Bayesian Linear Regression and Natural Logarithmic Correction for Digital Image-Based Extraction of Linear and Tridimensional Zoometrics in Dromedary Camels
Abstract
1. Introduction
2. Materials and Methods
2.1. Zoometric Parameter Definition
2.2. Animal Sample
2.3. Sampling
2.3.1. On-Field Zoometrics
2.3.2. Digital Imaging
2.4. Statistical Analysis
2.4.1. Method Comparison/Reproducibility and Repeatability: Interobserver Correlation Coefficient (ICC)
2.4.2. Scale Reliability and Repeatability: Cronbach’s Alpha (Cα)
2.4.3. Parametric Assumptions Testing and Approach Decision
2.4.4. Perimeters and Girths Calculation
2.4.5. Bayesian Linear Regression Modelling and Natural Logarithmic Correction for Perimeters and Girths
2.4.6. Jeffrey–Zellner–Siow (JZS) Mixture of g-Priors
2.4.7. Bayesian Modelling of Factor and Covariate Effects (FCEBM)
2.4.8. Factors and Covariate Effect Bayesian Interpretation (CEBI)
2.4.9. Convergence Criterion
2.4.10. Model Validity and Explanatory Power of Present Data, and Predictive Power of Future Data
3. Results
3.1. Parametric Assumptions Testing and Approach Decision
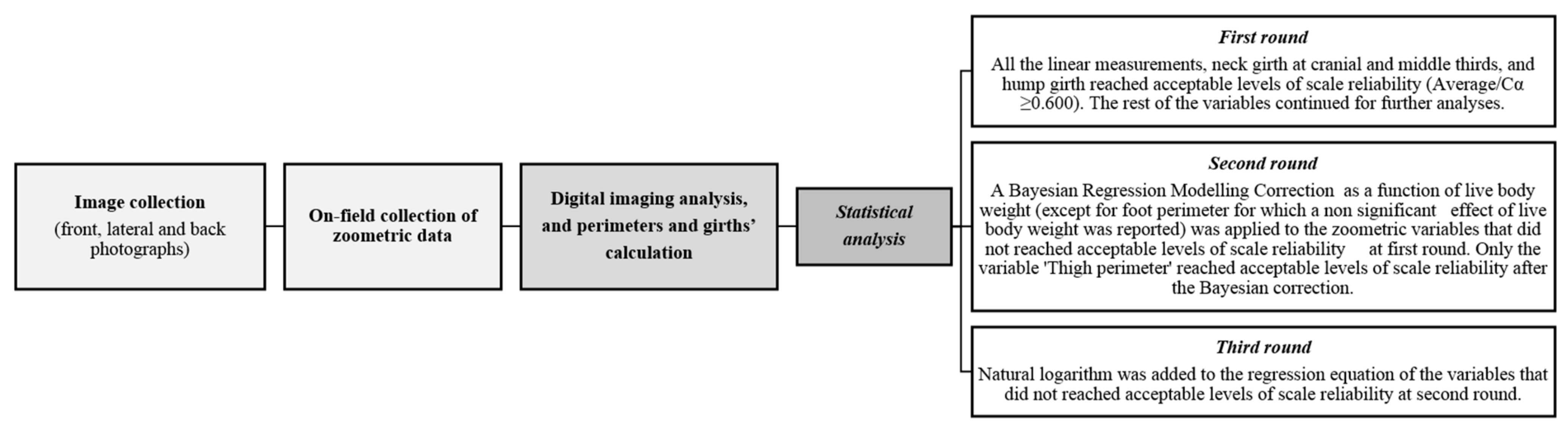
3.2. Initial/First Round of Method Comparison/Repeatability and Scale Reliability
3.3. Bayesian Linear Regression Modelling and Second Round of Method Comparison/Repeatability and Scale Reliability
3.4. Natural Logarithmic Correction for Perimeters and Girths and Third Round of Method Comparison/Repeatability and Scale Reliability
3.5. Final Outputs for Method Comparison/Repeatability and Scale Reliability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alhajeri, B.H.; Alaqeely, R.; Alhaddad, H. Classifying camel breeds using geometric morphometrics: A case study in Kuwait. Livest. Sci. 2019, 230, 103824. [Google Scholar] [CrossRef]
- Dorantes-Coronado, E.; Torres-Hernández, G.; Hernández-Mendo, O.; Rojo-Rubio, R. Zoometric measures and their utilization in prediction of live weight of local goats in southern México. SpringerPlus 2015, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Alhajeri, B.H.; Alhaddad, H.; Alaqeely, R.; Alaskar, H.; Dashti, Z.; Maraqa, T. Camel breed morphometrics: Current methods and possibilities. Trans. R. Soc. South Aust. 2021, 145, 90–111. [Google Scholar] [CrossRef]
- Parés, C.; Sañudo, C. Valoración Morfológica de Animales Domésticos; Ministerio de Medio Ambiente y Medio Rural y Marino: Madrid, Spain, 2009. [Google Scholar]
- Gaudioso, V.; Sanz-Ablanedo, E.; Lomillos, J.; Alonso, M.; Javares-Morillo, L.; Rodríguez, P. “Photozoometer”: A new photogrammetric system for obtaining morphometric measurements of elusive animals. Livest. Sci. 2014, 165, 147–156. [Google Scholar] [CrossRef]
- Rahagiyanto, A.; Adhyatma, M. A Review of Morphometric Measurements Techniques on Animals Using Digital Image Processing. Food Agric. Sci. Polije Proc. Ser. 2021, 3, 67–72. [Google Scholar]
- White, R.; Schofield, C.; Green, D.; Parsons, D.; Whittemore, C. The effectiveness of a visual image analysis (VIA) system for monitoring the performance of growing/finishing pigs. Anim. Sci. 2004, 78, 409–418. [Google Scholar] [CrossRef]
- Negretti, P.; Bianconi, G.; Finzi, A. Visual image analysis to estimate morphological and weight measurements in rabbits. World Rabbit. Sci. 2007, 15, 37–41. [Google Scholar] [CrossRef][Green Version]
- De Wet, L.; Vranken, E.; Chedad, A.; Aerts, J.-M.; Ceunen, J.; Berckmans, D. Computer-assisted image analysis to quantify daily growth rates of broiler chickens. Br. Poult. Sci. 2003, 44, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.; Peacock, A.; Lewis, O.; Boyce, R.; Roberts, D.; Coffey, M.; Kenyon, S.; Schutz, M. Potential for estimation of body condition scores in dairy cattle from digital images. J. Dairy Sci. 2008, 91, 3439–3453. [Google Scholar] [CrossRef] [PubMed]
- Azzaro, G.; Caccamo, M.; Ferguson, J.D.; Battiato, S.; Farinella, G.M.; Guarnera, G.C.; Puglisi, G.; Petriglieri, R.; Licitra, G. Objective estimation of body condition score by modeling cow body shape from digital images. J. Dairy Sci. 2011, 94, 2126–2137. [Google Scholar] [CrossRef] [PubMed]
- Cervantes, I.; Baumung, R.; Molina, A.; Druml, T.; Gutiérrez, J.; Sölkner, J.; Valera, M. Size and shape analysis of morphofunctional traits in the Spanish Arab horse. Livest. Sci. 2009, 125, 43–49. [Google Scholar] [CrossRef]
- Negretti, P.; Bianconi, G.; Bartocci, S.; Terramoccia, S.; Verna, M. Determination of live weight and body condition score in lactating Mediterranean buffalo by Visual Image Analysis. Livest. Sci. 2008, 113, 1–7. [Google Scholar] [CrossRef]
- O’Malley, B.P.; Schmitt, J.D.; Holden, J.P.; Weidel, B.C. Comparison of Specimen-and Image-Based Morphometrics for Cisco. J. Fish Wildl. Manag. 2021, 12, 208–215. [Google Scholar] [CrossRef]
- Shrader, A.M.; Ferreira, S.M.; Van Aarde, R.J. Digital photogrammetry and laser rangefinder techniques to measure African elephants. South Afr. J. Wildl. 2006, 36, 1–7. [Google Scholar]
- De Kock, M.E.; O’Donovan, D.; Khafaga, T.; Hejcmanová, P. Zoometric data extraction from drone imagery: The Arabian oryx (Oryx leucoryx). Environ. Conserv. 2021, 48, 295–300. [Google Scholar] [CrossRef]
- Proffitt, K.M.; Garrott, R.A.; Rotella, J.J.; Lele, S. Using form analysis techniques to improve photogrammetric mass-estimation methods. Mar. Mammal Sci. 2008, 24, 147–158. [Google Scholar] [CrossRef]
- De Bruyn, P.; Bester, M.; Carlini, A.; Oosthuizen, W. How to weigh an elephant seal with one finger: A simple three-dimensional photogrammetric application, Aquat. Biol. 2009, 5, 31–39. [Google Scholar] [CrossRef]
- Waite, J.N.; Schrader, W.J.; Mellish, J.-A.E.; Horning, M. Three-dimensional photogrammetry as a tool for estimating morphometrics and body mass of Steller sea lions (Eumetopias jubatus). Can. J. Fish. Aquat. Sci. 2007, 64, 296–303. [Google Scholar] [CrossRef]
- Bell, C.M.; Hindell, M.A.; Burton, H.R. Estimation of body mass in the southern elephant seal, Mirounga leonina, by photogrammetry and morphometrics. Mar. Mammal Sci. 1997, 13, 669–682. [Google Scholar] [CrossRef]
- Iglesias Pastrana, C.; Navas González, F.J.; Ciani, E.; Barba Capote, C.J.; Delgado Bermejo, J.V. Effect of research impact on emerging camel husbandry, welfare and social-related awareness. Animals 2020, 10, 780. [Google Scholar] [CrossRef]
- Al-Atiyat, R.M.; Suliman, G.; AlSuhaibani, E.; El-Waziry, A.; Al-Owaimer, A.; Basmaeil, S. The differentiation of camel breeds based on meat measurements using discriminant analysis. Trop. Anim. Health Prod. 2016, 48, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Bitaraf Sani, M.; Hosseini, S.A.; Asadzadeh, N.; Ghavipanje, N.; Afshin, M.; Jasouri, M.; Banabazi, M.H.; Esmaeilkhanian, S.; Zare Harofte, J.; Shafei Naderi, A. A New Approach in the Evaluation of Dairy Camels: Using Test Day Milk and Morphometric Records. Dairy 2022, 3, 78-86. [Google Scholar] [CrossRef]
- Abdallah, H.R.; Faye, B. Phenotypic classification of Saudi Arabian camel (Camelus dromedarius) by their body measurements. Emir. J. Food Agric. 2012, 24, 272–280. [Google Scholar]
- Meghelli, I.; Kaouadji, Z.; Yilmaz, O.; Cemal, İ.; Karaca, O.; Gaouar, S. Morphometric characterization and estimating body weight of two Algerian camel breeds using morphometric measurements. Trop. Anim. Health Prod. 2020, 52, 2505–2512. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, M.; Aljumaah, R.S.; Samara, E.M.; Faye, B.; Caja, G. A proposal of linear assessment scheme for the udder of dairy camels (Camelus dromedarius L.). Trop. Anim. Health Prod. 2016, 48, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Abdullahi, I.A.; Musa, H.A.-H.; Jibril, A. Scrotal circumference and testicular morphometric characteristics of the camel (Camelus dromedarius) in the semi-arid environment of northern Nigeria. Int. J. Morphol. 2012, 30, 1369–1372. [Google Scholar] [CrossRef]
- Köhler-Rollefson, I. Camel biodiversity—and how to conserve it. Anim. Front. 2022, 12, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Iglesias Pastrana, C.; Navas González, F.J.; Ciani, E.; González Ariza, A.; Delgado Bermejo, J.V. A tool for functional selection of leisure camels: Behaviour breeding criteria may ensure long-term sustainability of a European unique breed. Res. Vet. Sci. 2021, 140, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Çağlı, A.; Yılmaz, M. Determination of some body measurements of camels with three-dimensional modeling method (3D). Trop. Anim. Health Prod. 2021, 53, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gherissi, D.E.; Lamraoui, R.; Chacha, F.; Gaouar, S.B.S. Accuracy of Image Analysis for Linear Zoometric Measurements in Dromedary Camels. Trop. Anim. Health Prod. 2022, 54, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Padalino, B.; Monaco, D.; Lacalandra, G.M. Male camel behavior and breeding management strategies: How to handle a camel bull during the breeding season? Emir. J. Food Agric. 2015, 27, 338–349. [Google Scholar] [CrossRef]
- Iglesias, C.; Navas, F.; Ciani, E.; Arbulu, A.A.; González, A.; Marín, C.; Mérida, S.N.; Bermejo, J.V.D. Zoometric characterization and body condition score in Canarian camel breed. Arch. Zootec. 2020, 69, 102–107. [Google Scholar] [CrossRef][Green Version]
- Yakubu, A.; Ladokun, A.; Adua, M. Bioprediction of body weight from zoometrical traits of non-descript goats using linear and non-linear models in North Central Nigeria. Livest. Res. Rural Dev. 2011, 23, 6. [Google Scholar]
- Boujenane, I. Comparison of body weight estimation equations for camels (Camelus dromedarius). Trop. Anim. Health Prod. 2019, 51, 1003–1007. [Google Scholar] [CrossRef]
- Babu, K. Natural textile fibres: Animal and silk fibres. In Textiles and Fashion; Elsevier: Amsterdam, The Netherlands, 2015; pp. 57–78. [Google Scholar]
- Iglesias, C.; Navas, F.; Ciani, E.; Arando, A.; González, A.; Marín, C.; Nogales, S.; Delgado, J. Análisis biocinemático de locomoción y termografía aplicada en la raza camellar canaria. Arch. Zootec. 2020, 69, 102–107. [Google Scholar] [CrossRef][Green Version]
- Puig-Diví, A.; Escalona-Marfil, C.; Padullés-Riu, J.M.; Busquets, A.; Padullés-Chando, X.; Marcos-Ruiz, D. Validity and reliability of the Kinovea program in obtaining angles and distances using coordinates in 4 perspectives. PLoS ONE 2019, 14, e0216448. [Google Scholar] [CrossRef]
- Bunting, K.V.; Steeds, R.P.; Slater, L.T.; Rogers, J.K.; Gkoutos, G.V.; Kotecha, D. A Practical Guide to Assess the Reproducibility of Echocardiographic Measurements. J. Am. Soc. Echocardiogr. 2019, 32, 1505–1515. [Google Scholar] [CrossRef]
- Fleiss, J.L.; Cohen, J. The equivalence of weighted kappa and the intraclass correlation coefficient as measures of reliability. Educ. Psychol. Meas. 1973, 33, 613–619. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef]
- Nunnally, J.C.; Bernstein, I. The Role of University in the Development of Entrepreneurial Vocations: A Spanish Study; McGraw-Hill: New York, NY, USA, 1994; pp. 387–405. [Google Scholar]
- Beanland, C.; Schneider, Z.; LoBiondo-Wood, G.; Haber, J. Nursing Research: Methods, Critical Appraisal and Utilization; Mosby: Maryland Heights, MI, USA, 1999. [Google Scholar]
- Creswell, J.W. Educational Research: Planning, Conducting, and Evaluating Quantitative, 4th ed.; Prentice Hall Press: Upper Saddle River, NJ, USA, 2010. [Google Scholar]
- George, D.; Mallery, P. Reliability analysis. In SPSS for Windows, Step by Step: A Simple Guide and Reference, 14th ed.; Allyn & Bacon: Boston, MA, USA, 2003; pp. 222–232. [Google Scholar]
- Pallant, J. SPSS Survival Manual: A Step by Step Guide to Data Analysis Using IBM SPSS; Routledge: London, UK, 2020. [Google Scholar]
- González Ariza, A.; Arando Arbulu, A.; Navas González, F.J.; Ruíz Morales, F.d.A.; León Jurado, J.M.; Barba Capote, C.J.; Camacho Vallejo, M.E. Sensory preference and professional profile affinity definition of endangered native breed eggs compared to commercial laying lineages’ eggs. Animals 2019, 9, 920. [Google Scholar] [CrossRef]
- StataCorp. Stata Statistical Software, 15; StataCorp: College Station, TX, USA, 2017. [Google Scholar]
- IBM Corp. IBM SPSS Statistics for Windows, 25.0; IBM Corp: Armonk, NY, USA, 2017. [Google Scholar]
- Singaraju, G.S.; Js, Y.P.; Mandava, P.; Ganugapanta, V.R.; Teja, N.R.; Jn, P.R. Data Set for Computation of Maxillary Arch Perimeter with Ramanujan’s Equation for Ellipse in Different Skeletal Malocclusions. Data Br. 2020, 32, 106079. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.; Kusleika, R. Excel 2016 Formulas; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Chandrupatla, T.R.; Osler, T.J. The perimeter of an ellipse. Math. Sci. 2010, 35, 122–131. [Google Scholar]
- Energy Information Administration of the United States. Commercial Buildings Energy Consumption Survey: Commercial Buildings Energy Consumption and Expenditures; Michigan State University: East Lansing, MI, USA, 1989. [Google Scholar]
- Bao, L.; Gneiting, T.; Grimit, E.P.; Guttorp, P.; Raftery, A.E. Bias correction and Bayesian model averaging for ensemble forecasts of surface wind direction. Mon. Weather Rev. 2010, 138, 1811–1821. [Google Scholar] [CrossRef]
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Carlin, J. Solutions to some Exercises from Bayesian Data Analysis; Gelman, Carlin, Stern, and Rubin: Boca Raton, FL, USA, 2019. [Google Scholar]
- Koehrsen, W. Introduction to Bayesian Linear Regression. An explanation of the Bayesian approach to linear modeling. Towards Data Sci. 2018, 4, 1. [Google Scholar]
- Kundu, D. Bayesian inference and life testing plan for the Weibull distribution in presence of progressive censoring. Technometrics 2008, 50, 144–154. [Google Scholar] [CrossRef]
- IBM Corp. IBM SPSS Statistics Algorithms; IBM Corp: Armonk, NY, USA, 2017; p. 110. [Google Scholar]
- MacKay, D. Information theory, pattern recognition and neural networks. In Proceedings of the 1st International Conference on Evolutionary Computation, Chicago, IL, USA, 12–16 July 2003; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Brewer, K.R. Combined Survey Sampling Inference: Weighing Basu’s Elephants; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Hayes, A.F.; Glynn, C.J.; Huge, M.E. Cautions Regarding the Interpretation of Regression Coefficients and Hypothesis Tests in Linear Models with Interactions. Commun. Methods Meas. 2012, 6, 1–11. [Google Scholar] [CrossRef]
- Liang, F.; Paulo, R.; Molina, G.; Clyde, M.A.; Berger, J.O. Mixtures of g priors for Bayesian variable selection. J. Am. Stat. Assoc. 2008, 103, 410–423. [Google Scholar] [CrossRef]
- Heck, D. A Caveat on the Savage-Dickey Density Ratio: The Case of Computing Bayes Factors for Regression Parameters. Br. J. Math. Stat. Psychol. 2019, 72, 316–333. [Google Scholar] [CrossRef]
- Zellner, A.; Siow, A. Posterior odds ratios for selected regression hypotheses. Trab. Estadística Y Investig. Oper. 1980, 31, 585–603. [Google Scholar] [CrossRef]
- Rouder, J.N.; Morey, R.D.; Speckman, P.L.; Province, J.M. Default Bayes factors for ANOVA designs. J. Math. Psychol. 2012, 56, 356–374. [Google Scholar] [CrossRef]
- Bayarri, M.J.; Berger, J.O.; Forte, A.; García-Donato, G. Criteria for Bayesian model choice with application to variable selection. Ann. Stat. 2012, 40, 1550–1577. [Google Scholar] [CrossRef]
- Rouder, J.N.; Morey, R.D. Default Bayes factors for model selection in regression. Multivar. Behav. Res. 2012, 47, 877–903. [Google Scholar] [CrossRef] [PubMed]
- Morey, R.; Rouder, J. BayesFactor 0.9. 12-2. Comprehensive R Archive Network. 2015. Available online: https://cran.r-project.org/web/packages/BayesFactor/index.html (accessed on 20 July 2022).
- Rouder, J.N.; Speckman, P.L.; Sun, D.; Morey, R.D.; Iverson, G. Bayesian t tests for accepting and rejecting the null hypothesis. Psychon Bull. Rev. 2009, 16, 225–237. [Google Scholar] [CrossRef]
- Depaoli, S.; Van de Schoot, R. Improving transparency and replication in Bayesian statistics: The WAMBS-Checklist. Psychol. Methods 2017, 22, 240. [Google Scholar] [CrossRef]
- Kass, R.E.; Raftery, A.E. Bayes factors. J. Am. Stat. Assoc. 1995, 90, 773–795. [Google Scholar] [CrossRef]
- Arora, J.S. Chapter 14—Practical Applications of Optimization. In Introduction to Optimum Design, 14th ed.; Academic Press: Boston, MA, USA, 2017; pp. 601–680. [Google Scholar]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Bayesian Analysis of the Association between Casein Complex Haplotype Variants and Milk Yield, Composition, and Curve Shape Parameters in Murciano-Granadina Goats. Animals 2020, 10, 1845. [Google Scholar] [CrossRef] [PubMed]
- Geweke, J. Variable Selection and Model Comparison in Regression; Working Papers 539; Federal Reserve Bank of Minneapolis: Minnesota, MI, USA, 1996. [Google Scholar]
- Analla, M. Model validation through the linear regression fit to actual versus predicted values. Agric. Syst. 1998, 57, 115–119. [Google Scholar] [CrossRef]
- Doğan, N.Ö. Bland-Altman analysis: A paradigm to understand correlation and agreement. Turk. J. Emerg. Med. 2018, 18, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez Martínez, M.d.A. Software-Automatized Individual Lactation Model Fitting, Peak and Persistence and Bayesian Criteria Comparison for Milk Yield Genetic Studies in Murciano-Granadina Goats. Mathematics 2020, 8, 1505. [Google Scholar] [CrossRef]
- Pizarro Inostroza, M.G.; Navas González, F.J.; Landi, V.; León Jurado, J.M.; Delgado Bermejo, J.V.; Fernández Álvarez, J.; Martínez, M.d.A.M. Goat Milk Nutritional Quality Software-Automatized Individual Curve Model Fitting, Shape Parameters Calculation and Bayesian Flexibility Criteria Comparison. Animals 2020, 10, 1693. [Google Scholar] [CrossRef] [PubMed]
- Hall, P.; Maiti, T. Nonparametric estimation of mean-squared prediction error in nested-error regression models. Ann. Stat. 2006, 34, 1733–1750. [Google Scholar] [CrossRef]
- Jeffreys, H. Theory of Probability, 3rd ed.; Oxford University Press: Oxford, UK, 1961. [Google Scholar]
- Lee, M.; Wagenmakers, E. Bayesian Data Analysis for Cognitive Science: A Practical Course; Cambridge University Press: New York, NY, USA, 2013. [Google Scholar]
- Kaplan, D.; Depaoli, S. Bayesian structural equation modeling. In Handbook of Structural Equation Modeling; Hoyle, R.H., Ed.; The Guilford Press: New York, NY, USA, 2012; pp. 650–673. [Google Scholar]
- Gelman, A.; Carlin, J.B.; Stern, H.S.; Dunson, D.B.; Vehtari, A.; Rubin, D.B. Bayesian Data Analysis; CRC Press: Boca Ratón, FL, USA, 2013. [Google Scholar]
- Drton, M.; Plummer, M. A Bayesian information criterion for singular models. J. R. Stat. Soc. Ser. B Stat. Methodol. 2017, 79, 323–380. [Google Scholar] [CrossRef]
- Clyde, M.; Cetinkaya-Rundel, M.; Rundel, C.; Banks, D.; Chai, C.; Huang, L. Chapter 7. Bayesian Model Choice. In An Introduction to Bayesian Thinking; BookDown; CRC Press: Boca Ratón, FL, USA,, 2019. [Google Scholar]
- Gelman, A.; Goodrich, B.; Gabry, J.; Vehtari, A. R-squared for Bayesian regression models. Am. Stat. 2019, 73, 307–309. [Google Scholar] [CrossRef]
- Huang, L.; Li, S.; Zhu, A.; Fan, X.; Zhang, C.; Wang, H. Non-contact body measurement for qinchuan cattle with LiDAR sensor. Sensors 2018, 18, 3014. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ruiz, M.; Tarrat-Martín, D.; Sánchez-Guerrero, M.J.; Valera, M. Advances in horse morphometric measurements using LiDAR. Comput. Electron. Agric. 2020, 174, 105510. [Google Scholar] [CrossRef]
- Muir, A.; Vecsei, P.; Pratt, T.; Krueger, C.; Power, M.; Reist, J. Ontogenetic shifts in morphology and resource use of cisco Coregonus artedi. J. Fish Biol. 2013, 82, 600–617. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.S.; Gazley, M.F. Does my posterior look big in this? The effect of photographic distortion on morphometric analyses. Paleobiology 2017, 43, 508–520. [Google Scholar] [CrossRef]
- Colli, L.; Perrotta, G.; Negrini, R.; Bomba, L.; Bigi, D.; Zambonelli, P.; Verini Supplizi, A.; Liotta, L.; Ajmone-Marsan, P. Detecting population structure and recent demographic history in endangered livestock breeds: The case of the Italian autochthonous donkeys. Anim. Genet. 2013, 44, 69–78. [Google Scholar] [CrossRef]
- Abu-Zidan, F.M.; Eid, H.O.; Hefny, A.F.; Bashir, M.O.; Branicki, F. Camel bite injuries in United Arab Emirates: A 6 year prospective study. Injury 2012, 43, 1617–1620. [Google Scholar] [CrossRef]
- Pezzuolo, A.; Guarino, M.; Sartori, L.; Marinello, F. A feasibility study on the use of a structured light depth-camera for three-dimensional body measurements of dairy cows in free-stall barns. Sensors 2018, 18, 673. [Google Scholar] [CrossRef] [PubMed]
- Jensen, R.B.; Danielsen, S.H.; Tauson, A.-H. Body condition score, morphometric measurements and estimation of body weight in mature Icelandic horses in Denmark. Acta Vet. Scand. 2016, 58, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Albertí, P.; Ripoll, G.; Goyache, F.; Lahoz, F.; Olleta, J.; Panea, B.; Sañudo, C. Carcass characterisation of seven Spanish beef breeds slaughtered at two commercial weights. Meat Sci. 2005, 71, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Lakie, M. The influence of muscle tremor on shooting performance. Exp. Physiol. 2010, 95, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Marsden, C.; Meadows, J.; Lange, G.; Watson, R. The role of the ballistocardiac impulse in the genesis of physiological tremor. Brain 1969, 92, 647–662. [Google Scholar] [CrossRef] [PubMed]

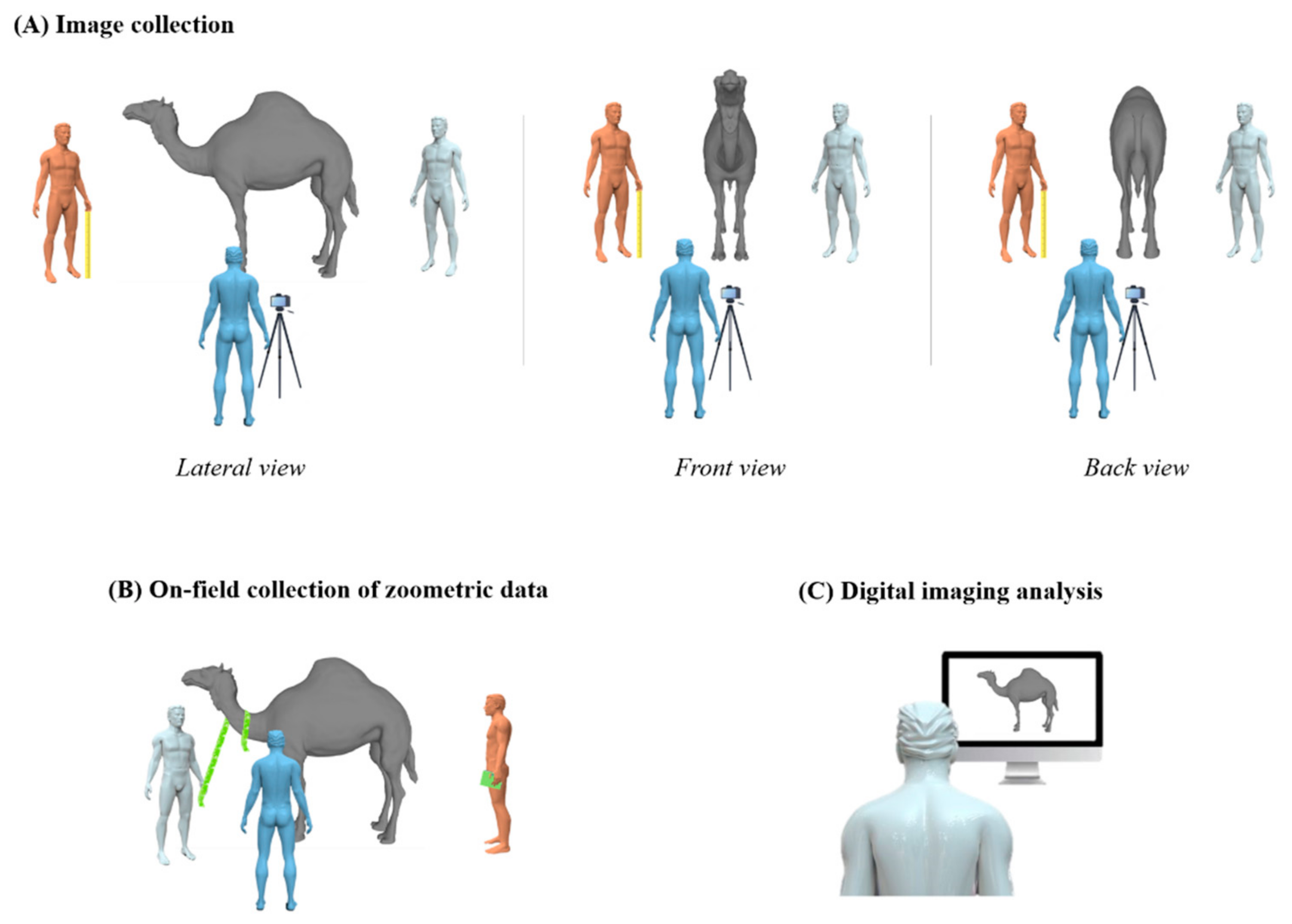

| Area | Measurements | Description |
|---|---|---|
| Head | Head length | Distance from the anterior edge of the nasal bones to the nuchal crest. |
| Head width | Distance between the midpoint of both orbits. | |
| Ear length | Distance from the base to the tip of the ear. | |
| Ear width | Widest distance perpendicular to ear length. | |
| Neck | Neck length: dorsal line | Distance from the base of the neck (cervicothoracic junction) to the base of the head (atlanto-occipital joint) following the upper line of the neck. |
| Neck length: ventral line | Distance from the base of the neck (cervicothoracic junction) to the base of the head (jaw angle) following the lower line of the neck. | |
| Neck girth: cranial third | Circular perimeter of the neck measured at its insertion to the base of the head. | |
| Neck girth: middle third | Circular perimeter of the neck measured at its middle part. | |
| Neck girth: caudal third | Circular perimeter of the neck measured at its insertion to the chest. | |
| Thorax and Dorsum | Chest width | Distance between the medial point of the front of the forelimbs, measured at the base of their insertion into the torso. |
| Heart girth | Circular perimeter of the chest measured directly behind the sternal callosity and before the hump. | |
| Height at withers (stature) | Distance from the withers to the ground. | |
| Body length | Distance between the shoulder to the point of the hip. | |
| Hump | Hump-to-tail distance | Distance between the most caudal point of the base of the hump to the base of the tail. |
| Hump length | Distance between the front and the back of the hump passing through the top of it. | |
| Hump width | Distance between the front and the back of the hump passing through one of its laterals. | |
| Hump height | Distance between the middle points of the base of the hump at each lateral, passing through the top of the hump. | |
| Hump girth | Circular perimeter of the hump measured at its base | |
| Rump and Tail | Rump length | Distance from the coxal to the ischial tuberosity. |
| Rump width | Distance between the right and left coxal tuberosity. | |
| Tail length | Distance from the base to the tip of the tail, excluding the tail skirt. | |
| Width at the base of the tail | Distance between the most lateral points of the base of the tail. | |
| Extremities | Thigh perimeter | Circular perimeter of the thigh measured at its middle part. |
| Hock perimeter | Circular perimeter of the hock measured at its middle part. | |
| Fore cannon bone perimeter | Circular perimeter of the front cannon bone measured at its middle part. | |
| Rear cannon bone perimeter | Circular perimeter of the hind cannon bone measured at its middle part. | |
| Feet | Sole length | Distance from the front to the back of the sole, measured at the planter surface. |
| Length of toe dorsal line | Distance from the external reference of the fetlock to the upper edge of the hoof. | |
| Heel height | Distance from the caudal point of the fetlock to the ground. | |
| Foot perimeter | Circular perimeter of the foot measured at its plantar edge. |
| Area | Measurements | Parameters | Intraclass Correlation | 95% Confidence Interval Lower Bound | 95% Confidence Interval Higher Bound | F Test with True Value 1 | df1 | df2 | Sig | Round a |
|---|---|---|---|---|---|---|---|---|---|---|
| Head | Head length | Single | 0.975 | 0.965 | 0.982 | 78.434 | 129 | 129 | 0.001 | 1st |
| Average/Cα | 0.987 | 0.982 | 0.991 | 78.434 | 129 | 129 | 0.001 | |||
| Head width | Single | 0.738 | 0.649 | 0.808 | 6.64 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.849 | 0.787 | 0.894 | 6.64 | 129 | 129 | 0.001 | |||
| Ear length | Single | 0.854 | 0.800 | 0.895 | 12.697 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.921 | 0.889 | 0.944 | 12.697 | 129 | 129 | 0.001 | |||
| Ear width | Single | 0.875 | 0.827 | 0.910 | 14.973 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.933 | 0.906 | 0.953 | 14.973 | 129 | 129 | 0.001 | |||
| Neck | Neck length: dorsal line | Single | 0.847 | 0.791 | 0.890 | 12.108 | 129 | 129 | 0.001 | 1st |
| Average/Cα | 0.917 | 0.883 | 0.942 | 12.108 | 129 | 129 | 0.001 | |||
| Neck length: ventral line | Single | 0.992 | 0.989 | 0.994 | 247.029 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.996 | 0.994 | 0.997 | 247.029 | 129 | 129 | 0.001 | |||
| Neck girth: cranial third | Single | 0.556 | 0.425 | 0.664 | 3.504 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.715 | 0.596 | 0.798 | 3.504 | 129 | 129 | 0.001 | |||
| Neck girth: middle third | Single | 0.664 | 0.555 | 0.750 | 4.947 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.798 | 0.714 | 0.857 | 4.947 | 129 | 129 | 0.001 | |||
| Neck girth: caudal third | Single | 0.541 | 0.407 | 0.652 | 3.357 | 129 | 129 | 0.001 | 3rd | |
| Average/Cα | 0.702 | 0.579 | 0.789 | 3.357 | 129 | 129 | 0.001 | |||
| Thorax and Dorsum | Chest width | Single | 0.779 | 0.701 | 0.838 | 8.036 | 129 | 129 | 0.001 | 1st |
| Average/Cα | 0.876 | 0.824 | 0.912 | 8.036 | 129 | 129 | 0.001 | |||
| Heart girth | Single | 0.682 | 0.578 | 0.764 | 5.285 | 129 | 129 | 0.001 | 3rd | |
| Average/Cα | 0.811 | 0.732 | 0.866 | 5.285 | 129 | 129 | 0.001 | |||
| Height at withers (stature) | Single | 0.986 | 0.979 | 0.990 | 152.735 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.993 | 0.989 | 0.995 | 152.735 | 129 | 129 | 0.001 | |||
| Body length | Single | 0.865 | 0.814 | 0.902 | 13.78 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.927 | 0.897 | 0.949 | 13.78 | 129 | 129 | 0.001 | |||
| Hump | Hump-to-tail distance | Single | 0.984 | 0.977 | 0.989 | 123.989 | 129 | 129 | 0.001 | 1st |
| Average/Cα | 0.992 | 0.989 | 0.994 | 123.989 | 129 | 129 | 0.001 | |||
| Hump length | Single | 0.905 | 0.868 | 0.932 | 20.049 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.950 | 0.929 | 0.965 | 20.049 | 129 | 129 | 0.001 | |||
| Hump width | Single | 0.989 | 0.985 | 0.992 | 182.709 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.995 | 0.992 | 0.996 | 182.709 | 129 | 129 | 0.001 | |||
| Hump height | Single | 0.995 | 0.992 | 0.996 | 375.396 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.997 | 0.996 | 0.998 | 375.396 | 129 | 129 | 0.001 | |||
| Hump girth | Single | 0.587 | 0.461 | 0.689 | 3.838 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.739 | 0.632 | 0.816 | 3.838 | 129 | 129 | 0.001 | |||
| Rump and Tail | Rump length | Single | 0.938 | 0.914 | 0.956 | 31.371 | 129 | 129 | 0.001 | 1st |
| Average/Cα | 0.968 | 0.955 | 0.977 | 31.371 | 129 | 129 | 0.001 | |||
| Rump width | Single | 0.984 | 0.978 | 0.989 | 124.652 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.992 | 0.989 | 0.994 | 124.652 | 129 | 129 | 0.001 | |||
| Tail length | Single | 0.984 | 0.977 | 0.988 | 121.041 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.992 | 0.988 | 0.994 | 121.041 | 129 | 129 | 0.001 | |||
| Width at the base of the tail | Single | 0.842 | 0.783 | 0.885 | 11.642 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.914 | 0.879 | 0.939 | 11.642 | 129 | 129 | 0.001 | |||
| Extremities | Thigh perimeter | Single | 0.618 | 0.500 | 0.714 | 4.239 | 129 | 129 | 0.001 | 2nd |
| Average/Cα | 0.764 | 0.666 | 0.833 | 4.239 | 129 | 129 | 0.001 | |||
| Hock perimeter | Single | 0.753 | 0.668 | 0.819 | 7.107 | 129 | 129 | 0.001 | 3rd | |
| Average/Cα | 0.859 | 0.801 | 0.901 | 7.107 | 129 | 129 | 0.001 | |||
| Fore cannon bone perimeter | Single | 0.612 | 0.492 | 0.709 | 4.156 | 129 | 129 | 0.001 | 3rd | |
| Average/Cα | 0.759 | 0.660 | 0.830 | 4.156 | 129 | 129 | 0.001 | |||
| Rear cannon bone perimeter | Single | 0.529 | 0.393 | 0.642 | 3.243 | 129 | 129 | 0.001 | 3rd | |
| Average/Cα | 0.692 | 0.564 | 0.782 | 3.243 | 129 | 129 | 0.001 | |||
| Feet | Sole length | Single | 0.718 | 0.623 | 0.792 | 6.097 | 129 | 129 | 0.001 | 1st |
| Average/Cα | 0.836 | 0.768 | 0.884 | 6.097 | 129 | 129 | 0.001 | |||
| Length of toe dorsal line | Single | 0.894 | 0.853 | 0.924 | 17.832 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.944 | 0.921 | 0.960 | 17.832 | 129 | 129 | 0.001 | |||
| Heel height | Single | 0.977 | 0.968 | 0.984 | 86.753 | 129 | 129 | 0.001 | 1st | |
| Average/Cα | 0.988 | 0.984 | 0.992 | 86.753 | 129 | 129 | 0.001 | |||
| Foot perimeter | Single | 0.486 | 0.343 | 0.607 | 2.894 | 129 | 129 | 0.001 | 3rd | |
| Average/Cα | 0.654 | 0.511 | 0.756 | 2.894 | 129 | 129 | 0.001 |
| Zoometric Trait (=y) | Parameter | Posterior | 95% Credible Interval | |||
|---|---|---|---|---|---|---|
| Mode | Mean | Variance | Lower Bound | Upper Bound | ||
| Corrected Neck girth: caudal third | Intercept | 40.713 | 40.713 | 214.839 | 11.938 | 69.488 |
| Neck girth: caudal third | 0.347 | 0.347 | 0.013 | 0.125 | 0.570 | |
| Live body weight | 0.088 | 0.088 | 0.000 | 0.054 | 0.121 | |
| Corrected Heart girth | Intercept | 76.511 | 76.511 | 48.808 | 62.796 | 90.227 |
| Heart girth | −0.017 | −0.017 | 0.001 | −0.081 | 0.046 | |
| Live body weight | 0.111 | 0.111 | 0.000 | 0.092 | 0.130 | |
| Corrected Thigh circumference | Intercept | 44.180 | 44.180 | 30.676 | 33.307 | 55.053 |
| Thigh circumference | 0.217 | 0.217 | 0.007 | 0.059 | 0.376 | |
| Live body weight | 0.039 | 0.039 | 0.000 | 0.021 | 0.058 | |
| Corrected Hock circumference | Intercept | 13.005 | 13.005 | 11.842 | 6.250 | 19.761 |
| Hock circumference | 0.335 | 0.335 | 0.008 | 0.162 | 0.508 | |
| Live body weight | 0.025 | 0.025 | 0.000 | 0.015 | 0.036 | |
| Corrected Fore cannon bone perimeter | Intercept | 10.646 | 10.646 | 2.169 | 7.755 | 13.537 |
| Fore cannon bone perimeter | 0.247 | 0.247 | 0.005 | 0.104 | 0.390 | |
| Live body weight | 0.006 | 0.006 | 0.000 | 0.001 | 0.010 | |
| Corrected Rear cannon bone perimeter | Intercept | 9.392 | 9.392 | 1.684 | 6.844 | 11.939 |
| Rear cannon bone perimeter | 0.225 | 0.225 | 0.003 | 0.111 | 0.340 | |
| Live body weight | 0.009 | 0.009 | 0.000 | 0.005 | 0.013 | |
| Corrected Foot perimeter | Intercept | 33.356 | 33.356 | 21.631 | 24.226 | 42.487 |
| Foot perimeter | 0.260 | 0.260 | 0.016 | 0.015 | 0.506 | |
| Zoometric Trait (=y) | Corrected Neck Girth: Caudal Third | Corrected Heart Girth | Corrected Thigh Circumference | Corrected Hock Circumference | Corrected Fore Cannon Bone Perimeter | Corrected Rear Cannon Bone Perimeter | Corrected Foot Perimeter |
|---|---|---|---|---|---|---|---|
| Correction Method | Bayesian regression and Natural logarithm | Bayesian regression and Natural logarithm | Bayesian regression | Bayesian regression and Natural logarithm | Bayesian regression and Natural logarithm | Bayesian regression and Natural logarithm | Bayesian regression and Natural logarithm |
| Regressors | Live body weight and Digital Imaging measurement for neck girth: caudal third | Live body weight and Digital Imaging measurement for heart girth | Live body weight and Digital Imaging measurement for Thigh circumference | Live body weight and Digital Imaging measurement for Hock circumference | Live body weight and Digital Imaging measurement for Fore cannon bone perimeter | Live body weight and Digital Imaging measurement for Rear cannon bone perimeter | Digital Imaging measurement for Foot perimeter |
| Round | 3rd | 3rd | 2nd | 3rd | 3rd | 3rd | 3rd |
| Equation | y = 40.713 + 0.347 ∗ (Neck girth: caudal third) + 0.088 ∗ (Live body weight) + ln(Neck girth: caudal third) | y = 76.511 + (−0.017) ∗ (Heart girth) + 0.111 ∗ (Live body weight) + ln(Heart girth) | y = 44.18 + 0.217 ∗ (Thigh circumference) + 0.039 ∗ (Live body weight) | y = 13.005 + 0.335(Hock circumference) + 0.025 ∗ (Live body weight) + ln(Hock circumference) | y = 10.646 + 0.247 ∗ (Fore cannon bone perimeter) + 0.006 ∗ (Live body weight) + ln(Fore cannon bone perimeter) | y = 9.392 + 0.225 ∗ (Rear cannon bone perimeter) + 0.009*(Live body weight) + ln(Rear cannon bone perimeter) | y = 33.356 + 0.260 ∗ (Foot perimeter) + ln(Foot perimeter) |
| Regression Sum of Squares | 11,540.442 | 12,611.463 | 3226.575 | 1504.485 | 114.180 | 182.118 | 162.324 |
| Regression df | 2.000 | 2.000 | 2.000 | 2.000 | 2.000 | 2.000 | 1.000 |
| Regression Mean Square | 5770.221 | 6305.731 | 1613.288 | 752.242 | 57.090 | 91.059 | 162.324 |
| F | 20.168 | 67.975 | 23.580 | 33.454 | 11.409 | 23.354 | 4.409 |
| Sig. | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.038 |
| Residual Sum of Squares (RSS) | 36,336.003 | 11,781.220 | 8689.017 | 2855.697 | 635.487 | 495.174 | 4712.808 |
| Residual df | 127.000 | 127.000 | 127.000 | 127.000 | 127.000 | 127.000 | 128.000 |
| Residual Mean Square | 286.110 | 92.766 | 68.417 | 22.486 | 5.004 | 3.899 | 36.819 |
| Bayes Factor (BF) | 269,937.547 | 6.39 × 1017 | 3,356,913.557 | 2.93 × 109 | 252.807 | 2,850,184.198 | 0.597 |
| R | 0.491 | 0.719 | 0.520 | 0.587 | 0.390 | 0.519 | 0.182 |
| Bayesian R Square | 0.241 | 0.517 | 0.271 | 0.345 | 0.152 | 0.269 | 0.033 |
| Bayesian Adjusted R Square | 0.229 | 0.509 | 0.259 | 0.335 | 0.139 | 0.257 | 0.026 |
| SE | 16.915 | 9.631 | 8.271 | 4.742 | 2.237 | 1.975 | 6.068 |
| Monte Carlo standard error (MCSE) | 279.508 | 90.625 | 66.839 | 21.967 | 4.888 | 3.809 | 36.252 |
| Bayesian Information Criterion (BIC) | 95,207.940 | 76,173.431 | 71,028.278 | 52,222.907 | 26,827.621 | 22,611.369 | 60,684.398 |
| Variables for Which a Correction Was Issued | P(T >= T_obs) Predictive Posterior p-Values |
|---|---|
| Corrected Neck girth: caudal third | 0.496 |
| Corrected Heart girth | 0.459 |
| Corrected Thigh circumference | 0.495 |
| Corrected Hock circumference | 0.495 |
| Corrected Fore cannon bone perimeter | 0.494 |
| Corrected Rear cannon bone perimeter | 0.497 |
| Corrected Foot perimeter | 0.497 |
| P(T >= T_obs) close to 0 or 1 indicates a lack of fit. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iglesias Pastrana, C.; Navas González, F.J.; Ciani, E.; Camacho Vallejo, M.E.; Delgado Bermejo, J.V. Bayesian Linear Regression and Natural Logarithmic Correction for Digital Image-Based Extraction of Linear and Tridimensional Zoometrics in Dromedary Camels. Mathematics 2022, 10, 3453. https://doi.org/10.3390/math10193453
Iglesias Pastrana C, Navas González FJ, Ciani E, Camacho Vallejo ME, Delgado Bermejo JV. Bayesian Linear Regression and Natural Logarithmic Correction for Digital Image-Based Extraction of Linear and Tridimensional Zoometrics in Dromedary Camels. Mathematics. 2022; 10(19):3453. https://doi.org/10.3390/math10193453
Chicago/Turabian StyleIglesias Pastrana, Carlos, Francisco Javier Navas González, Elena Ciani, María Esperanza Camacho Vallejo, and Juan Vicente Delgado Bermejo. 2022. "Bayesian Linear Regression and Natural Logarithmic Correction for Digital Image-Based Extraction of Linear and Tridimensional Zoometrics in Dromedary Camels" Mathematics 10, no. 19: 3453. https://doi.org/10.3390/math10193453
APA StyleIglesias Pastrana, C., Navas González, F. J., Ciani, E., Camacho Vallejo, M. E., & Delgado Bermejo, J. V. (2022). Bayesian Linear Regression and Natural Logarithmic Correction for Digital Image-Based Extraction of Linear and Tridimensional Zoometrics in Dromedary Camels. Mathematics, 10(19), 3453. https://doi.org/10.3390/math10193453






