A Survey on Tools and Techniques for Localizing Abnormalities in X-ray Images Using Deep Learning
Abstract
:1. Introduction
2. Background
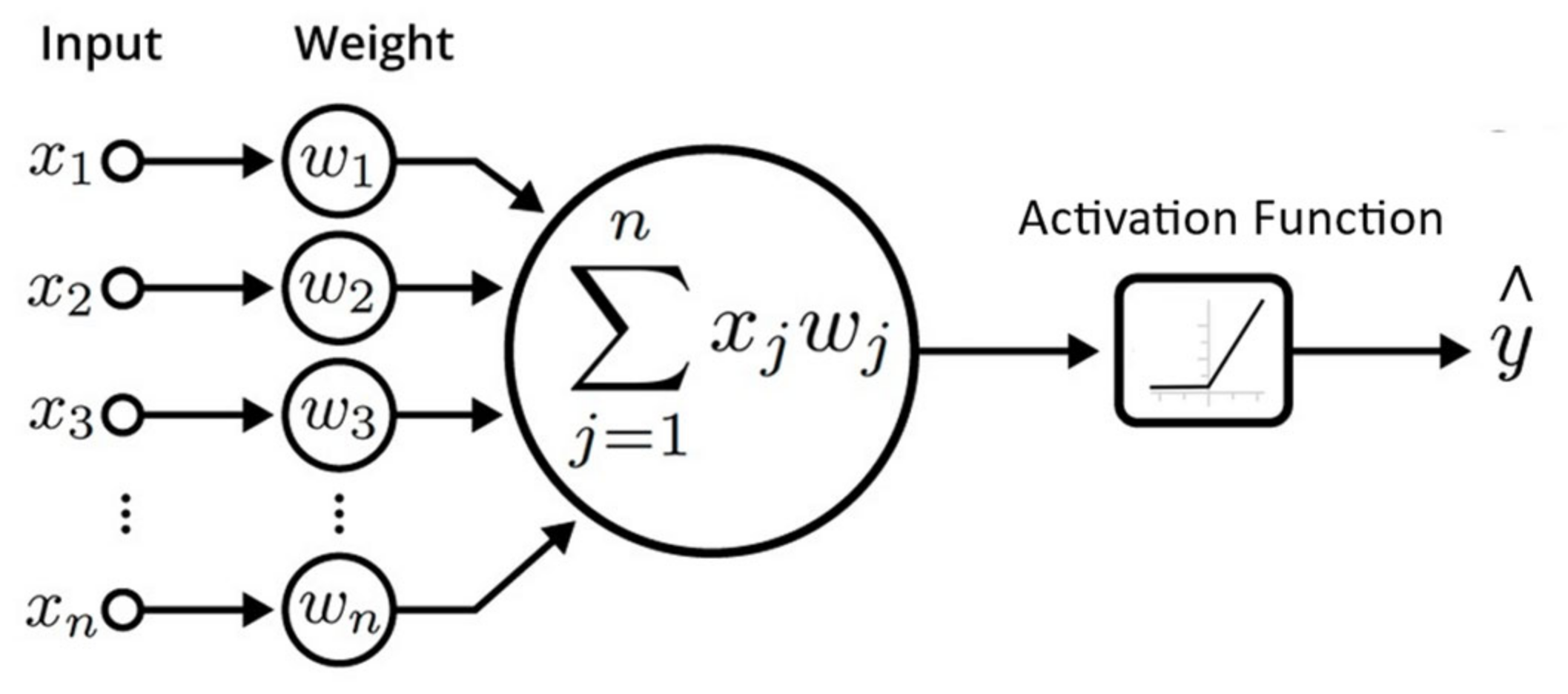
2.1. Artificial Neural Network
2.2. Multilayer Perceptron
2.3. Convolutional Neural Network
3. Method for Performance Analysis
3.1. Accuracy
- True Positive: output that correctly indicates the presence of a condition.
- True Negative: output that correctly indicates the absence of a condition.
- False Positive: output that wrongly indicates the presence of a condition.
- False Negative: output that wrongly indicates the absence of a condition.
3.2. Precision
3.3. Sensitivity
3.4. Specificity
3.5. Jaccard Index
 | (5) |
3.6. Evaluation Matrix for Medical Diagnosis
4. Chest X-ray Datasets
4.1. ChestXray8
4.2. CheXpert
4.3. CheXpert
4.4. VinDr-CXR
4.5. Montgomery County and Shenzhen Set
4.6. JSRT Database
4.7. JSRT Database
5. Diagnosis Using Chest Radiographs
5.1. Classification and Localization Using Supervised Learning
5.2. R-CNN
5.3. SPP-Net
5.4. Fast R-CNN
| S.No | Ref No | Methodology | Dataset |
|---|---|---|---|
| 1 | [72] | Using lung cropped CXR model with a CXR model to improve model performance | ChestX-ray14 JSRT + SCR, |
| 2 | [73] | Use of image-level prediction of Cardiomegaly and application for segmentation models | ChestX-ray14 |
| 3 | [74] | Classification of cardiomegaly using a network with DenseNet and U-Net | ChestX-ray14 |
| 4 | [75] | Employing lung cropped CXR model with CXR model using the segmentation quality | MIMIC-CXR |
| 5 | [76] | Improving Pneumonia detection by using of lung segmentation | Pneumonia |
| 6 | [77] | Segmentation of pneumonia using U-Net based model | RSNA-Pneumonia |
| 7 | [78] | To find similar studies, a database has been used for the intermediate ResNet-50 features | Montgomery, Shenzen |
| 8 | [79] | Detection and localization of COVID-19 through various networks and ensembling | COVID |
| 9 | [80] | GoogleNet has been trained with CXR patches and correlates with COVID-19 severity score | ChestX-ray14 |
| 10 | [81] | A segmentation and classification model proposed to compare with radiologist cohort | Private |
| 11 | [82] | A CNN model proposed for identification of abnormal CXRs and localization of abnormalities | Private |
| 12 | [83] | Localizing COVID-19 opacity and severity detection on CXRs | Private |
| 13 | [84] | Use of Lung cropped CXR in DenseNet for cardiomegaly detection | Open-I, PadChest |
| 14 | [85] | Applied multiple models and combinations of CXR datasets to detect COVID-19 | ChestX-ray14 JSRT + SCR, COVID-CXR |
| 15 | [86] | Multiple architectures evaluated for two-stage classification of pneumonia | Ped-pneumonia |
| 16 | [87] | Inception-v3 based pneumoconiosis detection and evaluation against two radiologists | Private |
| 17 | [88] | VGG-16 architecture adapted for classification of pediatric pneumonia types | Ped-pneumonia |
| 18 | [89] | Used ResNet-50 as backbone for segmentation model to detect healthy, pneumonia, and COVID-19 | COVID-CXR |
| 19 | [90] | CNN employed to detect the presence of subphrenic free air from CXR | Private |
| 20 | [91] | Binary classification vs One-class identification of viral pneumonia cases | Private |
| 21 | [92] | Applied a weighting scheme to improve abnormality for classification | ChestX-ray14 |
| 22 | [93] | To improve image-level classification, a Lesion detection network has been employed | Private |
| 23 | [94] | An ensemble scheme has been used for DenseNet-121 networks for COVID-19 classification | ChestX-ray14 |
5.5. Faster R-CNN
5.6. YOLO
5.7. SSD
6. Localization Using Weak-Supervised Learning
6.1. Class Activation Map (CAM) Based Localization
6.1.1. CAM (Vanilla Version)
6.1.2. Grad-CAM
6.1.3. Grad-CAM++
6.1.4. Score-CAM
6.1.5. Layer-CAM
6.1.6. Eigen-CAM
6.1.7. XGrad-CAM
6.1.8. Other Variants
6.2. Attention Models
6.2.1. Soft Attention
6.2.2. Hard Attention
6.3. Saliency Map
7. Challenges and Recommendations
7.1. Disclosure of Training Data
7.2. Source Code Sharing
7.3. Diversity in Data
7.4. Domain Adaption
7.5. Interpretability
7.6. Deriving Bounding Boxes and Segmentation Contour from Heatmaps
7.7. Comparative Analysis with Strong Annotation
7.8. Infer Diagnosis from Classification and Localization
7.9. Emerging Techniques
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shortliffe, E.H.; Buchanan, B.G. A model of inexact reasoning in medicine. Math. Biosci. 1975, 23, 351–379. [Google Scholar] [CrossRef]
- Miller, R.A.; Pople, H.E.; Myers, J.D. Internist-I, an Experimental Computer-Based Diagnostic Consultant for General Internal Medicine. N. Engl. J. Med. 1982, 307, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasan, M.J.; Uddin, J.; Pinku, S.N. A novel modified SFTA approach for feature extraction. In Proceedings of the 2016 3rd International Conference on Electrical Engineering and Information Communication Technology (ICEEICT), Dhaka, Bangladesh, 22–24 September 2016; pp. 1–5. [Google Scholar]
- Chan, H.; Hadjiiski, L.M.; Samala, R.K. Computer-aided diagnosis in the era of deep learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef]
- Çallı, E.; Sogancioglu, E.; van Ginneken, B.; van Leeuwen, K.G.; Murphy, K. Deep learning for chest X-ray analysis: A survey. Med. Image Anal. 2021, 72, 102125. [Google Scholar] [CrossRef]
- Wu, J.; Gur, Y.; Karargyris, A.; Syed, A.B.; Boyko, O.; Moradi, M.; Syeda-Mahmood, T. Automatic Bounding Box Annotation of Chest X-ray Data for Localization of Abnormalities. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Iowa City, IA, USA, 2020; pp. 799–803. [Google Scholar]
- Munawar, F.; Azmat, S.; Iqbal, T.; Gronlund, C.; Ali, H. Segmentation of Lungs in Chest X-ray Image Using Generative Adversarial Networks. IEEE Access 2020, 8, 153535–153545. [Google Scholar] [CrossRef]
- Ma, Y.; Niu, B.; Qi, Y. Survey of image classification algorithms based on deep learning. In Proceedings of the 2nd International Conference on Computer Vision, Image, and Deep Learning; Cen, F., bin Ahmad, B.H., Eds.; SPIE: Liuzhou, China, 2021; p. 9. [Google Scholar]
- Agrawal, T.; Choudhary, P. Segmentation and classification on chest radiography: A systematic survey. Vis. Comput. 2022, Online ahead of print. [Google Scholar] [CrossRef]
- Amarasinghe, K.; Rodolfa, K.; Lamba, H.; Ghani, R. Explainable Machine Learning for Public Policy: Use Cases, Gaps, and Research Directions. arXiv 2020, arXiv:2010.14374. [Google Scholar]
- Bhatt, D.; Patel, C.; Talsania, H.; Patel, J.; Vaghela, R.; Pandya, S.; Modi, K.; Ghayvat, H. CNN Variants for Computer Vision: History, Architecture, Application, Challenges and Future Scope. Electronics 2021, 10, 2470. [Google Scholar] [CrossRef]
- Shrestha, A.; Mahmood, A. Review of Deep Learning Algorithms and Architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Chen, C.; Wang, B.; Lu, C.X.; Trigoni, N.; Markham, A. A Survey on Deep Learning for Localization and Mapping: Towards the Age of Spatial Machine Intelligence. arXiv 2020, arXiv:2006.12567. [Google Scholar]
- Yang, R.; Yu, Y. Artificial Convolutional Neural Network in Object Detection and Semantic Segmentation for Medical Imaging Analysis. Front. Oncol. 2021, 11, 638182. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Niu, J.; Liu, X.; Chen, Z.; Tang, S. A Survey on Domain Knowledge Powered Deep Learning for Medical Image Analysis. arXiv 2004, arXiv:2004.12150. [Google Scholar]
- Maguolo, G.; Nanni, L. A Critic Evaluation of Methods for COVID-19 Automatic Detection from X-ray Images. arXiv 2020, arXiv:2004.12823. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, R.; Melekhov, I.; Lesonen, T.; Vaattovaara, E.; Tervonen, O.; Tiulpin, A. Bayesian Feature Pyramid Networks for Automatic Multi-Label Segmentation of Chest X-rays and Assessment of Cardio-Thoratic Ratio. arXiv 2019, arXiv:1908.02924. [Google Scholar]
- Ramos, A.; Alves, V. A Study on CNN Architectures for Chest X-rays Multiclass Computer-Aided Diagnosis. In Trends and Innovations in Information Systems and Technologies; Rocha, Á., Adeli, H., Reis, L.P., Costanzo, S., Orovic, I., Moreira, F., Eds.; Advances in Intelligent Systems and Computing; Springer International Publishing: Cham, Switzerland, 2020; Volume 1161, pp. 441–451. ISBN 978-3-030-45696-2. [Google Scholar]
- Bayer, J.; Münch, D.; Arens, M. A Comparison of Deep Saliency Map Generators on Multispectral Data in Object Detection. arXiv 2021, arXiv:2108.11767. [Google Scholar]
- Zhao, Z.-Q.; Zheng, P.; Xu, S.; Wu, X. Object Detection with Deep Learning: A Review. arXiv 2019, arXiv:1807.05511. [Google Scholar] [CrossRef] [Green Version]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. arXiv 2015, arXiv:151204150. [Google Scholar]
- Chattopadhyay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Improved Visual Explanations for Deep Convolutional Networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar]
- Srinivas, S.; Fleuret, F. Full-Gradient Representation for Neural Network Visualization. arXiv 2019, arXiv:1905.00780. [Google Scholar]
- Desai, S.; Ramaswamy, H.G. Ablation-CAM: Visual Explanations for Deep Convolutional Network via Gradient-free Localization. In Proceedings of the 2020 IEEE Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, CO, USA, 1–5 March 2020; pp. 972–980. [Google Scholar]
- Fu, R.; Hu, Q.; Dong, X.; Guo, Y.; Gao, Y.; Li, B. Axiom-based Grad-CAM: Towards Accurate Visualization and Explanation of CNNs. arXiv 2020, arXiv:2008.02312. [Google Scholar]
- Muhammad, M.B.; Yeasin, M. Eigen-CAM: Class Activation Map using Principal Components. arXiv 2020, arXiv:2008.00299. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, Z.; Du, M.; Yang, F.; Zhang, Z.; Ding, S.; Mardziel, P.; Hu, X. Score-CAM: Score-Weighted Visual Explanations for Convolutional Neural Networks. arXiv 2020, arXiv:1910.01279. [Google Scholar]
- Jiang, P.-T.; Zhang, C.-B.; Hou, Q.; Cheng, M.-M.; Wei, Y. LayerCAM: Exploring Hierarchical Class Activation Maps for Localization. IEEE Trans. Image Process. 2021, 30, 5875–5888. [Google Scholar] [CrossRef]
- Byun, S.-Y.; Lee, W. Recipro-CAM: Gradient-free reciprocal class activation map. arXiv 2022, arXiv:2209.14074. [Google Scholar]
- Englebert, A.; Cornu, O.; De Vleeschouwer, C. Poly-CAM: High resolution class activation map for convolutional neural networks. arXiv 2022, arXiv:2204.13359. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2017, arXiv:1610.02357. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:151203385. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. arXiv 2016, arXiv:1603.05027. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. arXiv 2018, arXiv:1707.07012. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2020, arXiv:1905.11946. [Google Scholar]
- Khan, E.; Rehman, M.Z.U.; Ahmed, F.; Alfouzan, F.A.; Alzahrani, N.M.; Ahmad, J. Chest X-ray Classification for the Detection of COVID-19 Using Deep Learning Techniques. Sensors 2022, 22, 1211. [Google Scholar] [CrossRef]
- Ponomaryov, V.I.; Almaraz-Damian, J.A.; Reyes-Reyes, R.; Cruz-Ramos, C. Chest x-ray classification using transfer learning on multi-GPU. In Proceedings of the Real-Time Image Processing and Deep Learning 2021; Kehtarnavaz, N., Carlsohn, M.F., Eds.; SPIE: Houston, TX, USA, 2021; p. 16. [Google Scholar]
- Tohka, J.; van Gils, M. Evaluation of machine learning algorithms for health and wellness applications: A tutorial. Comput. Biol. Med. 2021, 132, 104324. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Peng, Y.; Lu, L.; Lu, Z.; Bagheri, M.; Summers, R.M. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3462–3471. [Google Scholar]
- Sager, C.; Janiesch, C.; Zschech, P. A survey of image labelling for computer vision applications. J. Bus. Anal. 2021, 4, 91–110. [Google Scholar] [CrossRef]
- Ratner, A.; Bach, S.H.; Ehrenberg, H.; Fries, J.; Wu, S.; Ré, C. Snorkel: Rapid training data creation with weak supervision. Proc. VLDB Endow. 2017, 11, 269–282. [Google Scholar] [CrossRef] [Green Version]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv 2019, arXiv:1901.07031. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.Q.; Pham, H.H.; Linh, L.T.; Dao, M.; Khanh, L. VinDr-CXR: An open dataset of chest X-rays with radiologist annotations. PhysioNet 2021. [Google Scholar] [CrossRef] [PubMed]
- Oakden-Rayner, L. Exploring the ChestXray14 Dataset: Problems. Available online: https://laurenoakdenrayner.com/2017/12/18/the-chestxray14-dataset-problems/ (accessed on 8 August 2022).
- Bustos, A.; Pertusa, A.; Salinas, J.-M.; de la Iglesia-Vayá, M. PadChest: A large chest X-ray image dataset with multi-label annotated reports. Med. Image Anal. 2020, 66, 101797. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.-X.J.; Lu, P.-X.; Thoma, G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [PubMed]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a Digital Image Database for Chest Radiographs With and Without a Lung Nodule: Receiver Operating Characteristic Analysis of Radiologists’ Detection of Pulmonary Nodules. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef]
- Johnson, A.E.W.; Pollard, T.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.; Peng, Y.; Lu, Z.; Mark, R.G.; Berkowitz, S.J.; Horng, S. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. arXiv 2019, arXiv:1901.07042. [Google Scholar]
- Wong, K.C.L.; Moradi, M.; Wu, J.; Pillai, A.; Sharma, A.; Gur, Y.; Ahmad, H.; Chowdary, M.S.; J, C.; Polaka, K.K.R.; et al. A robust network architecture to detect normal chest X-ray radiographs. arXiv 2020, arXiv:2004.06147. [Google Scholar]
- Rozenberg, E.; Freedman, D.; Bronstein, A. Localization with Limited Annotation for Chest X-rays. arXiv 2019, arXiv:1909.08842. [Google Scholar]
- Hermoza, R.; Maicas, G.; Nascimento, J.C.; Carneiro, G. Region Proposals for Saliency Map Refinement for Weakly-supervised Disease Localisation and Classification. arXiv 2020, arXiv:200510550. [Google Scholar]
- Liu, J.; Zhao, G.; Fei, Y.; Zhang, M.; Wang, Y.; Yu, Y. Align, Attend and Locate: Chest X-Ray Diagnosis via Contrast Induced Attention Network With Limited Supervision. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 10631–10640. [Google Scholar]
- Avramescu, C.; Bogdan, B.; Iarca, S.; Tenescu, A.; Fuicu, S. Assisting Radiologists in X-ray Diagnostics. In IoT Technologies for HealthCare; Garcia, N.M., Pires, I.M., Goleva, R., Eds.; Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering; Springer: Cham, Switzerland, 2020; Volume 314, pp. 108–117. ISBN 978-3-030-42028-4. [Google Scholar]
- Cohen, J.P.; Viviano, J.D.; Bertin, P.; Morrison, P.; Torabian, P.; Guarrera, M.; Lungren, M.P.; Chaudhari, A.; Brooks, R.; Hashir, M.; et al. TorchXRayVision: A library of chest X-ray datasets and models. arXiv 2021, arXiv:2111.00595. [Google Scholar]
- Zhou, Z.-H. A brief introduction to weakly supervised learning. Natl. Sci. Rev. 2018, 5, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Oh, K.; Oh, I.-S. Accurate Landmark Localization for Medical Images Using Perturbations. Appl. Sci. 2021, 11, 10277. [Google Scholar] [CrossRef]
- Islam, M.T.; Aowal, M.A.; Minhaz, A.T.; Ashraf, K. Abnormality Detection and Localization in Chest X-rays using Deep Convolutional Neural Networks. arXiv 2017, arXiv:1705.09850. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. arXiv 2014, arXiv:1311.2524. [Google Scholar]
- Uijlings, J.R.R.; van de Sande, K.E.A.; Gevers, T.; Smeulders, A.W.M. Selective Search for Object Recognition. Int. J. Comput. Vis. 2013, 104, 154–171. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial Pyramid Pooling in Deep Convolutional Networks for Visual Recognition. In Computer Vision–ECCV 2014; Springer: Cham, Switzerland, 2014; Volume 8691, pp. 346–361. [Google Scholar]
- Girshick, R. Fast R-CNN. arXiv 2015, arXiv:1504.08083. [Google Scholar]
- Liu, H.; Wang, L.; Nan, Y.; Jin, F.; Wang, Q.; Pu, J. SDFN: Segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Comput. Med. Imaging Graph. 2019, 75, 66–73. [Google Scholar] [CrossRef] [Green Version]
- Sogancioglu, E.; Murphy, K.; Calli, E.; Scholten, E.T.; Schalekamp, S.; Van Ginneken, B. Cardiomegaly Detection on Chest Radiographs: Segmentation Versus Classification. IEEE Access 2020, 8, 94631–94642. [Google Scholar] [CrossRef]
- Que, Q.; Tang, Z.; Wang, R.; Zeng, Z.; Wang, J.; Chua, M.; Gee, T.S.; Yang, X.; Veeravalli, B. CardioXNet: Automated Detection for Cardiomegaly Based on Deep Learning. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 612–615. [Google Scholar]
- Moradi, M.; Madani, A.; Karargyris, A.; Syeda-Mahmood, T.F. Chest x-ray generation and data augmentation for cardiovascular abnormality classification. In Proceedings of the Medical Imaging 2018: Image Processing; Angelini, E.D., Landman, B.A., Eds.; SPIE: Houston, TX, USA, 2018; p. 57. [Google Scholar]
- E, L.; Zhao, B.; Guo, Y.; Zheng, C.; Zhang, M.; Lin, J.; Luo, Y.; Cai, Y.; Song, X.; Liang, H. Using deep-learning techniques for pulmonary-thoracic segmentations and improvement of pneumonia diagnosis in pediatric chest radiographs. Pediatr. Pulmonol. 2019, 54, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Hurt, B.; Yen, A.; Kligerman, S.; Hsiao, A. Augmenting Interpretation of Chest Radiographs With Deep Learning Probability Maps. J. Thorac. Imaging 2020, 35, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Owais, M.; Arsalan, M.; Mahmood, T.; Kim, Y.H.; Park, K.R. Comprehensive Computer-Aided Decision Support Framework to Diagnose Tuberculosis From Chest X-ray Images: Data Mining Study. JMIR Med. Inform. 2020, 8, e21790. [Google Scholar] [CrossRef]
- Rajaraman, S.; Sornapudi, S.; Alderson, P.O.; Folio, L.R.; Antani, S.K. Analyzing inter-reader variability affecting deep ensemble learning for COVID-19 detection in chest radiographs. PLoS ONE 2020, 15, e0242301. [Google Scholar] [CrossRef]
- Samala, R.K.; Hadjiiski, L.; Chan, H.-P.; Zhou, C.; Stojanovska, J.; Agarwal, P.; Fung, C. Severity assessment of COVID-19 using imaging descriptors: A deep-learning transfer learning approach from non-COVID-19 pneumonia. In Proceedings of the Medical Imaging 2021: Computer-Aided Diagnosis; Drukker, K., Mazurowski, M.A., Eds.; SPIE: Houston, TX, USA, 2021; p. 62. [Google Scholar]
- Park, S.; Lee, S.M.; Kim, N.; Choe, J.; Cho, Y.; Do, K.-H.; Seo, J.B. Application of deep learning–based computer-aided detection system: Detecting pneumothorax on chest radiograph after biopsy. Eur. Radiol. 2019, 29, 5341–5348. [Google Scholar] [CrossRef]
- Hwang, E.J.; Park, S.; Jin, K.-N.; Kim, J.I.; Choi, S.Y.; Lee, J.H.; Goo, J.M.; Aum, J.; Yim, J.-J.; Cohen, J.G.; et al. Development and Validation of a Deep Learning–Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw. Open 2019, 2, e191095. [Google Scholar] [CrossRef] [Green Version]
- Blain, M.; Kassin, M.T.; Varble, N.; Wang, X.; Xu, Z.; Xu, D.; Carrafiello, G.; Vespro, V.; Stellato, E.; Ierard, A.M.; et al. Determination of disease severity in COVID-19 patients using deep learning in chest X-ray images. Diagn. Interv. Radiol. 2021, 27, 20–27. [Google Scholar] [CrossRef]
- Ferreira-Junior, J.; Cardenas, D.; Moreno, R.; Rebelo, M.; Krieger, J.; Gutierrez, M. A general fully automated deep-learning method to detect cardiomegaly in chest X-rays. In Proceedings of the Medical Imaging 2021: Computer-Aided Diagnosis; Drukker, K., Mazurowski, M.A., Eds.; SPIE: Houston, TX, USA, 2021; p. 81. [Google Scholar]
- Tartaglione, E.; Barbano, C.A.; Berzovini, C.; Calandri, M.; Grangetto, M. Unveiling COVID-19 from CHEST X-ray with Deep Learning: A Hurdles Race with Small Data. Int. J. Environ. Res. Public. Health 2020, 17, 6933. [Google Scholar] [CrossRef]
- Narayanan, B.N.; Davuluru, V.S.P.; Hardie, R.C. Two-stage deep learning architecture for pneumonia detection and its diagnosis in chest radiographs. In Proceedings of the Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications; Deserno, T.M., Chen, P.-H., Eds.; SPIE: Houston, TX, USA, 2020; p. 15. [Google Scholar]
- Wang, X.; Yu, J.; Zhu, Q.; Li, S.; Zhao, Z.; Yang, B.; Pu, J. Potential of deep learning in assessing pneumoconiosis depicted on digital chest radiography. Occup. Environ. Med. 2020, 77, 597–602. [Google Scholar] [CrossRef]
- Ferreira, J.R.; Armando Cardona Cardenas, D.; Moreno, R.A.; de Fatima de Sa Rebelo, M.; Krieger, J.E.; Antonio Gutierrez, M. Multi-View Ensemble Convolutional Neural Network to Improve Classification of Pneumonia in Low Contrast Chest X-ray Images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1238–1241. [Google Scholar]
- Wang, Z.; Xiao, Y.; Li, Y.; Zhang, J.; Lu, F.; Hou, M.; Liu, X. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recognit. 2021, 110, 107613. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-Y.; Tsai, T.-Y.; Tseng, C.-Y.; Liu, K.-H.; Lee, C.-W. A Deep Learning Method for Alerting Emergency Physicians about the Presence of Subphrenic Free Air on Chest Radiographs. J. Clin. Med. 2021, 10, 254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xie, Y.; Pang, G.; Liao, Z.; Verjans, J.; Li, W.; Sun, Z.; He, J.; Li, Y.; Shen, C.; et al. Viral Pneumonia Screening on Chest X-rays Using Confidence-Aware Anomaly Detection. IEEE Trans. Med. Imaging 2021, 40, 879–890. [Google Scholar] [CrossRef]
- Nugroho, B.A. An aggregate method for thorax diseases classification. Sci. Rep. 2021, 11, 3242. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Shi, J.-X.; Yan, L.; Wang, Y.-G.; Zhang, X.-D.; Jiang, M.-S.; Wu, Z.-Z.; Zhou, K.-Q. Lesion-aware convolutional neural network for chest radiograph classification. Clin. Radiol. 2021, 76, 155.e1–155.e14. [Google Scholar] [CrossRef] [PubMed]
- Griner, D.; Zhang, R.; Tie, X.; Zhang, C.; Garrett, J.; Li, K.; Chen, G.-H. COVID-19 pneumonia diagnosis using chest X-ray radiograph and deep learning. In Proceedings of the Medical Imaging 2021: Computer-Aided Diagnosis; Drukker, K., Mazurowski, M.A., Eds.; SPIE: Houston, TX, USA, 2021; p. 3. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. arXiv 2016, arXiv:1506.01497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. arXiv 2016, arXiv:1506.02640. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLO9000: Better, Faster, Stronger. arXiv 2016, arXiv:1612.08242. [Google Scholar]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Jocher, G.; Chaurasia, A.; Stoken, A.; Borovec, J.; NanoCode012; Kwon, Y.; Xie, T.; Fang, J.; Imyhxy; Michael, K.; et al. Ultralytics/yolov5: V6.1—TensorRT, TensorFlow Edge TPU and OpenVINO Export and Inference. 2022. Available online: https://zenodo.org/record/7347926#.Y5qKLYdBxPY (accessed on 8 August 2022).
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Computer Vision—ECCV 2016; Springer: Cham, Switzerland, 2016; Volume 9905, pp. 21–37. [Google Scholar]
- Bazzani, L.; Bergamo, A.; Anguelov, D.; Torresani, L. Self-taught Object Localization with Deep Networks. arXiv 2016, arXiv:1409.3964. [Google Scholar]
- Donahue, J.; Jia, Y.; Vinyals, O.; Hoffman, J.; Zhang, N.; Tzeng, E.; Darrell, T. DeCAF: A Deep Convolutional Activation Feature for Generic Visual Recognition. arXiv 2013, arXiv:1310.1531. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and Transferring Mid-level Image Representations Using Convolutional Neural Networks. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Is object localization for free?—Weakly-supervised learning with convolutional neural networks. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 685–694. [Google Scholar]
- Basu, S.; Mitra, S.; Saha, N. Deep Learning for Screening COVID-19 using Chest X-ray Images. arXiv 2020, arXiv:2004.10507. [Google Scholar]
- Wehbe, R.M.; Sheng, J.; Dutta, S.; Chai, S.; Dravid, A.; Barutcu, S.; Wu, Y.; Cantrell, D.R.; Xiao, N.; Allen, B.D.; et al. DeepCOVID-XR: An Artificial Intelligence Algorithm to Detect COVID-19 on Chest Radiographs Trained and Tested on a Large U.S. Clinical Data Set. Radiology 2021, 299, E167–E176. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Peng, K.; Yang, X.; Huang, P.; Luo, Y.; Feng, P.; Wei, B. E-TBNet: Light Deep Neural Network for Automatic Detection of Tuberculosis with X-ray DR Imaging. Sensors 2022, 22, 821. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Bu, S. Transfer-Learning-Based Approach for the Diagnosis of Lung Diseases from Chest X-ray Images. Entropy 2022, 24, 313. [Google Scholar] [CrossRef]
- Li, K.; Wu, Z.; Peng, K.-C.; Ernst, J.; Fu, Y. Tell Me Where to Look: Guided Attention Inference Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Yang, X. An Overview of the Attention Mechanisms in Computer Vision. J. Phys. Conf. Ser. 2020, 1693, 012173. [Google Scholar] [CrossRef]
- Datta, S.K.; Shaikh, M.A.; Srihari, S.N.; Gao, M. Soft Attention Improves Skin Cancer Classification Performance. In Interpretability of Machine Intelligence in Medical Image Computing, and Topological Data Analysis and Its Applications for Medical Data; Reyes, M., Henriques Abreu, P., Cardoso, J., Hajij, M., Zamzmi, G., Rahul, P., Thakur, L., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12929, pp. 13–23. ISBN 978-3-030-87443-8. [Google Scholar]
- Yang, H.; Kim, J.-Y.; Kim, H.; Adhikari, S.P. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans. Med. Imaging 2019, 39, 1306–1315. [Google Scholar] [CrossRef]
- Truong, T.; Yanushkevich, S. Relatable Clothing: Soft-Attention Mechanism for Detecting Worn/Unworn Objects. IEEE Access 2021, 9, 108782–108792. [Google Scholar] [CrossRef]
- Petrovai, A.; Nedevschi, S. Fast Panoptic Segmentation with Soft Attention Embeddings. Sensors 2022, 22, 783. [Google Scholar] [CrossRef]
- Ren, X.; Huo, J.; Xuan, K.; Wei, D.; Zhang, L.; Wang, Q. Robust Brain Magnetic Resonance Image Segmentation for Hydrocephalus Patients: Hard and Soft Attention. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 385–389. [Google Scholar]
- Chen, C.; Gong, D.; Wang, H.; Li, Z.; Wong, K.-Y.K. Learning Spatial Attention for Face Super-Resolution. IEEE Trans. Image Process. 2021, 30, 1219–1231. [Google Scholar] [CrossRef] [PubMed]
- Jaderberg, M.; Simonyan, K.; Zisserman, A.; Kavukcuoglu, K. Spatial Transformer Networks. arXiv 2016, arXiv:1506.02025. [Google Scholar]
- Sønderby, S.K.; Sønderby, C.K.; Maaløe, L.; Winther, O. Recurrent Spatial Transformer Networks. arXiv 2015, arXiv:1509.05329. [Google Scholar]
- Bastidas, A.A.; Tang, H. Channel Attention Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Long Beach, CA, USA, 15–20 June 2019. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. arXiv 2018, arXiv:1807.06521. [Google Scholar]
- Choi, M.; Kim, H.; Han, B.; Xu, N.; Lee, K.M. Channel Attention Is All You Need for Video Frame Interpolation. Proc. AAAI Conf. Artif. Intell. 2020, 34, 10663–10671. [Google Scholar] [CrossRef]
- Zhou, T.; Canu, S.; Ruan, S. Automatic COVID-19 CT segmentation using U-NET integrated spatial and channel attention mechanism. Int. J. Imaging Syst. Technol. 2021, 31, 16–27. [Google Scholar] [CrossRef]
- Papadopoulos, A.; Korus, P.; Memon, N. Hard-Attention for Scalable Image Classification. In Proceedings of the Advances in Neural Information Processing Systems; Ranzato, M., Beygelzimer, A., Dauphin, Y., Liang, P.S., Vaughan, J.W., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2021; Volume 34, pp. 14694–14707. [Google Scholar]
- Elsayed, G.F.; Kornblith, S.; Le, Q.V. Saccader: Improving Accuracy of Hard Attention Models for Vision. arXiv 2019, arXiv:1908.07644. [Google Scholar]
- Wang, D.; Haytham, A.; Pottenburgh, J.; Saeedi, O.; Tao, Y. Hard Attention Net for Automatic Retinal Vessel Segmentation. IEEE J. Biomed. Health Inform. 2020, 24, 3384–3396. [Google Scholar] [CrossRef]
- Simons, D.J.; Chabris, C.F. Gorillas in Our Midst: Sustained Inattentional Blindness for Dynamic Events. Perception 1999, 28, 1059–1074. [Google Scholar] [CrossRef]
- Indurthi, S.R.; Chung, I.; Kim, S. Look Harder: A Neural Machine Translation Model with Hard Attention. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019; Association for Computational Linguistics: Florence, Italy, 2019; pp. 3037–3043. [Google Scholar]
- OpenCV. Saliency API. Available online: https://docs.opencv.org/4.x/d8/d65/group__saliency.html (accessed on 12 July 2022).
- Hou, X.; Zhang, L. Saliency Detection: A Spectral Residual Approach. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; pp. 1–8. [Google Scholar]
- Wang, B.; Dudek, P. A Fast Self-Tuning Background Subtraction Algorithm. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; pp. 401–404. [Google Scholar]
- Cheng, M.-M.; Zhang, Z.; Lin, W.-Y.; Torr, P. BING: Binarized Normed Gradients for Objectness Estimation at 300fps. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 3286–3293. [Google Scholar]
- Min, K.; Corso, J.J. TASED-Net: Temporally-Aggregating Spatial Encoder-Decoder Network for Video Saliency Detection. arXiv 2019, arXiv:1908.05786. [Google Scholar]
- Tsiami, A.; Koutras, P.; Maragos, P. STAViS: Spatio-Temporal AudioVisual Saliency Network. arXiv 2020, arXiv:2001.03063. [Google Scholar]
- Yao, L.; Prosky, J.; Poblenz, E.; Covington, B.; Lyman, K. Weakly Supervised Medical Diagnosis and Localization from Multiple Resolutions. arXiv 2018, arXiv:1803.07703. [Google Scholar]
- Tu, W.-C.; He, S.; Yang, Q.; Chien, S.-Y. Real-Time Salient Object Detection with a Minimum Spanning Tree. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2334–2342. [Google Scholar]
- Yang, J.; Yang, M.-H. Top-Down Visual Saliency via Joint CRF and Dictionary Learning. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zakir, A. Top–Down Saliency Detection Based on Deep-Learned Features. Int. J. Comput. Intell. Appl. 2019, 18, 1950009. [Google Scholar] [CrossRef]
- Wang, L.; Lu, H.; Wang, Y.; Feng, M.; Wang, D.; Yin, B.; Ruan, X. Learning to Detect Salient Objects with Image-Level Supervision. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 3796–3805. [Google Scholar]
- Zhang, J.; Zhang, T.; Dai, Y.; Harandi, M.; Hartley, R. Deep Unsupervised Saliency Detection: A Multiple Noisy Labeling Perspective. arXiv 2018, arXiv:1803.10910. [Google Scholar]
- Yao, C.; Kong, Y.; Feng, L.; Jin, B.; Si, H. Contour-Aware Recurrent Cross Constraint Network for Salient Object Detection. IEEE Access 2020, 8, 218739–218751. [Google Scholar] [CrossRef]
- Abouelmehdi, K.; Beni-Hessane, A.; Khaloufi, H. Big healthcare data: Preserving security and privacy. J. Big Data 2018, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- van Egmond, M.B.; Spini, G.; van der Galien, O.; IJpma, A.; Veugen, T.; Kraaij, W.; Sangers, A.; Rooijakkers, T.; Langenkamp, P.; Kamphorst, B.; et al. Privacy-preserving dataset combination and Lasso regression for healthcare predictions. BMC Med. Inform. Decis. Mak. 2021, 21, 266. [Google Scholar] [CrossRef]
- Dyda, A.; Purcell, M.; Curtis, S.; Field, E.; Pillai, P.; Ricardo, K.; Weng, H.; Moore, J.C.; Hewett, M.; Williams, G.; et al. Differential privacy for public health data: An innovative tool to optimize information sharing while protecting data confidentiality. Patterns 2021, 2, 100366. [Google Scholar] [CrossRef]
- Murphy, K.; Smits, H.; Knoops, A.J.G.; Korst, M.B.J.M.; Samson, T.; Scholten, E.T.; Schalekamp, S.; Schaefer-Prokop, C.M.; Philipsen, R.H.H.M.; Meijers, A.; et al. COVID-19 on Chest Radiographs: A Multireader Evaluation of an Artificial Intelligence System. Radiology 2020, 296, E166–E172. [Google Scholar] [CrossRef]
- Gong, Z.; Zhong, P.; Hu, W. Diversity in Machine Learning. IEEE Access 2019, 7, 64323–64350. [Google Scholar] [CrossRef]
- Redko, I.; Habrard, A.; Morvant, E.; Sebban, M.; Bennani, Y. Advances in Domain Adaption Theory; Elsevier: Amsterdam, The Netherlands, 2019; ISBN 978-1-78548-236-6. [Google Scholar]
- Sun, S.; Shi, H.; Wu, Y. A survey of multi-source domain adaptation. Inf. Fusion 2015, 24, 84–92. [Google Scholar] [CrossRef]
- Petch, J.; Di, S.; Nelson, W. Opening the Black Box: The Promise and Limitations of Explainable Machine Learning in Cardiology. Can. J. Cardiol. 2022, 38, 204–213. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Stiglic, G.; Kocbek, P.; Fijacko, N.; Zitnik, M.; Verbert, K.; Cilar, L. Interpretability of machine learning based prediction models in healthcare. WIREs Data Min. Knowl. Discov. 2020, 10. [Google Scholar] [CrossRef]
- Preechakul, K.; Sriswasdi, S.; Kijsirikul, B.; Chuangsuwanich, E. Improved image classification explainability with high-accuracy heatmaps. iScience 2022, 25, 103933. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. arXiv 2016, arXiv:1512.02325. [Google Scholar]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-Image Translation with Conditional Adversarial Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar]
- Aggarwal, R.; Ringold, S.; Khanna, D.; Neogi, T.; Johnson, S.R.; Miller, A.; Brunner, H.I.; Ogawa, R.; Felson, D.; Ogdie, A.; et al. Distinctions Between Diagnostic and Classification Criteria? Diagnostic Criteria in Rheumatology. Arthritis Care Res. 2015, 67, 891–897. [Google Scholar] [CrossRef]














| Model | Size (MB) | Top-1 Accuracy | Top-5 Accuracy | Parameters | Depth |
|---|---|---|---|---|---|
| Xception | 88 | 0.790 | 0.945 | 22,910,480 | 126 |
| VGG16 | 528 | 0.713 | 0.901 | 138,357,544 | 23 |
| ResNet50 | 98 | 0.749 | 0.921 | 25,636,712 | - |
| ResNet152V2 | 232 | 0.780 | 0.942 | 60,380,648 | - |
| InceptionV3 | 92 | 0.779 | 0.937 | 23,851,784 | 159 |
| InceptionResNetV2 | 215 | 0.803 | 0.953 | 55,873,736 | 572 |
| MobileNet | 16 | 0.704 | 0.895 | 4,253,864 | 88 |
| MobileNetV2 | 14 | 0.713 | 0.901 | 3,538,984 | 88 |
| DenseNet121 | 33 | 0.750 | 0.923 | 8,062,504 | 121 |
| NASNetMobile | 23 | 0.744 | 0.919 | 5,326,716 | - |
| EfficientNetB0 | 29 | - | - | 5,330,571 | - |
| Initiator | Name | Total | Frontal View | Geographic |
|---|---|---|---|---|
| National Institute of Health | ChestX-ray8 | 112,120 | 112,120 | Northeast USA |
| Stanford University | CheXpert | 223,141 | 191,010 | Western USA |
| University of Alicante | PadChest | 160,868 | 67,000 | Spain |
| VinBrain | VinDr-CXR | 15,000 | 15,000 | Vietnam |
| National Library of Medicine | Tuberculosis | 800 | 800 | China + USA |
| Approach | Variants | Description |
|---|---|---|
| Class Activation Maps | Grad-CAM | Weight the 2-Dimension activations by averaging gradients |
| Score-CAM | Perburate the input image by scaling activations to estimate how the output drops | |
| Full-Grad | Calculates the biases’ gradients from all over the network before summing them | |
| Attention models | RAN | In Residual Attention Networks, several attention modules have been employed to the backbone network for learning the mask in each convolutional layer |
| ADL | Attention based Dropout Layer utilizes the self-attention to process the feature maps of the model | |
| STN | Spatial Transformation Network explicitly allows the spatial manipulation of data within the CNN. | |
| Saliency Maps | ISL | Image Level Supervision first classifier is trained with foreground features and then generate saliency maps in top-down scheme. |
| DUS | Deep Unsupervised Saliency that works collaboratively with a latent saliency prediction module and a noise modeling module. | |
| C2S | Contour2Saliency exploits a coarse-to-fine architecture that generates saliency maps and contour maps simultaneously |
| Variant | Mechanism |
|---|---|
| CAM | Replaced the first fully-connected layer in the image classifiers with a global average pooling layer |
| Grad-CAM | Weight the activations using the average gradient |
| Grad-CAM++ | Extension of Grad-CAM that uses second order gradients |
| XGrad-CAM | Extension Grad-CAM that scales the gradients by the normalized activations |
| Ablation-CAM | Measure how the output drops after zeroing out activations |
| Score-CAM | Perbutate the image by the scaled activations and measure how the output drops |
| Eigen-CAM | Takes the first main element of the 2D activations and increases outcomes without utilizing class discrimination. |
| Layer-CAM | Spatially weight the activations by positive gradients. Works better especially in lower layers |
| Full-Grad | Computes the gradients of the biases from all over the network, and then sums them |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aasem, M.; Iqbal, M.J.; Ahmad, I.; Alassafi, M.O.; Alhomoud, A. A Survey on Tools and Techniques for Localizing Abnormalities in X-ray Images Using Deep Learning. Mathematics 2022, 10, 4765. https://doi.org/10.3390/math10244765
Aasem M, Iqbal MJ, Ahmad I, Alassafi MO, Alhomoud A. A Survey on Tools and Techniques for Localizing Abnormalities in X-ray Images Using Deep Learning. Mathematics. 2022; 10(24):4765. https://doi.org/10.3390/math10244765
Chicago/Turabian StyleAasem, Muhammad, Muhammad Javed Iqbal, Iftikhar Ahmad, Madini O. Alassafi, and Ahmed Alhomoud. 2022. "A Survey on Tools and Techniques for Localizing Abnormalities in X-ray Images Using Deep Learning" Mathematics 10, no. 24: 4765. https://doi.org/10.3390/math10244765







