MS-CheXNet: An Explainable and Lightweight Multi-Scale Dilated Network with Depthwise Separable Convolution for Prediction of Pulmonary Abnormalities in Chest Radiographs
Abstract
:1. Introduction
Contribution
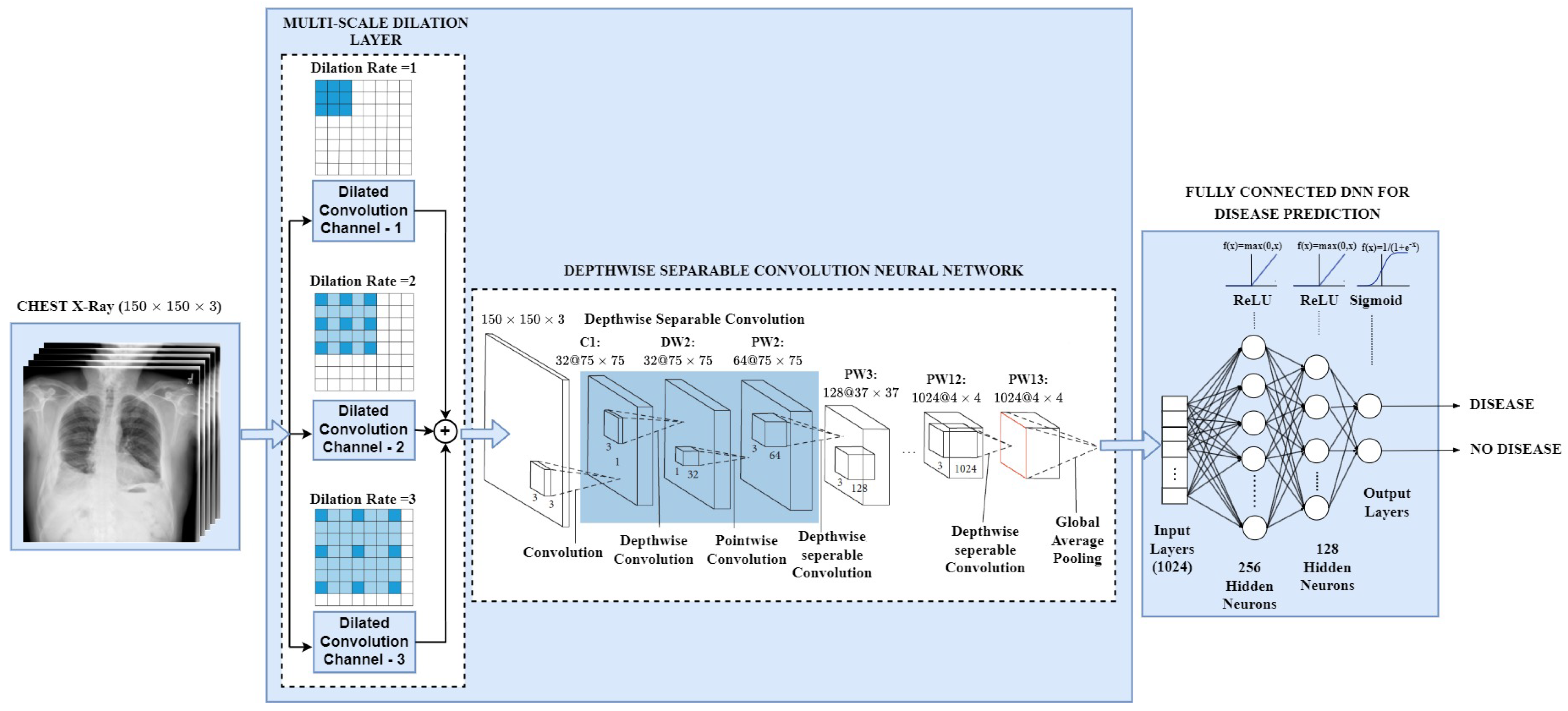
- With the focus of designing an effective deep learning network suitable to employ in cloud computing, mobile vision, and embedding system applications, we present an explainable and lightweight Multi-scale Chest X-ray Network (MS-CheXNet) to predict abnormal diseases from chest radiographs.
- To enlarge the receptive field and capture the discriminative multi-scale feature without increasing convolution parameters, we propose an effective Multi-Scale Dilation Layer (MSDL), which is conducive to learning varied-sized pulmonary abnormalities and boosts the prediction performance.
- We adopt a lightweight Depthwise Separable Convolution Neural Network (DS-CNN) to learn the dense imaging features by adjusting lesser network parameters than the conventional CNN. We employed a fully connected Deep Neural Network to predict the abnormalities from the chest radiographs.
- We incorporated Gradient-weighted Class Activation Mapping (Grad-CAM) technique to visualize and localize the abnormalities in the chest region. This makes our network explainable by checking the decision model’s transparency and understanding their ability to arrive at a decision.
- We compared the proposed MS-CheXNet with the existing state-of-the-art deep learning strategies. We assessed our model’s competence by applying it on two datasets: the publicly available Open-I dataset and Real-time diagnostic data collected from the private hospital.
2. Literature Review
2.1. Disease Detection and Localization Task
2.2. Disease Classification and Prediction Task
2.3. Outcome of the Literature Review
- We have found that deep convolution neural networks perform better in classifying medical images from the above literature.
- The transfer learning strategy with well-established deep learning models trained on ImageNet weights where initial learning is transferred during training addresses the problem of the enormous dataset needed for deep learning training [20,24,28]. The usage of imageNet weights yields good performance and solves the problem of an enormous dataset to train deep learning models.
- The existing deep learning strategies lack capturing the more discriminative features from the receptive field. Medical CXRs come with varied-sized abnormalities; thus, most of the existing techniques do not focus on multiscale features.
3. Materials and Methods
3.1. Data Augmentation
3.2. Multi-Scale Dilation Layer (MSDL)
3.3. Depthwise Separable Convolution Neural Network (DS-CNN)
3.4. Fully Connected Deep Neural Network for Abnormality Prediction
3.5. Disease Visualization Using Grad-CAM Technique
4. Experimental Setup
4.1. Parameter Configurations of Proposed MS-CheXNet and State-of-the-Art Deep Learning Models
4.2. Radiology Cohort Selection
4.3. Evaluation Criteria
- True Positive () indicates the CXR sample belonging to the abnormal class is being accurately categorized as an abnormal class
- True Negative () indicates the CXR sample belonging to the normal class is being accurately categorized as an normal class
- False Positive () indicates the CXR sample belonging to the normal class is being wrongly categorized as a abnormal class
- False Negative () indicates the CXR sample belonging to the abnormal class is being wrongly categorized as a normal class
5. Results and Discussions
5.1. Quantitative Analysis of Proposed MS-CheXNet with the Fine-Tuned Pre-Trained Deep Learning Models
5.2. Performance Analysis of Proposed MS-CheXNet with the Existing State-of-the-Art Deep Learning Strategies on Open-I Dataset
5.3. Qualitative Analysis of Proposed MS-CheXNet
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CXR | Chest X-ray |
| CT | Computed Tomography |
| MRI | Magnetic Resonance Imaging |
| CNN | Convolution Neural Network |
| DNN | Deep Neural Network |
| KMC | Kasturba Medical College |
| MeSH | Medical Subject Heading |
| MS-CheXNet | Multi Scale Chest X-ray Network |
| MSDL | Multi-Scale Dilation Layer |
| DS-CNN | Depthwise Separable Convolution Neural Network |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
References
- W.H.O. Chronic Respiratory Diseases. Available online: https://www.who.int/gard/publications/chronic_respiratory_diseases.pdf (accessed on 21 December 2021).
- UN. United Nations Scientific Committee on the Effects of Atomic Radiation (UNSCEAR). 2008. Available online: http://www.unscear.org/docs/publications/2008/UNSCEAR_2008_Annex-A-CORR.pdf (accessed on 14 August 2021).
- Abiyev, R.; Ma’aitah, M. Deep Convolutional Neural Networks for Chest Diseases Detection. J. Healthc. Eng. 2018, 2018, 4168538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Ren, F.; Li, Y.; Na, L.; Ma, Y. Pneumonia Detection from Chest X-ray Images Based on Convolutional Neural Network. Electronics 2021, 10, 1512. [Google Scholar] [CrossRef]
- Shetty, S.; Ananthanarayana, V.S.; Mahale, A. Medical Knowledge-Based Deep Learning Framework for Disease Prediction on Unstructured Radiology Free-Text Reports under Low Data Condition. In Proceedings of the 21st EANN (Engineering Applications of Neural Networks) 2020 Conference, Halkidiki, Greece, 5–7 June 2020; Springer International Publishing: Cham, Switzerland, 2020; pp. 352–364. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: New York, NY, USA, 2012; Volume 25. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-Scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Kermany, D. Labeled Optical Coherence Tomography (OCT) and Chest X-ray Images for Classification. Mendeley Data 2018, 2. [Google Scholar] [CrossRef]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Proc. AAAI Conf. Artif. Intell. 2019, 33, 590–597. [Google Scholar] [CrossRef] [Green Version]
- Johnson, A.E.W.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.Y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef] [Green Version]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 2015, 23, 304–310. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.X.J.; Lu, P.X.; Thoma, G.R. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014, 46, 475–477. [Google Scholar]
- Ryoo, S.; Kim, H.J. Activities of the Korean Institute of Tuberculosis. Osong Public Health Res. Perspect. 2014, 5, S43–S49. [Google Scholar] [CrossRef]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray: Hospital-Scale Chest X-ray Database and Benchmarks on Weakly Supervised Classification and Localization of Common Thorax Diseases. In Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics; Springer International Publishing: Cham, Switzerland, 2019; pp. 369–392. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef] [Green Version]
- Rajpurkar, P.; Irvin, J.; Ball, R.L.; Zhu, K.; Yang, B.; Mehta, H.; Duan, T.; Ding, D.; Bagul, A.; Langlotz, C.P.; et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018, 15, e1002686. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef] [Green Version]
- Candemir, S.; Rajaraman, S.; Thoma, G.; Antani, S. Deep Learning for Grading Cardiomegaly Severity in Chest X-rays: An Investigation. In Proceedings of the 2018 IEEE Life Sciences Conference (LSC), Montreal, QC, Canada, 28–30 October 2018. [Google Scholar] [CrossRef]
- Hwang, E.J.; Park, S.; Jin, K.N.; Kim, J.I.; Choi, S.Y.; Lee, J.H.; Goo, J.M.; Aum, J.; Yim, J.J.; Park, C.M.; et al. Development and Validation of a Deep Learning–based Automatic Detection Algorithm for Active Pulmonary Tuberculosis on Chest Radiographs. Clin. Infect. Dis. 2018, 69, 739–747. [Google Scholar] [CrossRef] [Green Version]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef] [Green Version]
- Pasa, F.; Golkov, V.; Pfeiffer, F.; Cremers, D.; Pfeiffer, D. Efficient Deep Network Architectures for Fast Chest X-ray Tuberculosis Screening and Visualization. Sci. Rep. 2019, 9, 6268. [Google Scholar] [CrossRef] [Green Version]
- Zou, X.L.; Ren, Y.; Feng, D.Y.; He, X.Q.; Guo, Y.F.; Yang, H.L.; Li, X.; Fang, J.; Li, Q.; Ye, J.J.; et al. A promising approach for screening pulmonary hypertension based on frontal chest radiographs using deep learning: A retrospective study. PLoS ONE 2020, 15, e0236378. [Google Scholar] [CrossRef]
- Hashmi, M.F.; Katiyar, S.; Keskar, A.G.; Bokde, N.D.; Geem, Z.W. Efficient Pneumonia Detection in Chest X-ray Images Using Deep Transfer Learning. Diagnostics 2020, 10, 417. [Google Scholar] [CrossRef]
- Lee, M.S.; Kim, Y.S.; Kim, M.; Usman, M.; Byon, S.S.; Kim, S.H.; Lee, B.I.; Lee, B.D. Evaluation of the feasibility of explainable computer-aided detection of cardiomegaly on chest radiographs using deep learning. Sci. Rep. 2021, 11, 16885. [Google Scholar] [CrossRef]
- Rajkomar, A.; Lingam, S.; Taylor, A.G.; Blum, M.; Mongan, J. High-Throughput Classification of Radiographs Using Deep Convolutional Neural Networks. J. Digit. Imaging 2016, 30, 95–101. [Google Scholar] [CrossRef]
- Chaudhary, A.; Hazra, A.; Chaudhary, P. Diagnosis of Chest Diseases in X-ray images using Deep Convolutional Neural Network. In Proceedings of the 2019 10th International Conference on Computing, Communication and Networking Technologies (ICCCNT), Kanpur, India, 6–8 July 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Tang, Y.X.; Tang, Y.B.; Peng, Y.; Yan, K.; Bagheri, M.; Redd, B.; Brandon, C.; Lu, Z.; Han, M.; Xiao, J.; et al. Automated abnormality classification of chest radiographs using deep convolutional neural networks. NPJ Digit. Med. 2020, 3, 70. [Google Scholar] [CrossRef]
- Cohen, J.P.; Hashir, M.; Brooks, R.; Bertrand, H. On the limits of cross-domain generalization in automated X-ray prediction. Proc. Mach. Learn. Res. 2020, 121, 136–155. [Google Scholar]
- Li, D.; Liu, Z.; Luo, L.; Tian, S.; Zhao, J. Prediction of Pulmonary Fibrosis Based on X-rays by Deep Neural Network. J. Healthc. Eng. 2022, 2022, 3845008. [Google Scholar] [CrossRef]
- Aydin, F.; Zhang, M.; Ananda-Rajah, M.; Haffari, G. Medical Multimodal Classifiers under Scarce Data Condition. arXiv 2019, arXiv:1902.08888. [Google Scholar]
- Lopez, K.; Fodeh, S.J.; Allam, A.; Brandt, C.A.; Krauthammer, M. Reducing Annotation Burden Through Multimodal Learning. Front. Big Data 2020, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Summers, R.M. TieNet: Text-Image Embedding Network for Common Thorax Disease Classification and Reporting in Chest X-rays. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 9049–9058. [Google Scholar]
- Griner, D.; Zhang, R.; Tie, X.; Zhang, C.; Garrett, J.W.; Li, K.; Chen, G.H. COVID-19 pneumonia diagnosis using chest X-ray radiograph and deep learning. In Medical Imaging 2021: Computer-Aided Diagnosis; Mazurowski, M.A., Drukker, K., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, WA, USA, 2021; Volume 11597, pp. 18–24. [Google Scholar] [CrossRef]
- Kusakunniran, W.; Karnjanapreechakorn, S.; Siriapisith, T.; Borwarnginn, P.; Sutassananon, K.; Tongdee, T.; Saiviroonporn, P. COVID-19 detection and heatmap generation in chest X-ray images. J. Med. Imaging 2021, 8, 014001. [Google Scholar] [CrossRef] [PubMed]
- Helal Uddin, M.; Hossain, M.N.; Islam, M.S.; Zubaer, M.A.A.; Yang, S.H. Detecting COVID-19 Status Using Chest X-ray Images and Symptoms Analysis by Own Developed Mathematical Model: A Model Development and Analysis Approach. COVID 2022, 2, 117–137. [Google Scholar] [CrossRef]
- Giełczyk, A.; Marciniak, A.; Tarczewska, M.; Lutowski, Z. Pre-processing methods in chest X-ray image classification. PLoS ONE 2022, 17, e0265949. [Google Scholar] [CrossRef]
- Gouda, W.; Almurafeh, M.; Humayun, M.; Jhanjhi, N.Z. Detection of COVID-19 Based on Chest X-rays Using Deep Learning. Healthcare 2022, 10, 343. [Google Scholar] [CrossRef]
- Bloice, M.; Stocker, C.; Holzinger, A. Augmentor: An Image Augmentation Library for Machine Learning. J. Open Source Softw. 2017, 2, 432. [Google Scholar] [CrossRef]
- Araujo, A.; Norris, W.D.; Sim, J. Computing Receptive Fields of Convolutional Neural Networks. Distill 2019, 4, e21. [Google Scholar] [CrossRef]
- Holschneider, M.; Kronland-Martinet, R.; Morlet, J.; Tchamitchian, P. A Real-Time Algorithm for Signal Analysis with the Help of the Wavelet Transform. In Wavelets—Time-Frequency Methods and Phase Space; Springer: Berlin/Heidelberg, Germany, 1989; Volume 1, p. 286. [Google Scholar] [CrossRef]
- Wang, P.; Chen, P.; Yuan, Y.; Liu, D.; Huang, Z.; Hou, X.; Cottrell, G. Understanding Convolution for Semantic Segmentation. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 1451–1460. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [Green Version]
- Yu, F.; Koltun, V. Multi-Scale Context Aggregation by Dilated Convolutions. In Proceedings of the International Conference on Learning Representations (ICLR), San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar] [CrossRef] [Green Version]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In Proceedings of the 32nd International Conference on International Conference on Machine Learning, CML’15, Lille, France, 7–9 July 2015; Volume 37, pp. 448–456. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified Linear Units Improve Restricted Boltzmann Machines. In Proceedings of the 27th International Conference on International Conference on Machine Learning, ICML’10, Haifa, Israel, 21–24 June 2010; Omnipress: Madison, WI, USA, 2010; pp. 807–814. [Google Scholar]
- Narayan, S. The generalized sigmoid activation function: Competitive supervised learning. Inf. Sci. 1997, 99, 69–82. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Chen, Q.; Yan, S. Network In Network. arXiv 2014, arXiv:1312.4400. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: https://www.tensorflow.org/ (accessed on 17 December 2021).
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Yao, Y.; Rosasco, L.; Caponnetto, A. On Early Stopping in Gradient Descent Learning. Constr. Approx. 2007, 26, 289–315. [Google Scholar] [CrossRef]
- Jing, B.; Xie, P.; Xing, E.P. On the Automatic Generation of Medical Imaging Reports. arXiv 2017, arXiv:1711.08195. [Google Scholar]
- Xue, Y.; Xu, T.; Rodney Long, L.; Xue, Z.; Antani, S.; Thoma, G.R.; Huang, X. Multimodal Recurrent Model with Attention for Automated Radiology Report Generation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2018, Granada, Spain, 16–20 September 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 457–466. [Google Scholar]
- Pino, P.; Parra, D.; Besa, C.; Lagos, C. Clinically Correct Report Generation from Chest X-rays Using Templates. In Proceedings of the Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Conjunction with MICCAI 2021, Strasbourg, France, 27 September 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 654–663. [Google Scholar] [CrossRef]
- Wissler, L.; Almashraee, M.; Monett, D.; Paschke, A. The Gold Standard in Corpus Annotation. In Proceedings of the 5th IEEE Germany Student Conference, Passau, Germany, 26–27 June 2014. [Google Scholar] [CrossRef]












| Dataset | Dataset Description | Predictable Disease |
|---|---|---|
| NIH Chest X-ray14 [10] | 112,120 images of 14 diseases gathered from 30,805 patient | Atelectasis, Cardiomegaly, Effusion, Infiltration, Mass, Pneumonia, Nodule, Pneumothorax, Edema, Emphysema, Fibrosis, Pleural Thickening and Hernia |
| Pediatric CXR [11] | 5856 CXR images in which 3883 are Pneumonia images | Pneumonia |
| CheXper [12] | 224,316 CXR of 65,240 cases | 14 Chest Diseases |
| MIMIC CXR [13] | 227,827 images with 14 chest disease images | 14 Chest Diseases |
| Open-I [14] | 7470 chest radiographs with frontal and lateral view | Pulmonary Edema, Cardiac Hypertrophy, Pleural effusion and Opacity |
| MC dataset [15] | 138 Chest images, 58 from Tuberculosis patient | Tuberculosis |
| Shenzhen [15] | 662 Chest images, 336 from Tuberculosis patient | Tuberculosis |
| KIT dataset [16] | 10,848 chest images, 3828 from Tuberculosis patient | Tuberculosis |
| Author & Year | Methodology | Task | Medical Domain | Abnormality | Imaging Data | Dataset |
|---|---|---|---|---|---|---|
| Rajkomar et al. [29], 2017 | The GoogleNet architecture is used to classify the CXRs into frontal and lateral. | Classification | Radiology | Pulmonary diseases | Chest X-ray | Private Dataset (909 Patients) |
| Pranav et al. [20], 2017 | The 121-layered Dense Convolutional Network named CheXNet to predict pneumonia pathology from CXRs and for the binary classification of pneumonia detection pretrained, ImageNet weights were utilized. | Detection | Radiology | Pnuemonia | Chest X-ray | NIH Chest X-ray14 (112,120 from 30,805 patients) |
| Candemir et al. [22], 2018 | Deep CNN models such as AlexNet, VGG-16, VGG-19 and Inception V3 is utilized to detect cardiomegaly from the CXRs. | Detection | Radiology | Cardiomegaly | Chest X-ray | Open-i (283 Cardiomegaly cases from 3683 patients) |
| Hwang et al. [23], 2018 | The ResNet based model with 27 layers and 12 residual connections is utilized to detect active pulmonary tuberculosis from the large private CXR cohort. | Detection | Radiology | Pulmonary Tuberculosis | Chest X-ray | Private Dataset (54,221 Normal CXRs and 6768 tuberculosis CXRs) |
| John et al. [24], 2019 | The DenseNet121 model pretrained with ImageNet weights is further trained and tested across different data cohorts to detect the pneumonia abnormality. | Detection | Radiology | Pnuemonia | Chest X-ray | 1. NIH Chest X-ray14 (112,120 from 30,805 patients) 2. MSH (42,396 from 12,904 patients) 3. Open-I (3807 from 3683 patients) |
| Pasa et al. [25], 2019 | CNN-based model is proposed for faster diagnosis of tuberculosis diseases and Grad-CAM technique is incorporated for disease visualization. | Detection and Visualization | Radiology | Tuberculosis | Chest X-ray | 1. NIH Tuberculosis CXR (138 and 662 patients) 2. Belarus Tuberculosis Portal dataset (304 patients) |
| Chaudhary et al. [30], 2019 | The CNN-based deep learning model with three convolution, ReLU, pooling, and fully connected layers was proposed to diagnose chest diseases from CXRs. | Classification | Radiology | Pulmonary diseases | Chest X-ray | NIH Chest X-ray14 (1,12,120 CXRs) |
| Tang et al. [31], 2020 | Identifying abnormality using Deep CNN models and comparison with the radiologist labels. | Classification | Radiology | Pulmonary diseases | Chest X-ray | 1. NIH ChestX-ray14 (112,120 from 30,805 patients) 2. Open-I (3807 CXRs from 3683 patients) 3. RSNA Dataset (21,152 patients) |
| Cohen et al. [32], 2020 | Investigative study to find discrepancies while generalizing the models with multiple chest X-ray datasets. | Classification | Radiology | Pulmonary diseases | Chest X-ray | 1. NIH Chest X-ray14 (112,120 from 30,805 patients) 2. PadChest (160,000 from 67,000 patients) 3. MIMIC-CXR (227,827 CXRs) 4. Open-I (3807 CXRs from 3683 patients) 5. RSNA Dataset (21,152 patients) |
| Zou et al. [26], 2020 | Detection and screening of pulmonary hypertension using three deep learning models (Resnet50, Xception, and Inception V3) | Detection and Visualization | Radiology | Pulmonary hypertension | Chest X-ray | Private dataset (762 patients from three institute in China) |
| Hashmi et al. [27], 2020 | A weighted classifier combining the weighted predictions of the state-of-the-art deep learning model is introduced to detect pneumonia in CXRs | Detection and Visualization | Radiology | Pnuemonia | Chest X-ray | Private dataset (7022 CXRs) |
| Dalton et al. [37], 2021 | The classification of COVID-19 abnormality is performed using ensemble of DenseNet-121 Networks | Classification | Radiology | COVID-19 | Chest X-ray | Private dataset (12,000 patients) |
| Lee et al. [28], 2021 | The ResNet 101 and U-Net pretrained on ImageNet is used to segment and detect the cardiomegaly diseases from the CXRs | Segmentation and Detection | Radiology | Cardiomegaly | Chest X-ray | 1. JSRT dataset (247 patients) 2. Montgomery dataset (138 patients) 3. Private dataset (408 patients) |
| Worapan et al. [38], 2021 | The ResNet101 model is utilized to detect COVID-19 and heatmap is produced for segmented lung area. | Detection and Visualization | Radiology | COVID-19 | Chest X-ray | Private dataset (5743 CXRs) |
| Helal et al. [39], 2022 | The CNN-based deep learning model named SymptomNet is proposed to detect COVID-19 and heatmap is generated to visualize the disease. | Detection and Visualization | Radiology | COVID-19 | Chest X-ray | Private dataset (500 CXRs from Bangladesh) |
| Agata et al. [40], 2022 | The CNN-based deep learning method is used to classify between COVID-19 and pneumonia. Furthermore, examined some preprocessing strategies such as blurring, thresholding, and histogram equalization. | Classification | Radiology | Pneumonia and COVID-19 | Chest X-ray | Pooled data from various cohorts (6939 CXRs) |
| Gouda et al. [41], 2022 | The ResNet-50-based two different deep learning approaches have been proposed to detect COVID-19. | Detection | Radiology | COVID-19 | Chest X-ray | Pooled data from various cohorts (2790 CXRs) |
| Li et al. [33], 2022 | The U-Net and ResNet based model was proposed to segment, classify and predict pulmonary fibrosis from CXRs. | Segmentation, Classification and Prediction | Radiology | Pulmonary Fibrosis | Chest X-ray | NIH Chest X-ray14 (Pulmonary fibrosis CXRs from 112,120 images) |
| Type | Filter Shape | Stride | Input Size | Output Size | |
|---|---|---|---|---|---|
| Dilated Convolution ( = 1) | 3 × 3 × 1 | 1 | 150 × 150 × 3 | 150 × 150 × 1 | |
| Dilated Convolution ( = 2) | 3 × 3 × 1 | 1 | 150 × 150 × 3 | 150 × 150 × 1 | |
| Dilated Convolution ( = 3) | 3 × 3 × 1 | 1 | 150 × 150 × 3 | 150 × 150 × 1 | |
| Concatenation (Merge Layer) | - | - |
150 × 150 × 1 ( = 1) 150 × 150 × 1 ( = 2) 150 × 150 × 1 ( = 3) | 150 × 150 × 3 | |
| Convolution | 3 × 3 × 32 | 2 | 150 × 150 × 3 | 75 × 75 × 32 | |
| Depthwise Convolution | 3 × 3 × 32 | 1 | 75 × 75 × 32 | 75 × 75 × 32 | |
| Seperable Convolution | 1 × 1 × 64 | 1 | 75 × 75 × 32 | 75 × 75 × 64 | |
| Zero Padding | - | - | 75 × 75 × 64 | 76 × 76 × 64 | |
| Depthwise Convolution | 3 × 3 × 64 | 2 | 76 × 76 × 64 | 37 × 37 × 64 | |
| Seperable Convolution | 1 × 1 × 128 | 1 | 37 × 37 × 64 | 37 × 37 × 128 | |
| Depthwise Convolution | 3 × 3 × 128 | 1 | 37 × 37 × 128 | 37 × 37 × 128 | |
| Seperable Convolution | 1 × 1 × 128 | 1 | 37 × 37 × 128 | 37 × 37 × 128 | |
| Zero Padding | - | - | 37 × 37 × 128 | 38 × 38 × 128 | |
| Depthwise Convolution | 3 × 3 × 128 | 2 | 38 × 38 × 128 | 18 × 18 × 128 | |
| Seperable Convolution | 1 × 1 × 256 | 1 | 18 × 18 × 128 | 18 × 18 × 256 | |
| Depthwise Convolution | 3 × 3 × 256 | 1 | 18 × 18 × 256 | 18 × 18 × 256 | |
| Seperable Convolution | 1 × 1 × 256 | 1 | 18 × 18 × 256 | 18 × 18 × 256 | |
| Zero Padding | - | - | 18 × 18 × 256 | 19 × 19 × 256 | |
| Depthwise Convolution | 3 × 3 × 256 | 2 | 19 × 19 × 256 | 9 × 9 × 256 | |
| Seperable Convolution | 1 × 1 × 512 | 1 | 9 × 9 × 256 | 9 × 9 × 512 | |
| 5 × |
Depthwise Convolution Seperable Convolution |
3 × 3 × 512 1 × 1 × 512 |
1 1 |
9 × 9 × 512 9 × 9 × 512 |
9 × 9 × 512 9 × 9 × 512 |
| Zero Padding | - | - | 9 × 9 × 512 | 10 × 10 × 512 | |
| Depthwise Convolution | 3 × 3 × 512 | 2 | 10 × 10 × 512 | 4 × 4 × 512 | |
| Seperable Convolution | 1 × 1 × 1024 | 1 | 4 × 4 × 512 | 4 × 4 × 1024 | |
| Depthwise Convolution | 3 × 3 × 1024 | 2 | 4 × 4 × 1024 | 4 × 4 × 1024 | |
| Seperable Convolution | 1 × 1 × 1024 | 1 | 4 × 4 × 1024 | 4 × 4 × 1024 | |
| Global Average Pooling | Pool 4 × 4 | 1 | 4 × 4 × 1024 | 1 × 1 × 1024 | |
| Dataset Description | Open-I Cohort | KMC Cohort |
|---|---|---|
| Tot. # of CXR images | 3996 | 502 |
| Tot. # of CXR images after removal of missing reports | 3638 | 502 |
| Tot. # of CXR after standard data augmentation | 6229 | 1498 |
| Tot. # of Training/Validation Set | 5606 | 1348 |
| Tot. # of Test Set | 623 | 150 |
| Tot. % of Normal cases (i.e., No Pulmonary diseases) | 38% | 52% |
| Tot. % of Abnormal cases (i.e., Pulmonary diseases) | 62% | 48% |
| Augmentation Strategies | Value |
|---|---|
| Rotation range | [−5, +5] |
| Zoom range | 0.95 |
| Shear range | [−5, +5] |
| Brightness range | [0.5, 1.5] |
| Models | Total Parameters (in Millions) |
|---|---|
| MobileNet | 3.2289 |
| VGG16 | 14.7147 |
| EfficientNetB1 | 6.5752 |
| VGG19 | 20.0244 |
| ResNet50 | 23.5877 |
| Xception | 20.8615 |
| InceptionV3 | 21.8028 |
| DenseNet121 | 25.1283 |
| Proposed MS-CheXNet | 4.8105 |
| Models | Accuracy | Precision | Recall | F1-Score | MCC | AUROC |
|---|---|---|---|---|---|---|
| MobileNet | 0.7675 | 0.7670 | 0.767 | 0.7668 | 0.5339 | 0.8108 |
| VGG16 | 0.6357 | 0.6361 | 0.64 | 0.64 | 0.5605 | 0.8418 |
| EfficientNetB1 | 0.7805 | 0.7803 | 0.7801 | 0.7802 | 0.5605 | 0.8418 |
| VGG19 | 0.6357 | 0.6361 | 0.64 | 0.647 | 0.2722 | 0.6357 |
| ResNet50 | 0.7465 | 0.7436 | 0.745 | 0.746 | 0.492 | 0.7901 |
| Xception | 0.77 | 0.776 | 0.77 | 0.76 | 0.573 | 0.8109 |
| InceptionV3 | 0.7473 | 0.7471 | 0.748 | 0.746 | 0.4993 | 0.8004 |
| DenseNet121 | 07336 | 0.74 | 0.7354 | 0.7346 | 0.4688 | 0.8003 |
| Proposed MS-CheXNet | 0.7922 | 0.7926 | 0.7928 | 0.7927 | 0.5855 | 0.8572 |
| Models | Accuracy | Precision | Recall | F1-Score | MCC | AUROC |
|---|---|---|---|---|---|---|
| MobileNet | 0.7804 | 0.7801 | 0.7801 | 0.7803 | 0.5604 | 0.8228 |
| VGG16 | 0.6623 | 0.6621 | 0.6623 | 0.6622 | 0.5731 | 0.8314 |
| EfficientNet | 0.7945 | 0.7943 | 0.7942 | 0.7941 | 0.5858 | 0.8330 |
| VGG19 | 0.6642 | 0.6641 | 0.6641 | 0.6653 | 0.3822 | 0.6642 |
| ResNet50 | 0.7657 | 0.7656 | 0.7656 | 0.7654 | 0.5102 | 0.8012 |
| Xception | 0.7821 | 0.7823 | 0.7822 | 0.7821 | 0.5168 | 0.8351 |
| InceptionV3 | 0.7741 | 0.7743 | 0.7743 | 0.7741 | 0.4963 | 0.8103 |
| DenseNet121 | 0.7511 | 0.7513 | 0.7513 | 0.7511 | 0.4826 | 0.8099 |
| Proposed MS-CheXNet | 0.8225 | 0.8201 | 0.8200 | 0.8200 | 0.6401 | 0.8793 |
| Reference | Accuracy | Precision | Recall | F1-Score | MCC | AUROC |
|---|---|---|---|---|---|---|
| John et al. [24] (2018) | - | - | - | - | - | 0.725 |
| Faik et al. [34] (2019) | 0.74 | - | - | - | - | - |
| Wang et al. [36] (2019) | - | - | - | - | - | 0.741 |
| Lopez et al. [35] (2020) | - | 0.52 | 0.42 | 0.46 | - | 0.61 |
| Proposed MS-CheXNet | 0.7922 | 0.7926 | 0.7928 | 0.7927 | 0.5855 | 0.8572 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shetty, S.; S., A.V.; Mahale, A. MS-CheXNet: An Explainable and Lightweight Multi-Scale Dilated Network with Depthwise Separable Convolution for Prediction of Pulmonary Abnormalities in Chest Radiographs. Mathematics 2022, 10, 3646. https://doi.org/10.3390/math10193646
Shetty S, S. AV, Mahale A. MS-CheXNet: An Explainable and Lightweight Multi-Scale Dilated Network with Depthwise Separable Convolution for Prediction of Pulmonary Abnormalities in Chest Radiographs. Mathematics. 2022; 10(19):3646. https://doi.org/10.3390/math10193646
Chicago/Turabian StyleShetty, Shashank, Ananthanarayana V S., and Ajit Mahale. 2022. "MS-CheXNet: An Explainable and Lightweight Multi-Scale Dilated Network with Depthwise Separable Convolution for Prediction of Pulmonary Abnormalities in Chest Radiographs" Mathematics 10, no. 19: 3646. https://doi.org/10.3390/math10193646








