Deep Red Lesion Classification for Early Screening of Diabetic Retinopathy
Abstract
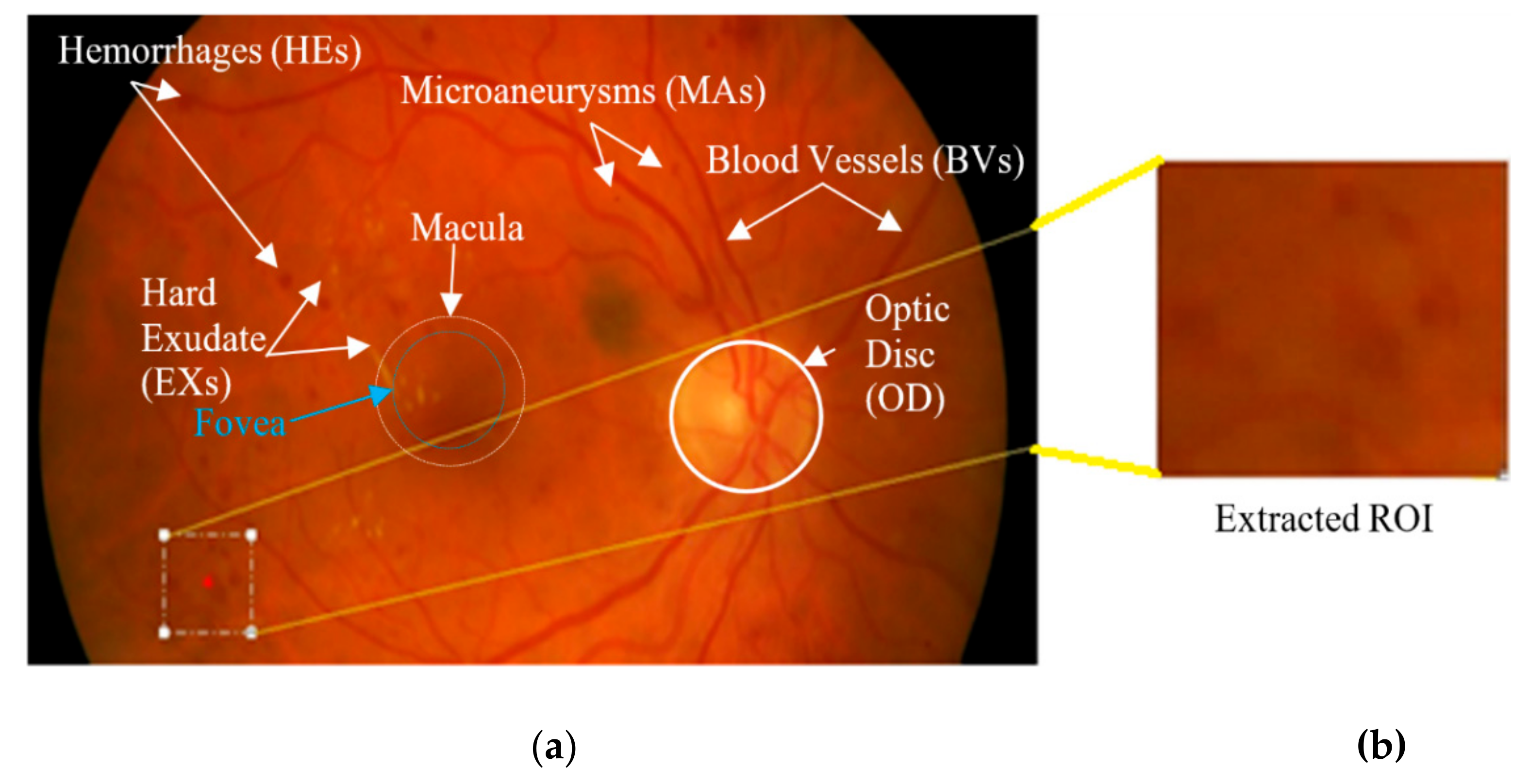
:1. Introduction
2. Materials and Methods
2.1. Datasets
2.1.1. e-Ophtha_MA
2.1.2. Retinopathy Online Challenge (ROC)
2.1.3. Standard Diabetic Retinopathy Database Calibration Level 1 (DiaRetDB1) v2.1
2.1.4. Indian Diabetic Retinopathy Image Dataset (IDRiD)
2.1.5. Messidor
2.2. The Proposed Method
2.2.1. Preprocessing
2.2.2. Patch Generation
2.2.3. Dataset Distribution (Train/Development/Test Sets)
2.2.4. CNN Training
2.2.5. Baseline ResNet Architecture
2.2.6. Freezing Initial Layers
2.2.7. Architectural Enhancement
Reinforced Skip Connections
Layers Modification
2.2.8. Evaluation Protocol
2.2.9. Experimental Setup
3. Results
3.1. Fine-Tuning
3.2. DR-ResNet50 Architecture
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| FOV Extraction |
| Small patch generation |
| Extract patches centroid at the lesions from unhealthy images |
| Extract adjacent patches centered within FOVs from healthy images |
| Data augmentation and distribution into train, validation, and test sets using Scheme 1 or 2 |
| Choosing a suitable pre-trained CNN model through transfer learning |
| Development of DR-ResNet50 model |
| Training phase |
| Fine tunning through transfer learning coupled with |
| Hyper-parameters settings |
| Freezing initial layers |
| Architectural modifications |
| Reinforced Skip connections |
| Deep layers modifications |
| Replace GAP with GMP |
| Replace Softmax for a new task |
| Replace fc for binary classification |
| Replace output layer with SSE loss |
| Training and validation with performance monitoring |
| Testing phase |
| Load trained DR-ResNet50 model |
| Prediction on unseen Test instances |
| if ‘cross-validation’ then |
| Extract all overlapped patches within FOVs |
| Prediction to cross-validate on different datasets |
| Post-processing to evaluate performance by computing quantitative metrics and Grad-CAM |
References
- Baena-Díez, J.M.; Peñafiel, J.; Subirana, I.; Ramos, R.; Elosua, R.; Marín-Ibañez, A.; Guembe, M.J.; Rigo, F.; Tormo-Díaz, M.J.; Moreno-Iribas, C.; et al. Risk of Cause-Specific Death in Individuals With Diabetes: A Competing Risks Analysis. Diabetes Care 2016, 39, 1987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; da Rocha Fernandes, J.D.; Ohlrogge, A.W.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.Y.; Aiello, L.P.; Ferris, F.; Gupta, N.; Kawasaki, R.; Lansingh, V.; Maia, M.; Mathenge, W. International Council of Ophthalmology (ICO): Updated 2017 Guidelines for Diabetic Eye Care; International Council of Ophthalmology (ICO): San Francisco, CA, USA, 2017. [Google Scholar]
- Cheung, N.; Mitchell, P.; Wong, T.Y. Diabetic retinopathy. Lancet 2010, 376, 124–136. [Google Scholar] [CrossRef]
- Williams, R.; Colagiuri, S.; Almutairi, R.; Montoya, P.A.; Abdul, B.; Beran, D.; Besançon, S.; Bommer, C.; Borgnakke, W.; Boyko, E.; et al. International Diabetes Federation. IDF Diabetes Atlas, 9th ed.; International Diabetes Federation: Brussels, Belgium, 2019. [Google Scholar]
- Stiefelhagen, M.P. Mehr als eine Stoffwechselerkrankung: Der Diabetes verschont kaum ein Organ. MMW Fortschritte Medizin 2019, 161, 21–22. [Google Scholar]
- Klein, B.E.K. Overview of Epidemiologic Studies of Diabetic Retinopathy. Ophthalmic Epidemiol. 2007, 14, 179–183. [Google Scholar] [CrossRef]
- Scanlon, P.H. Diabetic retinopathy. Medicine 2015, 43, 13–19. [Google Scholar] [CrossRef]
- Butler, J.M.; Guthrie, S.M.; Koc, M.; Afzal, A.; Caballero, S.; Brooks, H.L.; Mames, R.N.; Segal, M.S.; Grant, M.B.; Scott, E.W. SDF-1 is both necessary and sufficient to promote proliferative retinopathy. J. Clin. Investig. 2005, 115, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Abràmoff, M.D.; Garvin, M.K.; Sonka, M. Retinal imaging and image analysis. IEEE Rev. Biomed. Eng. 2010, 3, 169–208. [Google Scholar] [CrossRef] [Green Version]
- Karadeniz, S.; Zimmet, P.; Aschner, P.; Belton, A.; Cavan, D.; Jalang’o, A.; Gandhi, N.; Hill, L.; Makaroff, L.; Mesurier, R.L.; et al. Diabetes Eye Health: A guide for Health Care Professionals; International Diabetes Federation: Brussels, Belgium, 2015. [Google Scholar]
- Klonoff, D.C.; Schwartz, D.M. An economic analysis of interventions for diabetes. Diabetes Care 2000, 23, 390. [Google Scholar] [CrossRef] [Green Version]
- Decencière, E.; Cazuguel, G.; Zhang, X.; Thibault, G.; Klein, J.C.; Meyer, F.; Marcotegui, B.; Quellec, G.; Lamard, M.; Danno, R.; et al. TeleOphta: Machine learning and image processing methods for teleophthalmology. IRBM 2013, 34, 196–203. [Google Scholar] [CrossRef]
- Niemeijer, M.; Ginneken, B.v.; Staal, J.; Suttorp-Schulten, M.S.A.; Abramoff, M.D. Automatic detection of red lesions in digital color fundus photographs. IEEE Trans. Med. Imaging 2005, 24, 584–592. [Google Scholar] [CrossRef] [Green Version]
- Seoud, L.; Hurtut, T.C.J.; Cheriet, F.J.M.; Langlois, P. Red lesion detection using dynamic shape features for diabetic retinopathy screening. IEEE Trans. Med. Imaging 2016, 35, 1116–1126. [Google Scholar] [CrossRef] [PubMed]
- Orlando, J.I.; Prokofyeva, E.; Del Fresno, M.; Blaschko, M.B. An ensemble deep learning based approach for red lesion detection in fundus images. Comput. Methods Programs Biomed. 2018, 153, 115–127. [Google Scholar] [CrossRef] [Green Version]
- Zago, G.T.; Andreão, R.V.; Dorizzi, B.; Teatini Salles, E.O. Diabetic retinopathy detection using red lesion localization and convolutional neural networks. Comput. Biol. Med. 2020, 116, 103537. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Erfurth, U.; Sadeghipour, A.; Gerendas, B.S.; Waldstein, S.M.; Bogunovic, H. Artificial intelligence in retina. Prog. Retin. Eye Res. 2018, 67, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Laÿ, B. Analyse Automatique Des Images Angio Fluorographiques Au Cours De La Retinopathie Diabetique. Ph.D. Thesis, Centre of Mathematical Morphology, Paris School of Mines, Paris, France, 1983. [Google Scholar]
- Baudoin, E.C.; Laÿ, B.J.; Klein, J.C. Automatic detection of microaneurysms in diabetic fluorescein angiographies. Rev. D’Épidémiol. Sante Publique 1984, 32, 254–261. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [Green Version]
- Azizpour, H.; Razavian, A.S.; Sullivan, J.; Maki, A.; Carlsson, S. From generic to specific deep representations for visual recognition. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; pp. 36–45. [Google Scholar]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Peng, Z. Rice diseases detection and classification using attention based neural network and bayesian optimization. Expert Syst. Appl. 2021, 178, 114770. [Google Scholar] [CrossRef]
- Kauppi, T.; Kalesnykiene, V.; Sorri, I.; Raninen, A.; Voutilainen, R.; Kamarainen, j.; Lensu, L.; Uusitalo, H. Diabetic Retinopathy Database and Evaluation Protocol (DiaRetDB1 V2.1); Machine Vision and Pattern Recognition Laboratory, Lappeenranta University of Technology: Lappeenranta, Finland, 2009. [Google Scholar]
- Niemeijer, M.; Ginneken, B.v.; Cree, M.J.; Mizutani, A.; Quellec, G.; Sanchez, C.I.; Zhang, B.; Hornero, R.; Lamard, M.; Muramatsu, C.; et al. Retinopathy Online Challenge: Automatic Detection of Microaneurysms in Digital Color Fundus Photographs. IEEE Trans. Med. Imaging 2010, 29, 185–195. [Google Scholar] [CrossRef]
- Decencière, E.; Zhang, X.; Cazuguel, G.; Lay, B.; Cochener, B.; Trone, C.; Gain, P.; Ordonez, R.; Massin, P.; Erginay, A.; et al. Feedback on a publicly distributed image database: The Messidor Database. Image Anal. Stereol. 2014, 33, 231–234. [Google Scholar] [CrossRef] [Green Version]
- Prasanna, P.; Samiksha, P.; Ravi, K.; Manesh, K.; Girish, D.; Vivek, S.; Fabrice, M. Indian Diabetic Retinopathy Image Dataset (IDRiD). IEEE Dataport 2018. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Ashraf, M.N.; Hussain, M.; Habib, Z. Review of Various Tasks Performed in the Preprocessing Phase of a Diabetic Retinopathy Diagnosis System. Curr. Med. Imaging 2020, 16, 397–426. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhu, W.; Shi, F.; Zhu, S.; Chen, X. Automatic detection of microaneurysms in retinal fundus images. Comput. Med. Imaging Graph. 2017, 55, 106–112. [Google Scholar] [CrossRef]
- Yadav, D.; Kumar Karn, A.; Giddalur, A.; Dhiman, A.; Sharma, S.; Muskan; Yadav, A.K. Microaneurysm Detection Using Color Locus Detection Method. Measurement 2021, 176, 109084. [Google Scholar] [CrossRef]
- Grinsven, M.J.J.P.v.; Ginneken, B.v.; Hoyng, C.B.; Theelen, T.; Sánchez, C.I. Fast Convolutional Neural Network Training Using Selective Data Sampling: Application to Hemorrhage Detection in Color Fundus Images. IEEE Trans. Med. Imaging 2016, 35, 1273–1284. [Google Scholar] [CrossRef]
- Sinthanayothin, C.; Boyce, J.F.; Williamson, T.H.; Cook, H.L.; Mensah, E.; Lal, S.; Usher, D. Automated detection of diabetic retinopathy on digital fundus images. Diabet. Med. 2002, 19, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Larsen, M.; Godt, J.; Larsen, N.; Lund-Andersen, H.; Sjølie, A.K.; Agardh, E.; Kalm, H.; Grunkin, M.; Owens, D.R. Automated Detection of Fundus Photographic Red Lesions in Diabetic Retinopathy. Investig. Ophthalmol. Vis. Sci. 2003, 44, 761–766. [Google Scholar] [CrossRef]
- Grisan, E.; Ruggeri, A. Segmentation of Candidate Dark Lesions in Fundus Images Based on Local Thresholding and Pixel Density. In Proceedings of the Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS2007), Lyon, France, 23–26 August 2007; pp. 6735–6738. [Google Scholar]
- García, M.; Sánchez, C.I.; López, M.I.; Díez, A.; Hornero, R. Automatic Detection of Red Lesions in Retinal Images Using a Multilayer Perceptron Neural Network. In Proceedings of the 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS2008), Vancouver, BC, Canada, 20–25 August 2008; pp. 5425–5428. [Google Scholar]
- Zhang, B.; Wu, X.; You, J.; Li, Q.; Karray, F. Hierarchical Detection of Red Lesions in Retinal Images by Multiscale Correlation Filtering; SPIE: Orlando, FL, USA, 2009; Volume 7260. [Google Scholar]
- Saleh, M.D.; Eswaran, C. An automated decision-support system for non-proliferative diabetic retinopathy disease based on MAs and HAs detection. Comput. Methods Programs Biomed. 2012, 108, 186–196. [Google Scholar] [CrossRef]
- Ashraf, M.N.; Habib, Z.; Hussain, M. Texture Feature Analysis of Digital Fundus Images for Early Detection of Diabetic Retinopathy. In Proceedings of the 11th International Conference on Computer Graphics, Imaging and Visualization: New Techniques and Trends, CGIV, Singapore, 6–8 August 2014; pp. 57–62. [Google Scholar]
- Ashraf, M.N.; Habib, Z.; Hussain, M. Computer Aided Diagnose of Diabetic Retinopathy; LAP LAMBERT Academic Publishing: Deutschland, Germany, 2015. [Google Scholar]
- Srivastava, R.; Wong, D.W.K.; Duan, L.; Liu, J.; Wong, T.Y. Red Lesion Detection In Retinal Fundus Images Using Frangi-Based Filters. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 5663–5666. [Google Scholar]
- Srivastava, R.; Duan, L.; Wong, D.W.K.; Liu, J.; Wong, T.Y. Detecting retinal microaneurysms and hemorrhages with robustness to the presence of blood vessels. Comput. Methods Programs Biomed. 2017, 138, 83–91. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhang, X.; Geng, L.; Zhang, F.; Wu, J.; Tong, J.; Ogunbona, P.O.; Shan, C. Automatic non-proliferative diabetic retinopathy screening system based on color fundus image. Biomed. Eng. Online 2017, 16, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colomer, A.; Igual, J.; Naranjo, V. Detection of Early Signs of Diabetic Retinopathy Based on Textural and Morphological Information in Fundus Images. Sensors 2020, 20, 1005. [Google Scholar] [CrossRef] [Green Version]
- Walter, T.; Massin, P.; Arginay, A.; Ordonez, R.; Jeulin, C.; Klein, J.C. Automatic detection of microaneurysms in color fundus images. Med. Image Anal. 2007, 11, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.; van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 2017, 10, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Asiri, N.; Hussain, M.; Al Adel, F.; Alzaidi, N. Deep learning based computer-aided diagnosis systems for diabetic retinopathy: A survey. Artif. Intell. Med. 2019, 99, 101701. [Google Scholar] [CrossRef] [Green Version]
- Haloi, M. Improved Microaneurysm Detection using Deep Neural Networks. arXiv 2015, arXiv:1505.04424. [Google Scholar]
- Pratt, H.; Coenen, F.; Broadbent, D.M.; Harding, S.P.; Zheng, Y. Convolutional Neural Networks for Diabetic Retinopathy. Procedia Comput. Sci. 2016, 90, 200–205. [Google Scholar] [CrossRef] [Green Version]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Abramoff, M.D.; Lou, Y.; Erginay, A.; Clarida, W.; Amelon, R.; Folk, J.C.; Niemeijer, M. Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep Learning. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5200–5206. [Google Scholar] [CrossRef] [Green Version]
- Gargeya, R.; Leng, T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017, 124, 962–969. [Google Scholar] [CrossRef]
- Quellec, G.; Charriere, K.; Boudi, Y.; Cochener, B.; Lamard, M. Deep image mining for diabetic retinopathy screening. Med. Image Anal. 2017, 39, 178–193. [Google Scholar] [CrossRef] [Green Version]
- Wan, S.; Liang, Y.; Zhang, Y. Deep convolutional neural networks for diabetic retinopathy detection by image classification. Comput. Electr. Eng. 2018, 72, 274–282. [Google Scholar] [CrossRef]
- Voets, M.; Møllersen, K.; Bongo, L.A. Reproduction study using public data of: Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. PLoS ONE 2019, 14, e0217541. [Google Scholar] [CrossRef]
- Chudzik, P.; Majumdar, S.; Caliva, F.; Al-Diri, B.; Hunter, A. Microaneurysm detection using fully convolutional neural networks. Comput. Methods Programs Biomed. 2018, 158, 185–192. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Boser, B.; Denker, J.S.; Henderson, D.; Howard, R.E.; Hubbard, W.; Jackel, L.D. Backpropagation Applied to Handwritten Zip Code Recognition. Neural Comput. 1989, 1, 541–551. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 2015 International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Panwar, H.; Gupta, P.K.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef] [PubMed]
- Cuadros, J.; Bresnick, G. EyePACS: An Adaptable Telemedicine System for Diabetic Retinopathy Screening. J. Diabetes Sci. Technol. 2009, 3, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Fernandez-Loaiza, P.; Sauma, J.; Hernandez-Bogantes, E.; Masis, M. Classification of diabetic retinopathy and diabetic macular edema. World J. Diabetes 2013, 4, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 6–8 December 2013; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Wei, L.; Yangqing, J.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper With Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.v.d.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef] [Green Version]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The Importance of Skip Connections in Biomedical Image Segmentation. In Deep Learning and Data Labeling for Medical Applications; Springer: Cham, Switzerland, 2016; pp. 179–187. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Hua, Y.; Mou, L.; Zhu, X.X. LAHNet: A Convolutional Neural Network Fusing Low- and High-Level Features for Aerial Scene Classification. In Proceedings of the IGARSS 2018–2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 4728–4731. [Google Scholar]
- Bishop, C.M. Neural Networks For Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks For Semantic Segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Liu, Y.; Guo, Y.; Georgiou, T.; Lew, M.S. Fusion that matters: Convolutional fusion networks for visual recognition. Multimed. Tools Appl. 2018, 77, 29407–29434. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, P.; Girshick, R.; Malik, J. Analyzing the Performance of Multilayer Neural Networks for Object Recognition. In Proceedings of the Computer Vision–ECCV 2014; Springer International Publishing: Cham, Switzerland, 2014; pp. 329–344. [Google Scholar]
- Yoo, D.; Park, S.; Lee, J.; In So, K. Multi-scale pyramid pooling for deep convolutional representation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015; pp. 71–80. [Google Scholar]
- Hu, K.; Zhang, Z.; Niu, X.; Zhang, Y.; Cao, C.; Xiao, F.; Gao, X. Retinal vessel segmentation of color fundus images using a multiscale convolutional neural network with an improved cross-entropy loss function. Neurocomputing 2018, 309, 179–191. [Google Scholar] [CrossRef]
- Stehman, S.V. Selecting and interpreting measures of thematic classification accuracy. Remote Sens. Environ. 1997, 62, 77–89. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Biophys. Acta (BBA) Protein Struct. 1975, 405, 442–451. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Swets, J.A. ROC analysis applied to the evaluation of medical imaging techniques. Investig. Radiol. 1979, 14, 109–121. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Chakrabarti, R.; Harper, C.A.; Keeffe, J.E. Diabetic retinopathy management guidelines. Expert Rev. Ophthalmol. 2012, 7, 417–439. [Google Scholar] [CrossRef]
- Sánchez, C.I.; Niemeijer, M.; Dumitrescu, A.V.; Suttorp-Schulten, M.S.A.; Abràmoff, M.D.; van Ginneken, B. Evaluation of a Computer-Aided Diagnosis System for Diabetic Retinopathy Screening on Public Data. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4866–4871. [Google Scholar] [CrossRef]









| Ref. | Major Aspects | Results | |||
|---|---|---|---|---|---|
| Sinthanayothin et al., 2002 [34] | A recursive region growing method to segment both vessels and red lesions. | Se.% | Spec.% | ACC.% | AUC% |
| 78 | 89 | -- | -- | ||
| Larsen et al., 2003 [35] | Fundus images were obtained after dilation of the pupil. | 96.7 | 71.4 | -- | -- |
| Niemeijer et al., 2005 [14] | Computed sixty-eight handcrafted features. Per-image and per-lesion achievements are reported in the 1st and 2nd row of results. | 100 | 87 | -- | -- |
| 30 | -- | -- | -- | ||
| Grisan and Ruggeri 2007 [36] | Local thresholding and pixel density-based detection. | -- | -- | 94 for HEs | -- |
| Garc’ıa et al., 2008 [37] | Introduced a feature selection and a neural network, suggested by Grisan and Ruggeri [36], for image-based classification. | -- | -- | 80 | -- |
| Zhang et al., 2009 [38] | A multi-scale correlation filtering and dynamic-thresholding technique. | -- | Avg. FP = 0.1856 | -- | -- |
| Saleh and Eswaran 2012 [39] | Size and shape-based features. Results in the 1st and 2nd row are of MAs and HEs, respectively. | 84.31 | 93.63 | -- | -- |
| 87.53 | 95.08 | -- | -- | ||
| Ashraf et al., 2014 [40] and 2015 [41] | Textures features analysis by local binary pattern [40], and higher-order statistical features [41]. | 87.48 | 85.99 | 86.15 | 87 |
| 87.92 | 87.54 | 87.44 | 87 | ||
| Srivastava et al., 2015 [42] and 2017 [43] | Frangi filters-based features. | AUC for MAs = 97 & HEs = 87 [42], and 0.92 for both lesions [43]. | |||
| Seoud et al., 2016 [15] | Dynamic shape-based features. | -- | -- | -- | 89.9 |
| Xiao et al., 2017 [44] | A phase congruency-based MAs detection. HEs detection by k- mean clustering and SVM. | ACC. 92 and 93 for MAs and HEs. | |||
| Colomer et al., 2020 [45] | DR lesions were differentiated by combining texture and morphological information. | 75.61 | 75.62 | -- | 83.30 |
| Ref. | Major Aspects | DR Screening Results | Datasets | |
|---|---|---|---|---|
| Orlando et al., 2018 [16] | Handcrafted and CNN-based features were combined to detect red lesions. | AUC% | Se. | e-Ophtha_MA Messidor |
| 90.31 | -- | |||
| 89.32 | 0.916 | |||
| Zago et al., 2020 [17] | Dual CNN models have trained over 65 × 65 image regions. | 91.2 | 0.940 | Messidor IDRiD |
| 81.8 | 0.841 | |||
| Dataset | Number of Images | Resolution | Special Procedures | Dataset Distribution | Purpose |
|---|---|---|---|---|---|
| e-Ophtha_MA [13] | 381 | 1440 × 960 and 2048 × 1360 | Preprocessing for FOV extraction. The 200 × 200 patch generation. Data augmentation | Whole set | Training, Validation and Testing |
| DiaRetDB1 v2.1 [25] | 89 | 1500 × 1152 | Preprocessing for FOV extraction. The 200 × 200 patch generation. | Test set | Cross-Validation |
| ROC [26] | 100 | 768 × 576, 1058 × 1061, and 1389 × 1383 | Training set | ||
| IDRiD [28] | 516 | 4288 × 2848 | Test set | ||
| Messidor [27] | 1200 | 1440 × 960, 2240 × 1488, and 2304 × 1536 | Test set |
| Sr. | Model | ACC (%) | Bias | Variance |
|---|---|---|---|---|
| 1 | AlexNet | 90.06 | Very High | Very High |
| 2 | VGG16 | 91.57 | High | High |
| 3 | GoogLeNet | 92.08 | Moderate | High |
| 4 | Inception-v3 | 90.93 | Moderate | High |
| 5 | ResNet50 | 93.45 | Mild | Moderate |
| 6 | DenseNet | 92.86 | Moderate | Moderate |
| Freeze up to | Residual Blocks | Training Time (Minutes) | Se. (%) in (95 % CI) | Spec. (%) in (95 % CI) | AUC (%) in (95 % CI) | ACC (%) in (95 % CI) |
|---|---|---|---|---|---|---|
| No Freeze | 0 | 2176 | 89.66 (89.60–89.72) | 94.99 (94.98–95) | 93.42 (93.40–93.44) | 93.45 (93.41–93.49) |
| Layer 36 | 3 | 2167 | 91.06 (91.02–91.10) | 97.99 (97.98–98) | 94.73 (94.70–94.76) | 94.71 (94.70–94.72) |
| Layer 48 | 4 | 2160 | 91.44 (91.35–91.53) | 98.13 (97.57–98.69) | 94.30 (93.76–94.84) | 95.02 (94.50–95.54) |
| Layer 58 | 5 | 2156 | 91.94 (91.58–92.30) | 98.66 (98.57–98.75) | 95.66 (95.32–96) | 95.68 (94.86–96.50) |
| Layer 68 | 6 | 2154 | 89.66 (82.82–96.5) | 98.27 (97.5–99.04) | 93.97 (94.5–93.44) | 93.94 (95.50–93.38) |
| Layer 78 | 7 | 2151 | 90.13 (88.26–92) | 97.41 (95–99.82) | 93.78 (92.56–95) | 93.97 (92.94–95) |
| Layer 90 | 8 | 2146 | 85.96 (85.92–86) | 97.51 (96.15–98.87) | 91.74 (91.48–92) | 91.71 (91.42–92) |
| Exp. | Evaluation Type | Data Split Scheme | Enhancement | Loss Function | Bias and Variance | Se. (95 % CI) | Spec. (95 % CI) | AUC 95 % CI) | ACC (95 % CI) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Per Lesion | 1 | RSK1 | Cross Entropy | Low | 0.9267 (0.925–0.9284) | 0.9890 (0.988–0.99) | 0.9578 (0.9576–0.958) | 0.9579 (0.9577–0.9581) |
| 2 | RSK1 and RSK2 | Very Low | 0.9349 (0.933–0.9368) | 0.9886 (0.9885–0.9887) | 0.9675 (0.967–0.968) | 0.9676 (0.968–0.9672) | |||
| 3 | RSK1, RSK2, and GMP | Very Low | 0.9496 (0.945–0.951) | 0.9890 (0.988–0.99) | 0.9678 (0.9666–0.969) | 0.9679 (0.966–0.969) | |||
| 4 | RSK1, RSK2, and GMP | SSE | None | 0.9815 (0.98–0.983) | 0.9972 (0.9971–0.9973) | 0.9893 (0.989–0.9896) | 0.9893 (0.989–0.9896) | ||
| 5 | Per Lesion | 2 | RSK1, RSK2, and GMP | SSE | None | 0.9850 (0.982–0.988) | 0.9911 (0.991–0.992) | 0.9910 (0.99–0.992) | 0.9910 (0.99–0.992) |
| 6 | Per Image | RSK1, RSK2, and GMP | SSE | None | 0.9851 (0.9821–0.9881) | 0.9910 (0.99–0.992) | 0.9910 (0.99–0.992) | 0.9910 (0.99–0.992) |
| Dataset | Evaluation Type | Se. (95 % CI) | Spec. (95 % CI) | AUC (95 % CI) | ACC (95 % CI) |
|---|---|---|---|---|---|
| DiaRetDB1 v2.1 | Per Lesion basis | 0.9498 (0.9410–0.9586) | 0.9660 (0.957–0.975) | 0.9578 (0.955–0.9606) | 0.9448 (0.9551–0.9603) |
| ROC | 0.9270 (0.917–0.937) | 0.9230 (0.917–0.929) | 0.925 (0.916–0.934) | 0.925 (0.916–0.934) | |
| IDRiD | 0.8665 (0.866–0.8670) | 0.8046 (0.8041–0.8051) | 0.8355 (0.8347–0.8363) | 0.8356 (0.8347–0.8365) | |
| Messidor | Per image basis | 0.9421 (0.9411–0.9431) | 0.8940 (0.891–0.897) | 0.9185 (0.905–0.932) | 0.9186 (0.906–0.9312) |
| IDRiD | 0.8426 (0.8398–0.8454) | 0.8044 (0.7907–0.8181) | 0.8235 (0.8176–0.8294) | 0.8235 (0.8076–0.8394) |
| Dataset | Method | Se. in (95 % CI) | AUC in (95 % CI) |
|---|---|---|---|
| e-Ophtha_MA | Orlando et al. [16] | -- | 0.9031 |
| This Work | 0.985 (0.982–0.988) | 0.991 (0.988–0.993) | |
| IDRiD | Zago et al. [17] | 0.841 (0.753-.948) | 0.818 (0.742–0.898) |
| This Work | 0.8426 (0.8398–0.8454) | 0.8235 (0.8176–0.8294) | |
| Messidor | Expert A [86] | 0.945 | 0.922 (0.902–0.936) |
| Expert B [86] | 0.912 | 0.865 (0.789–0.925) | |
| Orlando et al. [16] | 0.916 (0.894–0.943) | 0.893 (0.875–0.912) | |
| Zago et al. d [17] | 0.940 (0.921–0.959) | 0.912 (0.897–0.928) | |
| This Work | 0.942 (0.941–0.943) | 0.918 (0.905–0.932) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, M.N.; Hussain, M.; Habib, Z. Deep Red Lesion Classification for Early Screening of Diabetic Retinopathy. Mathematics 2022, 10, 686. https://doi.org/10.3390/math10050686
Ashraf MN, Hussain M, Habib Z. Deep Red Lesion Classification for Early Screening of Diabetic Retinopathy. Mathematics. 2022; 10(5):686. https://doi.org/10.3390/math10050686
Chicago/Turabian StyleAshraf, Muhammad Nadeem, Muhammad Hussain, and Zulfiqar Habib. 2022. "Deep Red Lesion Classification for Early Screening of Diabetic Retinopathy" Mathematics 10, no. 5: 686. https://doi.org/10.3390/math10050686
APA StyleAshraf, M. N., Hussain, M., & Habib, Z. (2022). Deep Red Lesion Classification for Early Screening of Diabetic Retinopathy. Mathematics, 10(5), 686. https://doi.org/10.3390/math10050686







