TwoViewDensityNet: Two-View Mammographic Breast Density Classification Based on Deep Convolutional Neural Network
Abstract
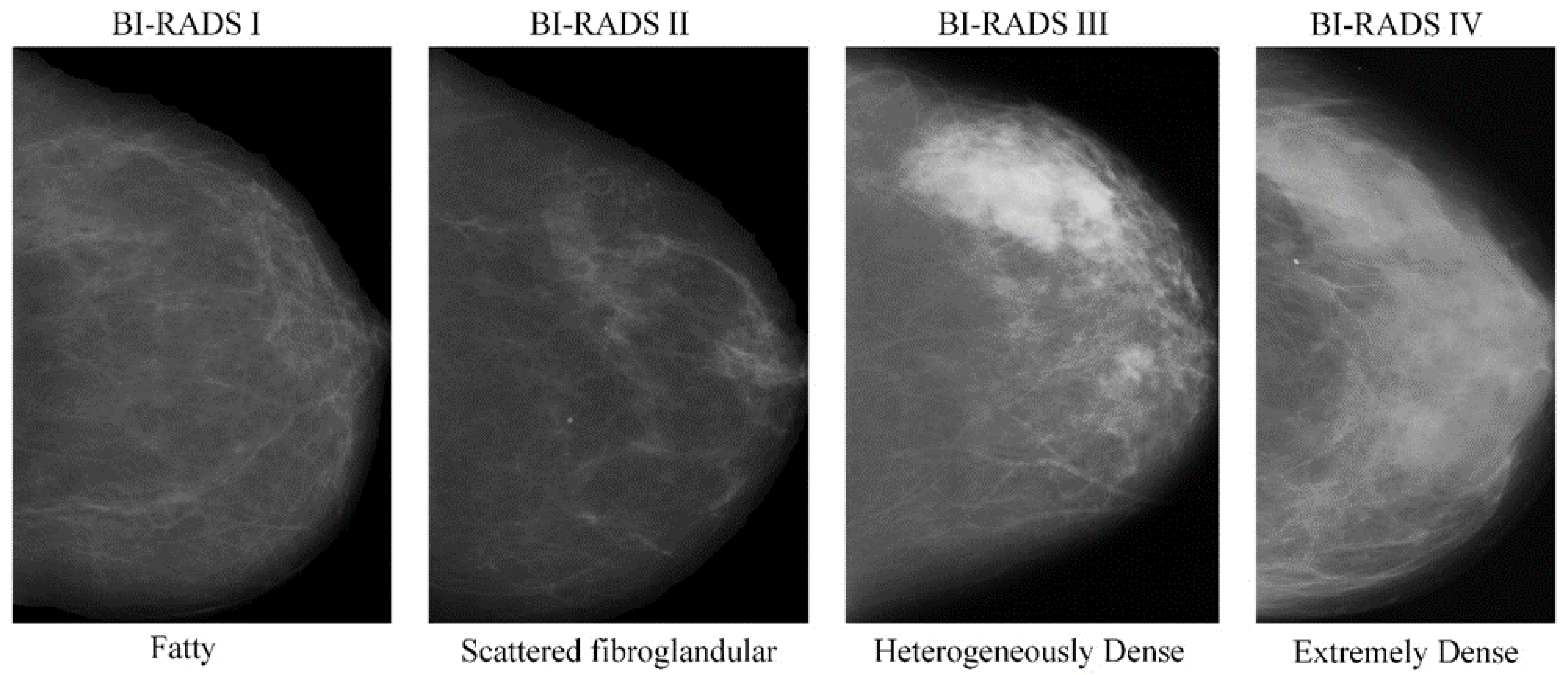
:1. Introduction
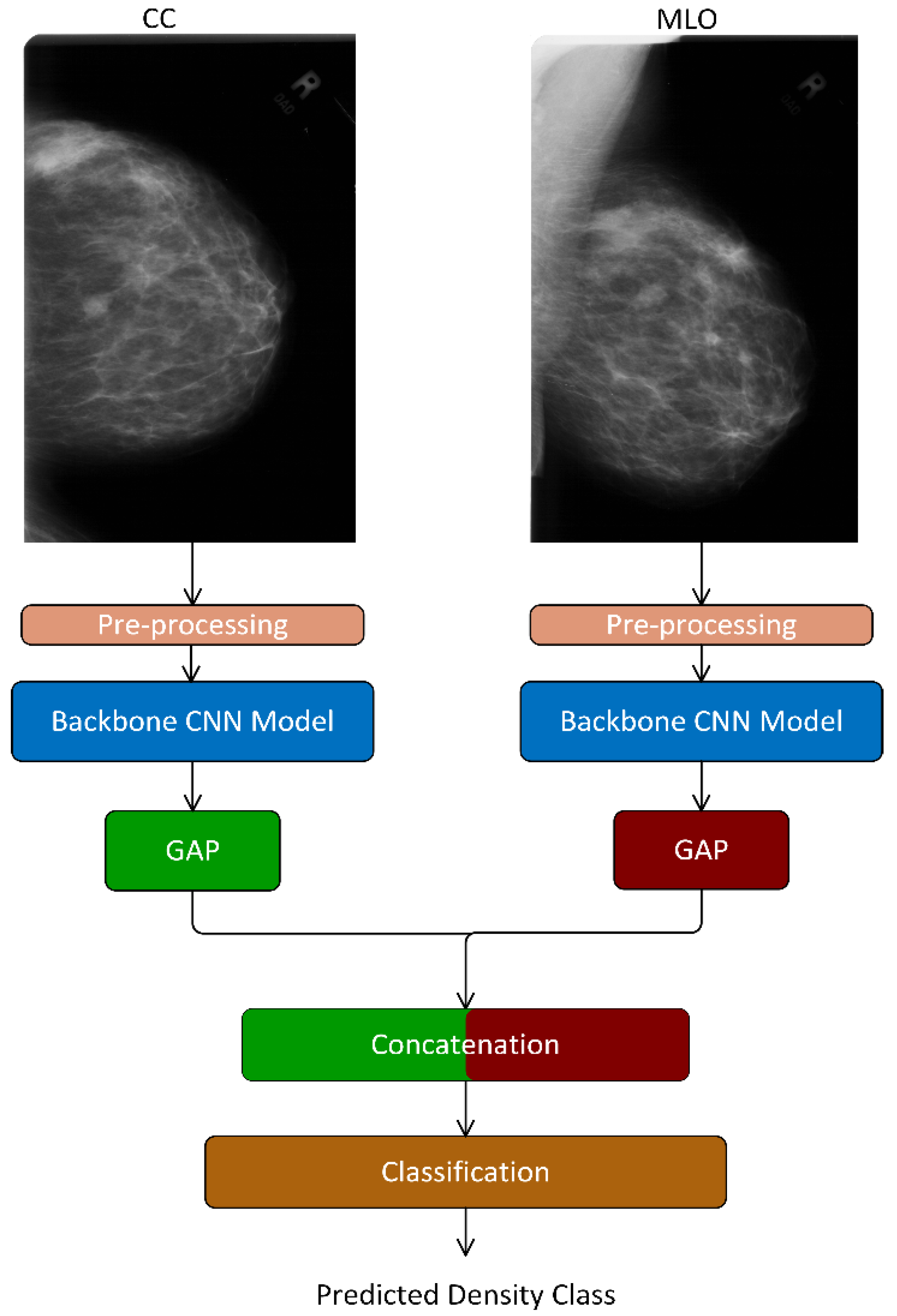
- We proposed an end-to-end deep learning-based model—TwoViewDensityNet—for the classification of breast density using dual mammogram views, i.e., craniocaudal (CC) view and mediolateral oblique (MLO) view. It combines the CC and MLO views by leveraging the relationship between views and using a CNN as the backbone model. First, it extracts the complementary information from each view using a CNN model, fuses them using a concatenation layer, and finally, predicts the density class using an FC layer with SoftMax activation.
- We evaluated different preprocessing techniques to enhance the mammogram image before feeding it to the CNN model and found the one that is best suited for the proposed model.
- We employed different loss functions and their valuable characteristics to tackle the class-imbalance problem.
2. Related Work
3. Proposed Method
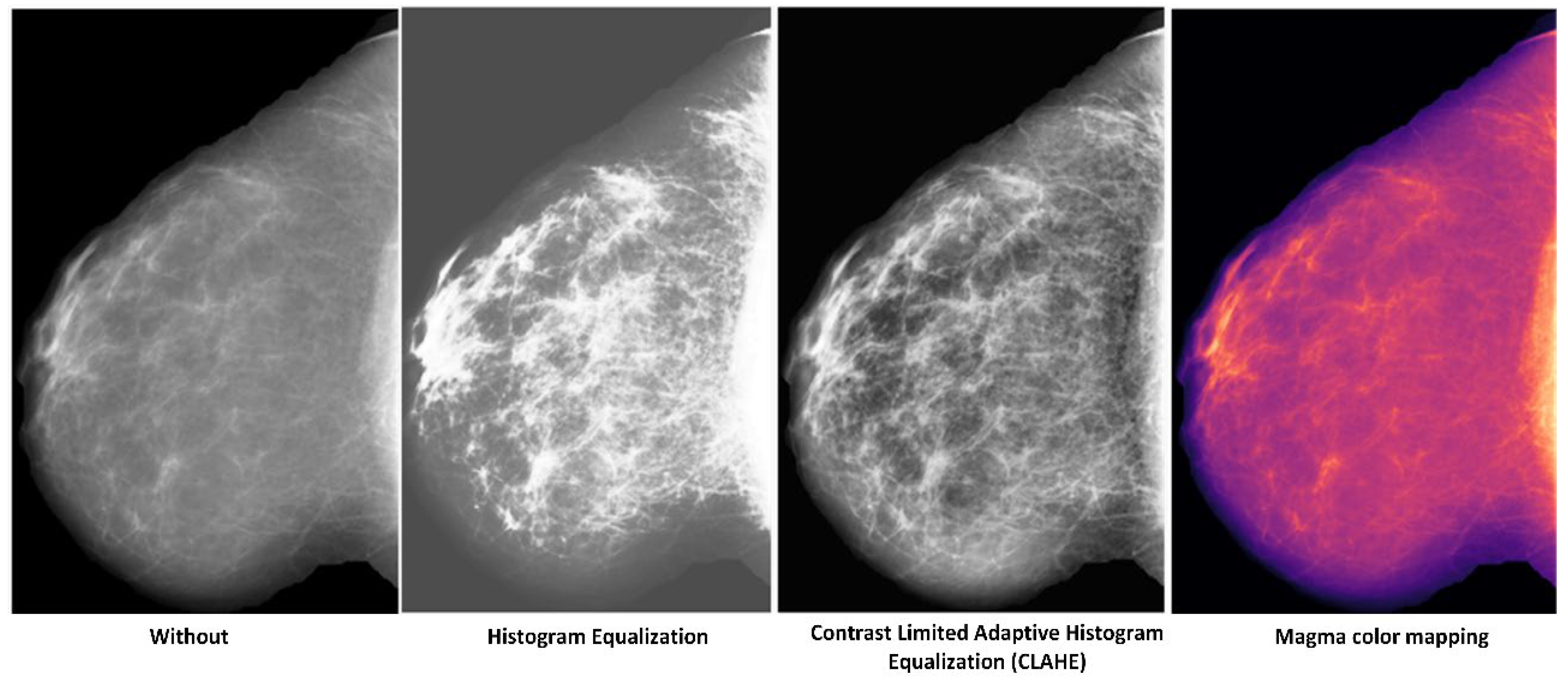
3.1. Preprocessing
3.2. Backbone Convolutional Neural Network (CNN) Model
3.3. Concatenation Layer
3.4. Classification Layer
3.5. Training the TwoViewDensityNet
3.5.1. Loss Functions
3.5.2. Algorithms Used for Training
3.5.3. Datasets
3.5.4. Data Augmentation
4. Evaluation Protocol
5. Experimental Results and Discussion
5.1. Ablation Study
5.1.1. Which Backbone Model?
5.1.2. Which Preprocessing Operation?
5.1.3. Single View or Dual View?
5.1.4. Which Loss Function?
5.2. Comparison with State-of-the-Art Methods for Breast Density Classification
5.3. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wolfe, J.N. Risk for breast cancer development determined by mammographic parenchymal pattern. Cancer 1976, 37, 2486–2492. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, J.N. Breast patterns as an index of risk for developing breast cancer. Am. J. Roentgenol. 1976, 126, 1130–1137. [Google Scholar] [CrossRef] [PubMed]
- McCormack, V.A.; dos Santos Silva, I. Breast Density and Parenchymal Patterns as Markers of Breast Cancer Risk: A Meta-analysis. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazari, S.S.; Mukherjee, P. An overview of mammographic density and its association with breast cancer. Breast Cancer 2018, 25, 259–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albeshan, S.M.; Hossain, S.Z.; Mackey, M.G.; Peat, J.K.; Al Tahan, F.M.; Brennan, P.C. Preliminary investigation of mammographic density among women in Riyadh: Association with breast cancer risk factors and implications for screening practices. Clin. Imaging 2019, 54, 138–147. [Google Scholar] [CrossRef]
- American College of Radiology (ACR). Illustrated Breast Imaging Reporting and Data System (BI-RADS); American College of Radiology: Reston, VA, USA, 2003. [Google Scholar]
- Li, C.; Xu, J.; Liu, Q.; Zhou, Y.; Mou, L.; Pu, Z.; Xia, Y.; Zheng, H.; Wang, S. Multi-View Mammographic Density Classification by Dilated and Attention-Guided Residual Learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 1003–1013. [Google Scholar] [CrossRef]
- Yi, P.H.; Lin, A.; Wei, J.; Yu, A.C.; Sair, H.I.; Hui, F.K.; Hager, G.D.; Harvey, S.C. Deep-Learning-Based Semantic Labeling for 2D Mammography and Comparison of Complexity for Machine Learning Tasks. J. Digit. Imaging 2019, 32, 565–570. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Geras, K.J.; Shen, Y.; Su, J.; Kim, S.G.; Kim, E.; Wolfson, S.; Moy, L.; Cho, K. Breast Density Classification with Deep Convolutional Neural Networks. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 6682–6686. [Google Scholar]
- Lehman, C.D.; Yala, A.; Schuster, T.; Dontchos, B.; Bahl, M.; Swanson, K.; Barzilay, R. Mammographic Breast Density Assessment Using Deep Learning: Clinical Implementation. Radiology 2019, 290, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Heath, M.; Bowyer, K.; Kopans, D.; Kegelmeyer, P.; Moore, R.; Chang, K.; Munishkumaran, S. Current Status of the Digital Database for Screening Mammography. In Digital Mammography; Springer: Dordrecht, The Netherlands, 1998. [Google Scholar]
- Zhao, W.; Wang, R.; Qi, Y.; Lou, M.; Wang, Y.; Yang, Y.; Deng, X.; Ma, Y. BASCNet: Bilateral adaptive spatial and channel attention network for breast density classification in the mammogram. Biomed. Signal Process. Control. 2021, 70, 103073. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Suleiman, M.E.; Demchig, D.; Brennan, P.C.; McEntee, M.F. BI-RADS Density Categorization Using Deep Neural Networks. In Medical Imaging 2019: Image Perception, Observer Performance, and Technology Assessment; SPIE: Bellingham, WA, USA, 2019. [Google Scholar]
- Deng, J.; Ma, Y.; Li, D.A.; Zhao, J.; Liu, Y.; Zhang, H. Classification of breast density categories based on SE-Attention neural networks. Comput. Methods Programs Biomed. 2020, 193, 105489. [Google Scholar] [CrossRef]
- Mohamed, A.A.; Berg, W.A.; Peng, H.; Luo, Y.; Jankowitz, R.C.; Wu, S. A deep learning method for classifying mammographic breast density categories. Med. Phys. 2017, 45, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Cogan, T.; Tamil, L.S. Deep Understanding of Breast Density Classification. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1140–1143. [Google Scholar]
- Jouirou, A.; Baâzaoui, A.; Barhoumi, W. Multi-view information fusion in mammograms: A comprehensive overview. Inf. Fusion 2019, 52, 308–321. [Google Scholar] [CrossRef]
- Wilms, M.; Krüger, J.; Marx, M.; Ehrhardt, J.; Bischof, A.; Handels, H. Estimation of Corresponding Locations in Ipsilateral Mammograms: A Comparison of Different Methods. In Medical Imaging 2015: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2015. [Google Scholar]
- Ma, Y.; Peng, Y. Simultaneous detection and diagnosis of mammogram mass using bilateral analysis and soft label based metric learning. Biocybern. Biomed. Eng. 2022, 42, 215–232. [Google Scholar] [CrossRef]
- Xian, J.; Wang, Z.; Cheng, K.-T.; Yang, X. Towards Robust Dual-View Transformation via Densifying Sparse Supervision for Mammography Lesion Matching. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September 2021. [Google Scholar]
- Kowsalya, S.; Priyaa, D.S. An Adaptive Behavioral Learning Technique based Bilateral Asymmetry Detection in Mammogram Images. Indian J. Sci. Technol. 2016, 9, S1. [Google Scholar] [CrossRef]
- Lyu, Q.; Namjoshi, S.V.; McTyre, E.; Topaloglu, U.; Barcus, R.; Chan, M.D.; Cramer, C.K.; Debinski, W.; Gurcan, M.N.; Lesser, G.J.; et al. A transformer-based deep learning approach for classifying brain metastases into primary organ sites using clinical whole brain MRI images. Patterns 2022, 3, 100613. [Google Scholar] [CrossRef]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. Fully Automated Classification of Mammograms Using Deep Residual Neural Networks. In Proceedings of the IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 310–314. [Google Scholar]
- Yi, D.; Sawyer, R.L.; Cohn, D.; Dunnmon, J.A.; Lam, C.K.; Xiao, X.; Rubin, D. Optimizing and Visualizing Deep Learning for Benign/Malignant Classification in Breast Tumors. arXiv 2017, arXiv:1705.06362. [Google Scholar]
- Cogan, T.; Cogan, M.; Tamil, L.S. RAMS: Remote and automatic mammogram screening. Comput. Biol. Med. 2019, 107, 18–29. [Google Scholar] [CrossRef]
- MatPlotLib Perceptually Uniform Colormaps. Available online: https://www.mathworks.com/matlabcentral/fileexchange/62729-matplotlibperceptually-uniform-colormaps (accessed on 25 November 2021).
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- KHe, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Veit, A.; Wilber, M.J.; Belongie, S. Residual Networks Behave Like Ensembles of Relatively Shallow Networks. In Proceedings of the Advances in Neural Information Processing Systems 29, Barcelona, Spain, 5–10 December 2016; pp. 550–558. [Google Scholar]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Glorot, X.; Bengio, Y. Understanding the Difficulty of Training Deep Feedforward Neural Networks. In Proceedings of the 13th International Conference on Artificial Intelligence and Statistics (AISTATS), Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- De Boer, P.T.; Kroese, D.P.; Mannor, S.; Rubinstein, R.Y. A Tutorial on the Cross-Entropy Method. Ann. Oper. Res. 2005, 134, 19–67. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE Transactions on Pattern Analysis and Machine Intelligence, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Bosman, A.S.; Engelbrecht, A.; Helbig, M. Visualising basins of attraction for the cross-entropy and the squared error neural network loss functions. Neurocomputing 2020, 400, 113–136. [Google Scholar] [CrossRef] [Green Version]
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. Inbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, E.; Fleisher, M. Accelerated learning in layered neural networks. Complex Syst. 1988, 2, 625–640. [Google Scholar]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. 1974, 36, 111–147. [Google Scholar] [CrossRef]
- Cetin, K.; Oktay, Y.; Sinan, A. Performance Analysis of Machine Learning Techniques in Intrusion Detection. In Proceedings of the 24th Signal Processing and Communication Application Conference (SIU), Zonguldak, Turkey, 16–19 May 2016; pp. 1473–1476. [Google Scholar]
- Ranganathan, P.; Pramesh, C.S.; Aggarwal, R. Common pitfalls in statistical analysis: Measures of agreement. Perspect. Clin. Res. 2017, 8, 187–191. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Hoo, Z.H.; Candlish, J.; Teare, D. What is an ROC curve? Emerg. Med. J. 2017, 34, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Sprague, B.L.; Conant, E.F.; Onega, T.; Garcia, M.P.; Beaber, E.F.; Herschorn, S.D.; Lehman, C.D.; Tosteson, A.N.; Lacson, R.; Schnall, M.D.; et al. Variation in Mammographic Breast Density Assessments Among Radiologists in Clinical Practice: A Multicenter Observational Study. Ann. Intern. Med. 2016, 165, 457–464. [Google Scholar] [CrossRef]




| References | Model | Dataset | ACC (%) | AUC (%) | F1-Score (%) |
|---|---|---|---|---|---|
| Single View | |||||
| Li et al. [7] (2021) | ResNet50 + DC + CA (DC: dilated convolutions. CA:channel-wise attention) | Private | 88.70 | 97.40 | 87.10 |
| INbreast | 70 | 84.70 | 63.50 | ||
| Jian et al. [14] (2020) | Inception-V4-SE- Attention | Private | 92.17 | - | - |
| ResNeXt-SE-Attention and | Private | 89.97 | |||
| DenseNet-SE-Attention | Private | 89.64 | |||
| Yi et al. [8] (2019) | ResNet-50 | DDSM | 68 | - | - |
| Lehman et al. [10] (2019) | ResNet-18 | Private | 77 | - | - |
| Gandomkar et al. [13] (2019) | Inception-V3 | Private | 83.33 | - | 77.50 |
| Mohamed et al. [15] (2018) | AlexNet | Private | - | 92 | - |
| Multi-View | |||||
| Zhao et al. [12] (2021) | BASCNet (ResNet) (Bilateral-view adaptive spatial and channel attention network) | DDSM | 85.10 | 91.54 | 78.92 |
| INbreast | 90.51 | 99.09 | 78.11 | ||
| Li et al. [7] (2021) | ResNet50 + DC + CA (DC: dilated convolutions. CA: channel-wise attention) | Private | 92.10 | 98.1 | 91.2 |
| 92.50 | 98.2 | 91.7 | |||
| 75.20 | 93.6 | 67.9 | |||
| Timothy and Lakshman [16] (2020) | DualViewNet | CBISDDSM | - | 89.70 | - |
| Wu et al. [9] (2018) | VGG Net | Private | 69.40 | 84.20 | - |
| Confusion Matrix | Actual Positive | Actual Negative |
|---|---|---|
| Predicted Positive | TP 1 | FP 3 |
| Predicted Negative | FN 4 | TN 2 |
| Model | Overall Classification Accuracy (OCA %) |
|---|---|
| ResNet 50 [28] | 74.94 |
| DenseNet201 [30] | 69.58 |
| EfficientNet b0 [29] | 64.06 |
| Model | Preprocessing | (OCA %) |
|---|---|---|
| ResNet50 | Without | 66.83 |
| Contrast-limited adaptive histogram equalization (CLAHE) | 67.41 | |
| Histogram equalization | 65.23 | |
| Magma color mapping | 74.94 |
| Model | Overall Classification Accuracy (OCA %) |
|---|---|
| Single View | |
| ResNet 50 | 74.94 |
| DenseNet201 | 69.58 |
| EfficientNet b0 | 64.06 |
| Dual View | |
| ResNet 50 | 91.36 |
| DenseNet201 | 86.16 |
| EfficientNet b0 | 73.97 |
| Loss Function | OCA | ||||
|---|---|---|---|---|---|
| ResNet-50 CE loss | 91.36 ± 3.29 | 96.28 ± 3.29 | 96.29 ± 20 | 89.44 ± 2.10 | 77.70 ± 16.4 |
| ResNet-50 focal loss | 95.83 ± 3.63 | 94.25 ± 5.72 | 99.14 ± 0.98 | 93.17 ± 6.95 | 93.83 ± 5.86 |
| ResNet-50 SSE loss | 94.01 ± 3.61 | 94.55 ± 5.98 | 98.10 ± 1.17 | 92.67 ± 4.76 | 85.38 ± 9.96 |
| Confusion Matrix | Accuracy (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold | Predicted | ||||||||||
| B-I | B-II | B-III | B-IV | OCA | |||||||
| Fold 1 | Actual | B-I | 63 | 8 | 0 | 0 | 94.03 | 88.73 | 98.10 | 93.17 | 89.61 |
| B-II | 4 | 206 | 0 | 0 | |||||||
| B-III | 1 | 6 | 150 | 4 | |||||||
| B-IV | 0 | 1 | 7 | 69 | |||||||
| Fold 2 | Actual | B-I | 69 | 0 | 0 | 0 | 99.38 | 100 | 100 | 100 | 96.20 |
| B-II | 0 | 210 | 0 | 0 | |||||||
| B-III | 0 | 0 | 161 | 0 | |||||||
| B-IV | 0 | 1 | 2 | 76 | |||||||
| Fold 3 | Actual | B-I | 61 | 8 | 0 | 0 | 91.89 | 88.41 | 98.10 | 88.20 | 85.90 |
| B-II | 4 | 206 | 0 | 0 | |||||||
| B-III | 0 | 0 | 142 | 19 | |||||||
| B-IV | 0 | 1 | 10 | 67 | |||||||
| Fold 4 | Actual | B-I | 69 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | 100 |
| B-II | 0 | 210 | 0 | 0 | |||||||
| B-III | 0 | 0 | 161 | 0 | |||||||
| B-IV | 0 | 0 | 0 | 78 | |||||||
| Fold 5 | Actual | B-I | 66 | 4 | 0 | 0 | 93.83 | 94.29 | 99.52 | 84.47 | 97.44 |
| B-II | 1 | 209 | 0 | 0 | |||||||
| B-III | 0 | 0 | 136 | 25 | |||||||
| B-IV | 0 | 0 | 2 | 76 | |||||||
| References | Model | Dataset | ACC (%) | AUC (%) | F1-score (%) | Kappa (%) |
|---|---|---|---|---|---|---|
| Single View | ||||||
| Li et al. [7], 2021 | ResNet50 + DC + CA (DC: dilated convolutions. CA:channel-wise attention) | INbreast | 70 | 84.70 | 63.50 | - |
| Yi et al. [8], 2019 | ResNet-50 | DDSM | 68 | - | - | - |
| Lehman et al. [10], 2019 | ResNet-18 | INbreast | 63.80 | 81.20 | 48.90 | - |
| Gandomkar et al. [13], 2019 | Inception-V3 | INbreast | 63.90 | 82.10 | 53.10 | - |
| Mohamed et al. [15], 2018 | AlexNet | INbreast | 59.60 | 82 | 35.4 | - |
| Multi-View | ||||||
| Zhao et al. [12], 2021 | BASCNet (ResNet) (Bilateral-view adaptive spatial and channel attention network) | DDSM | 85.10 | 91.54 | 78.92 | - |
| INbreast | 90.51 | 99.09 | 78.11 | - | ||
| Proposed system | TwoViewDensityNet | DDSM | 95.83 | 99.51 | 98.63 | 94.37 |
| INbreast | 96 | 97.44 | 97.14 | 94.31 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busaleh, M.; Hussain, M.; Aboalsamh, H.A.; Fazal-e-Amin; Al Sultan, S.A. TwoViewDensityNet: Two-View Mammographic Breast Density Classification Based on Deep Convolutional Neural Network. Mathematics 2022, 10, 4610. https://doi.org/10.3390/math10234610
Busaleh M, Hussain M, Aboalsamh HA, Fazal-e-Amin, Al Sultan SA. TwoViewDensityNet: Two-View Mammographic Breast Density Classification Based on Deep Convolutional Neural Network. Mathematics. 2022; 10(23):4610. https://doi.org/10.3390/math10234610
Chicago/Turabian StyleBusaleh, Mariam, Muhammad Hussain, Hatim A. Aboalsamh, Fazal-e-Amin, and Sarah A. Al Sultan. 2022. "TwoViewDensityNet: Two-View Mammographic Breast Density Classification Based on Deep Convolutional Neural Network" Mathematics 10, no. 23: 4610. https://doi.org/10.3390/math10234610
APA StyleBusaleh, M., Hussain, M., Aboalsamh, H. A., Fazal-e-Amin, & Al Sultan, S. A. (2022). TwoViewDensityNet: Two-View Mammographic Breast Density Classification Based on Deep Convolutional Neural Network. Mathematics, 10(23), 4610. https://doi.org/10.3390/math10234610







