnet [21] | 1 | 42 | 0.720 ± 0.127 | 0.576 ± 0.134 | 0.761 ± 0.041 | 0.616 ± 0.050 | 0.738 ± 0.049 | 0.587 ± 0.060 |
| SegNet [20] | 1 | 73 | 0.830 ± 0.086 | 0.748 ± 0.118 | 0.868 ± 0.035 | 0.769 ± 0.053 | 0.845 ± 0.037 | 0.733 ± 0.054 |
| Unet | 1 | 78 | 0.951 ± 0.038 | 0.909 ± 0.060 | 0.954 ± 0.022 | 0.912 ± 0.035 | 0.944 ± 0.027 | 0.894 ± 0.043 |
| Unet (modified) | 3 | 102 | 0.954 ± 0.040 | 0.915 ± 0.062 | 0.957 ± 0.022 | 0.919 ± 0.036 | 0.949 ± 0.028 | 0.903 ± 0.044 |
| Proposed method | 3 | 75 | 0.955 ± 0.038 | 0.916 ± 0.060 | 0.959 ± 0.021 | 0.921 ± 0.034 | 0.953 ± 0.026 | 0.911 ± 0.041 |
| Axial Plane | ||||||||
|---|---|---|---|---|---|---|---|---|
| Methods | Input Slices | Epochs | WM | GM | CSF | |||
| RVD(%) | VOE | RVD(%) | VOE | RVD(%) | VOE | |||
| Multiresnet [21] | 1 | 38 | −14.362 | 0.462 | −4.692 | 0.395 | −5.935 | 0.426 |
| SegNet [20] | 1 | 72 | 2.468 | 0.242 | −2.618 | 0.222 | 3.129 | 0.262 |
| Unet | 1 | 82 | 2.261 | 0.092 | −0.4114 | 0.086 | 0.4507 | 0.107 |
| Unet (modified) | 3 | 69 | 2.214 | 0.092 | 0.3998 | 0.083 | −1.073 | 0.100 |
| Proposed method | 3 | 67 | 0.4833 | 0.088 | −0.2854 | 0.077 | 0.9819 | 0.088 |
| Coronal plane | ||||||||
| Multiresnet [21] | 1 | 50 | −8.48 | 0.41 | −3.429 | 0.383 | −1.648 | 0.415 |
| SegNet [20] | 1 | 70 | −0.0132 | 0.197 | −1.647 | 0.204 | 4.21 | 0.242 |
| Unet | 1 | 64 | −0.0583 | 0.076 | −1.148 | 0.088 | 2.149 | 0.119 |
| Unet (modified) | 3 | 101 | 0.544 | 0.072 | −1.072 | 0.081 | 1.49 | 0.098 |
| Proposed method | 3 | 82 | −0.009 | 0.072 | −0.548 | 0.079 | 0.881 | 0.093 |
| Sagittal plane | ||||||||
| Multiresnet [21] | 1 | 42 | −10.46 | 0.424 | −0.724 | 0.384 | −4.511 | 0.413 |
| SegNet [20] | 1 | 73 | −0.665 | 0.252 | −1.132 | 0.231 | 1.297 | 0.267 |
| Unet | 1 | 78 | 1.978 | 0.091 | −0.669 | 0.088 | −0.337 | 0.106 |
| Unet (modified) | 3 | 102 | −0.779 | 0.085 | −0.4686 | 0.081 | 1.547 | 0.097 |
| Proposed method | 3 | 75 | −1.228 | 0.084 | 0.5545 | 0.079 | −0.3729 | 0.089 |
| Authors | Methods | DSC Score | ||
|---|---|---|---|---|
| WM | GM | CSF | ||
| Zhang et al. [43] | CNN | 86.4% | 85.2% | 83.5% |
| Nie et al. [34] | FCN | 88.7% | 87.3% | 85.5% |
| Khagi et al. [10] | SegNet | 81.9% | 74.6% | 72.2% |
| Lee et al. [41] | Patch-wise UNet | 94.33% | 93.33% | 92.67% |
| Yamanakkanavar et al. [42] | Patch-wise Mnet | 95.17% | 94.32% | 93.60% |
| Proposed method | SIP-UNet | 95.6% | 95.73% | 95.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prajapati, R.; Kwon, G.-R. SIP-UNet: Sequential Inputs Parallel UNet Architecture for Segmentation of Brain Tissues from Magnetic Resonance Images. Mathematics 2022, 10, 2755. https://doi.org/10.3390/math10152755
Prajapati R, Kwon G-R. SIP-UNet: Sequential Inputs Parallel UNet Architecture for Segmentation of Brain Tissues from Magnetic Resonance Images. Mathematics. 2022; 10(15):2755. https://doi.org/10.3390/math10152755
Chicago/Turabian StylePrajapati, Rukesh, and Goo-Rak Kwon. 2022. "SIP-UNet: Sequential Inputs Parallel UNet Architecture for Segmentation of Brain Tissues from Magnetic Resonance Images" Mathematics 10, no. 15: 2755. https://doi.org/10.3390/math10152755
APA StylePrajapati, R., & Kwon, G.-R. (2022). SIP-UNet: Sequential Inputs Parallel UNet Architecture for Segmentation of Brain Tissues from Magnetic Resonance Images. Mathematics, 10(15), 2755. https://doi.org/10.3390/math10152755







