A Connectome-Based Digital Twin Caenorhabditis elegans Capable of Intelligent Sensorimotor Behavior
Abstract
:1. Introduction
2. Materials and Methods
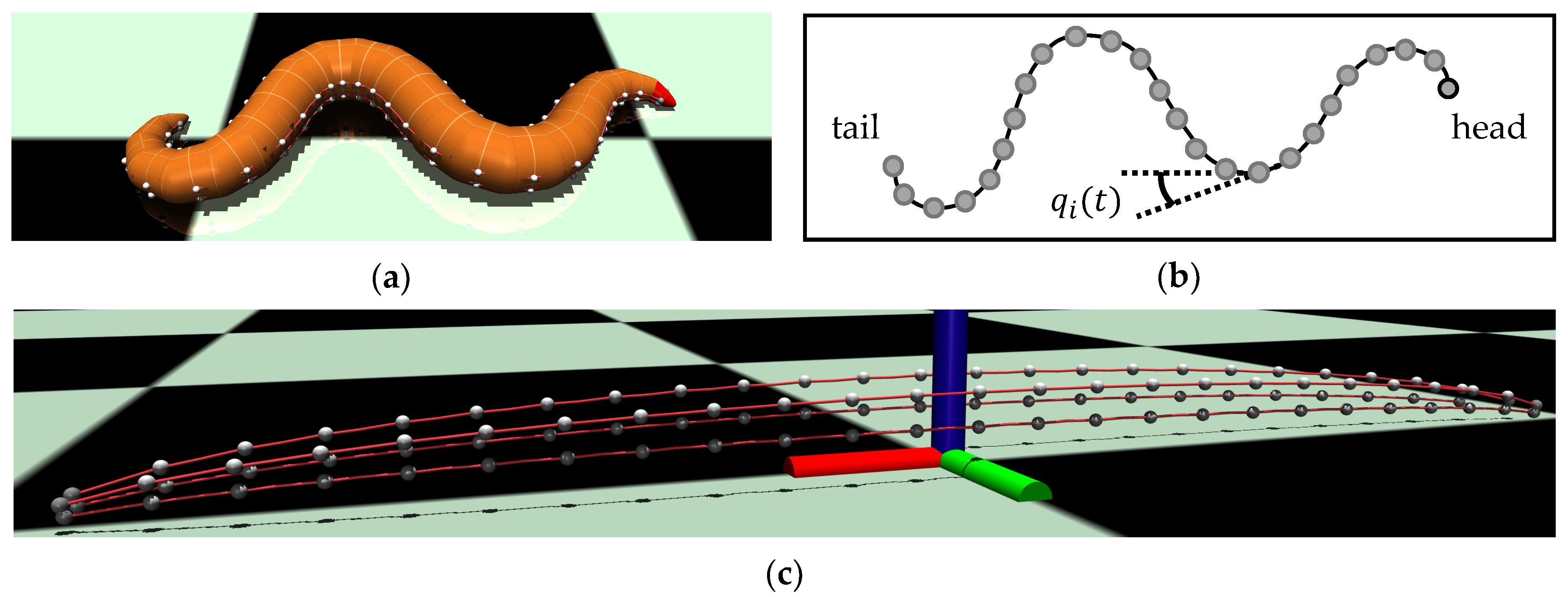
2.1. Digital C. elegans Body
2.2. Virtual Chemotaxis Environment and the Chemotaxis Task
2.3. C. elegans Neural Network Model
2.3.1. Nodes
2.3.2. Chemical Connections and Electrical Connections
2.3.3. Proprioceptive Feedback
2.3.4. Chemosensory Stimuli
2.4. Closed-Loop Simulation of Chemotaxis Behavior with the PID Controller
2.4.1. Posture of Chemotaxis Behavior
2.4.2. Closed-Loop Control with the PID Controller
2.5. Chemotaxis Behavioral Training of the C. elegans Neural Network Model
3. Results
3.1. Chemotaxis Simulation with the Digital Twin C. elegans
3.2. Behavioral Analysis of the Digital Twin C. elegans
3.2.1. Behavioral Performance
3.2.2. Behavioral Statistics
4. Discussion
4.1. Behavioral Mechanism of the Digital Twin C. elegans
4.1.1. Behavioral Mechanism of Sinusoidal Crawling
4.1.2. Behavioral Mechanism of Chemotaxis Navigation
4.2. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DL | dorsal left |
| DR | dorsal right |
| VL | ventral left |
| VR | ventral right |
| PID | proportional–integral–derivative |
| CENN | C. elegans neural network |
| BPTT | backpropagation through time |
| MSE | mean square error |
| SN | sensory neuron |
| IN | interneuron |
| MN | motor neuron |
| HMN | head motor neuron |
| SMN | sublateral motor neuron |
| VCMN | ventral cord motor neuron |
Appendix A. Schematic Drawing of C. elegans

Appendix B. Training and Testing Procedure of the C. elegans Neural Network Model
Appendix B.1. Training Procedure
| Parameter | Value | Comment |
|---|---|---|
| (0.9, 0.999) | Adam’s coefficients | |
| 0 | Adam’s weight decay | |
| 0.05 | learning rate | |
| B | 288 | batch size |
| L | 64 steps (2.56 s) | length of training sequence |
| E | 500 | maximum training epochs |
| 0.0015 | threshold of evaluation loss |
Appendix B.2. Online Testing Procedure
Appendix C. Details of Chemotaxis Simulation with the Digital C. elegans Body

Appendix D. Results of Chemotaxis Simulation with the PID Controller
Appendix D.1. Chemotaxis Simulation
Appendix D.2. Behavioral Analysis of Chemotaxis Simulation

Appendix E. Results of Node Ablation with the Digital Twin C. elegans
| Type | Not Yet Experimentally Proven | Experimentally Proven |
|---|---|---|
| SN6 | ADFR, ASGR, AWBR | |
| SN5 | ADLR, ASHR | |
| SN4 | ALNL, ALNR, PLNL, SDQL | |
| SN3 | ADEL, ALML, DVA | |
| SN1 | CEPDL, CEPVL, CEPVR, IL1DR, IL1R, IL1VL, IL1VR, IL2DR, OLLL, OLLR, OLQDL, OLQVR, URYDL, URYDR, URYVL, URYVR | |
| IN3 | AIAR, AIZR, AVFL, AVHL, AVHR, PVQR, PVR | |
| IN2 | AVJR, AVKR, RICR, RMGL, SAAVL, SAAVR | |
| IN1 | AVEL, AVER, RIAR, RIPR | AVAL, AVAR, AVBL, AVBR, PVCL |
| HMN | RIVL, RIVR, RMDDL, RMDDR, RMDL, RMDR, RMDVR, RMED, RMEL, RMER, RMEV, RMHR, URADR, URAVL, URAVR | |
| SMN | SABD, SABVR, SIAVL, SIBDL, SIBVL, SMBDL, SMBDR, SMBVR, SMDDL, SMDDR, SMDVL, SMDVR | |
| VCMN | AS03, AS11 | DA (DA01, DA02, DA03, DA04, DA07, DA08, DA09), DB (DB01, DB02, DB03, DB05, DB06, DB07), DD (DD01, DD04, DD05, DD06), PDB, VA (VA01, VA02, VA03, VA04, VA05, VA08, VA09, VA11, VA12), VB (VB01, VB02, VB03, VB09, VB10, VB11), VD (VD03, VD07, VD09, VD10, VD11, VD12, VD13) |
| Type | Not Yet Experimentally Proven | Experimentally Proven |
|---|---|---|
| Pharynx | I1R | |
| SN6 | AFDR, AWBL | ASEL, ASER |
| SN4 | SDQR | |
| SN3 | AVM | |
| SN1 | IL1L, IL2DL, IL2L, IL2R | |
| IN4 | AIML, AINL, RIH, RIR | |
| IN3 | ADAR, AIAL, AUAL, PVPL, RIS | |
| IN2 | AVJL, DVC, PVT, RIGL, RMGR | |
| IN1 | RIAL | |
| HMN | RMDVL | |
| SMN | SIAVR, SIBDR, SIBVR | |
| VCMN | AS02, VA06, VA07, VB04, VB07, VD02, VD06 | |
| Sex Motor Neurons | HSNR, VC01, VC06 |
References
- Corsi, A.K.; Wightman, B.; Chalfie, M. A transparent window into biology: A primer on Caenorhabditis elegans. Genetics 2015, 200, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, C.I. Chemosensation in C. elegans. WormBook 2006, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Cook, N.; Venkatachalam, V.; Martinez-Velazquez, L.A.; Zhang, X.; Calvo, A.C.; Hawk, J.; MacInnis, B.L.; Frank, M.; Ng, J.H.R.; et al. Bidirectional thermotaxis in Caenorhabditis elegans is mediated by distinct sensorimotor strategies driven by the AFD thermosensory neurons. Proc. Natl. Acad. Sci. USA 2014, 111, 2776–2781. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, X.; Xin, Q.; Hung, W.; Florman, J.; Huo, J.; Xu, T.; Xie, Y.; Alkema, M.J.; Zhen, M.; et al. Flexible motor sequence generation during stereotyped escape responses. eLife 2020, 9, e56942. [Google Scholar] [CrossRef]
- Susoy, V.; Hung, W.; Witvliet, D.; Whitener, J.E.; Wu, M.; Park, C.F.; Graham, B.J.; Zhen, M.; Venkatachalam, V.; Samuel, A.D. Natural sensory context drives diverse brain-wide activity during C. elegans mating. Cell 2021, 184, 5122–5137. [Google Scholar] [CrossRef]
- Kunitomo, H.; Sato, H.; Iwata, R.; Satoh, Y.; Ohno, H.; Yamada, K.; Iino, Y. Concentration memory-dependent synaptic plasticity of a taste circuit regulates salt concentration chemotaxis in Caenorhabditis elegans. Nat. Commun. 2013, 4, 2210. [Google Scholar] [CrossRef]
- Ha, H.I.; Hendricks, M.; Shen, Y.; Gabel, C.V.; Fang-Yen, C.; Qin, Y.; Colon-Ramos, D.; Shen, K.; Samuel, A.D.; Zhang, Y. Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron 2010, 68, 1173–1186. [Google Scholar] [CrossRef]
- Cook, S.J.; Jarrell, T.A.; Brittin, C.A.; Wang, Y.; Bloniarz, A.E.; Yakovlev, M.A.; Nguyen, K.C.; Tang, L.T.H.; Bayer, E.A.; Duerr, J.S.; et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 2019, 571, 63–71. [Google Scholar] [CrossRef]
- Witvliet, D.; Mulcahy, B.; Mitchell, J.K.; Meirovitch, Y.; Berger, D.R.; Wu, Y.; Liu, Y.; Koh, W.X.; Parvathala, R.; Holmyard, D.; et al. Connectomes across development reveal principles of brain maturation. Nature 2021, 596, 257–261. [Google Scholar] [CrossRef]
- Sarma, G.P.; Lee, C.W.; Portegys, T.; Ghayoomie, V.; Jacobs, T.; Alicea, B.; Cantarelli, M.; Currie, M.; Gerkin, R.C.; Gingell, S.; et al. OpenWorm: Overview and recent advances in integrative biological simulation of Caenorhabditis elegans. Philos. Trans. R. Soc. B 2018, 373, 20170382. [Google Scholar] [CrossRef]
- Gleeson, P.; Lung, D.; Grosu, R.; Hasani, R.; Larson, S.D. c302: A multiscale framework for modelling the nervous system of Caenorhabditis elegans. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170379. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Soh, Z.; Suzuki, M.; Iino, Y.; Tsuji, T. Forward and backward locomotion patterns in C. elegans generated by a connectome-based model simulation. Sci. Rep. 2021, 11, 13737. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Tsuji, T.; Ohtake, H. A model of motor control of the nematode C. elegans with neuronal circuits. Artif. Intell. Med. 2005, 35, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.H.; Berri, S.; Cohen, N. Gait modulation in C. elegans: An integrated neuromechanical model. Front. Comput. Neurosci. 2012, 6, 10. [Google Scholar] [CrossRef]
- Lechner, M.; Grosu, R.; Hasani, R.M. Worm-level control through search-based reinforcement learning. arXiv 2017, arXiv:1711.03467. [Google Scholar]
- Pierce-Shimomura, J.T.; Morse, T.M.; Lockery, S.R. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J. Neurosci. 1999, 19, 9557–9569. [Google Scholar] [CrossRef]
- Ward, A.; Liu, J.; Feng, Z.; Xu, X.S. Light-sensitive neurons and channels mediate phototaxis in C. elegans. Nat. Neurosci. 2008, 11, 916–922. [Google Scholar] [CrossRef]
- Iino, Y.; Yoshida, K. Parallel use of two behavioral mechanisms for chemotaxis in Caenorhabditis elegans. J. Neurosci. 2009, 29, 5370–5380. [Google Scholar] [CrossRef]
- Chalasani, S.H.; Chronis, N.; Tsunozaki, M.; Gray, J.M.; Ramot, D.; Goodman, M.B.; Bargmann, C.I. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 2007, 450, 63–70. [Google Scholar] [CrossRef]
- Luo, L.; Wen, Q.; Ren, J.; Hendricks, M.; Gershow, M.; Qin, Y.; Greenwood, J.; Soucy, E.R.; Klein, M.; Smith-Parker, H.K.; et al. Dynamic encoding of perception, memory, and movement in a C. elegans chemotaxis circuit. Neuron 2014, 82, 1115–1128. [Google Scholar] [CrossRef]
- Itskovits, E.; Ruach, R.; Kazakov, A.; Zaslaver, A. Concerted pulsatile and graded neural dynamics enables efficient chemotaxis in C. elegans. Nat. Commun. 2018, 9, 2866. [Google Scholar] [CrossRef] [PubMed]
- Pierce-Shimomura, J.T.; Dores, M.; Lockery, S.R. Analysis of the effects of turning bias on chemotaxis in C. elegans. J. Exp. Biol. 2005, 208, 4727–4733. [Google Scholar] [CrossRef] [PubMed]
- Appleby, P.A. A model of chemotaxis and associative learning in C. elegans. Biol. Cybern. 2012, 106, 373–387. [Google Scholar] [CrossRef]
- Sakamoto, K.; Soh, Z.; Suzuki, M.; Kurita, Y.; Tsuji, T. A neural network model of Caenorhabditis elegans and simulation of chemotaxis-related information processing in the neural network. In Proceedings of the 2015 SAI Intelligent Systems Conference (IntelliSys), London, UK, 10–11 November 2015; pp. 668–673. [Google Scholar]
- Soh, Z.; Sakamoto, K.; Suzuki, M.; Iino, Y.; Tsuji, T. A computational model of internal representations of chemical gradients in environments for chemotaxis of Caenorhabditis elegans. Sci. Rep. 2018, 8, 17190. [Google Scholar] [CrossRef] [PubMed]
- Palyanov, A.; Khayrulin, S.; Larson, S.D. Three-dimensional simulation of the Caenorhabditis elegans body and muscle cells in liquid and gel environments for behavioural analysis. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170376. [Google Scholar] [CrossRef] [PubMed]
- Palyanov, A.Y.; Khayrulin, S. Sibernetic: A software complex based on the PCI SPH algorithm aimed at simulation problems in biomechanics. Russ. J. Genet. Appl. Res. 2015, 5, 635–641. [Google Scholar] [CrossRef]
- Szigeti, B.; Gleeson, P.; Vella, M.; Khayrulin, S.; Palyanov, A.; Hokanson, J.; Currie, M.; Cantarelli, M.; Idili, G.; Larson, S. OpenWorm: An open-science approach to modeling Caenorhabditis elegans. Front. Comput. Neurosci. 2014, 8, 137. [Google Scholar] [CrossRef]
- Zador, A.; Escola, S.; Richards, B.; Ölveczky, B.; Bengio, Y.; Boahen, K.; Botvinick, M.; Chklovskii, D.; Churchland, A.; Clopath, C.; et al. Catalyzing next-generation Artificial Intelligence through NeuroAI. Nat. Commun. 2023, 14, 1597. [Google Scholar] [CrossRef]
- Todorov, E.; Erez, T.; Tassa, Y. Mujoco: A physics engine for model-based control. In Proceedings of the 2012 IEEE/RSJ International Conference on Intelligent Robots and Systems, Vilamoura-Algarve, Portugal, 7–12 October 2012; pp. 5026–5033. [Google Scholar]
- Girard, L.R.; Fiedler, T.J.; Harris, T.W.; Carvalho, F.; Antoshechkin, I.; Han, M.; Sternberg, P.W.; Stein, L.D.; Chalfie, M. WormBook: The online review of Caenorhabditis elegans biology. Nucleic Acids Res. 2007, 35, D472–D475. [Google Scholar] [CrossRef]
- Dong, X.; Kheiri, S.; Lu, Y.; Xu, Z.; Zhen, M.; Liu, X. Toward a living soft microrobot through optogenetic locomotion control of Caenorhabditis elegans. Sci. Robot. 2021, 6, eabe3950. [Google Scholar] [CrossRef]
- Ward, S. Chemotaxis by the nematode Caenorhabditis elegans: Identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. USA 1973, 70, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Fick, A.V. On liquid diffusion. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1855, 10, 30–39. [Google Scholar] [CrossRef]
- Crank, J. The Mathematics of Diffusion; Oxford University Press: Oxford, UK, 1979. [Google Scholar]
- Ferree, T.C.; Lockery, S.R. Computational rules for chemotaxis in the nematode C. elegans. J. Comput. Neurosci. 1999, 6, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Wicks, S.R.; Roehrig, C.J.; Rankin, C.H. A dynamic network simulation of the nematode tap withdrawal circuit: Predictions concerning synaptic function using behavioral criteria. J. Neurosci. 1996, 16, 4017–4031. [Google Scholar] [CrossRef]
- Lechner, M.; Hasani, R.; Amini, A.; Henzinger, T.A.; Rus, D.; Grosu, R. Neural circuit policies enabling auditable autonomy. Nat. Mach. Intell. 2020, 2, 642–652. [Google Scholar] [CrossRef]
- Kunert, J.; Shlizerman, E.; Kutz, J.N. Low-dimensional functionality of complex network dynamics: Neurosensory integration in the Caenorhabditis elegans connectome. Phys. Rev. E 2014, 89, 052805. [Google Scholar] [CrossRef]
- Kunert, J.M.; Proctor, J.L.; Brunton, S.L.; Kutz, J.N. Spatiotemporal feedback and network structure drive and encode Caenorhabditis elegans locomotion. PLoS Comput. Biol. 2017, 13, e1005303. [Google Scholar] [CrossRef]
- Harris, M.R.; Wytock, T.P.; Kovács, I.A. Computational Inference of Synaptic Polarities in Neuronal Networks. Adv. Sci. 2022, 9, 2104906. [Google Scholar] [CrossRef]
- Fenyves, B.G.; Szilágyi, G.S.; Vassy, Z.; Sőti, C.; Csermely, P. Synaptic polarity and sign-balance prediction using gene expression data in the Caenorhabditis elegans chemical synapse neuronal connectome network. PLoS Comput. Biol. 2020, 16, e1007974. [Google Scholar] [CrossRef]
- Wen, Q.; Po, M.D.; Hulme, E.; Chen, S.; Liu, X.; Kwok, S.W.; Gershow, M.; Leifer, A.M.; Butler, V.; Fang-Yen, C.; et al. Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron 2012, 76, 750–761. [Google Scholar] [CrossRef]
- Wen, Q.; Gao, S.; Zhen, M. Caenorhabditis elegans excitatory ventral cord motor neurons derive rhythm for body undulation. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170370. [Google Scholar] [CrossRef] [PubMed]
- Zhen, M.; Samuel, A.D. C. elegans locomotion: Small circuits, complex functions. Curr. Opin. Neurobiol. 2015, 33, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Thiele, T.R.; Faumont, S.; Ezcurra, M.; Lockery, S.R.; Schafer, W.R. Functional asymmetry in Caenorhabditis elegans taste neurons and its computational role in chemotaxis. Nature 2008, 454, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.P.; Linder, A.N.; Plummer, G.S.; Shaevitz, J.W.; Leifer, A.M. Automatically tracking neurons in a moving and deforming brain. PLoS Comput. Biol. 2017, 13, e1005517. [Google Scholar] [CrossRef]
- Soh, Z.; Suzuki, M.; Tsuji, T. An Estimation Method for Environmental Friction Based on Body Dynamic Model of Caenorhabditis elegans. J. Robot. Netw. Artif. Life 2017, 4, 32–40. [Google Scholar] [CrossRef]
- Doya, K. Recurrent networks: Learning algorithms. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambridge, MA, USA, 2003; pp. 955–960. [Google Scholar]
- Schafer, W.R. Deciphering the neural and molecular mechanisms of C. elegans behavior. Curr. Biol. 2005, 15, R723–R729. [Google Scholar] [CrossRef]
- Xu, T.; Huo, J.; Shao, S.; Po, M.; Kawano, T.; Lu, Y.; Wu, M.; Zhen, M.; Wen, Q. Descending pathway facilitates undulatory wave propagation in Caenorhabditis elegans through gap junctions. Proc. Natl. Acad. Sci. USA 2018, 115, E4493–E4502. [Google Scholar] [CrossRef]
- Yan, G.; Vértes, P.E.; Towlson, E.K.; Chew, Y.L.; Walker, D.S.; Schafer, W.R.; Barabási, A.L. Network control principles predict neuron function in the Caenorhabditis elegans connectome. Nature 2017, 550, 519–523. [Google Scholar] [CrossRef]
- Liu, P.; Chen, B.; Wang, Z.W. GABAergic motor neurons bias locomotor decision-making in C. elegans. Nat. Commun. 2020, 11, 5076. [Google Scholar] [CrossRef]
- Liu, Q.; Kidd, P.B.; Dobosiewicz, M.; Bargmann, C.I. C. elegans AWA olfactory neurons fire calcium-mediated all-or-none action potentials. Cell 2018, 175, 57–70. [Google Scholar] [CrossRef]
- Jiang, J.; Su, Y.; Zhang, R.; Li, H.; Tao, L.; Liu, Q. C. elegans enteric motor neurons fire synchronized action potentials underlying the defecation motor program. Nat. Commun. 2022, 13, 2783. [Google Scholar] [CrossRef] [PubMed]
- Lockery, S.R.; Goodman, M.B. The quest for action potentials in C. elegans neurons hits a plateau. Nat. Neurosci. 2009, 12, 377–378. [Google Scholar] [CrossRef] [PubMed]
- Yemini, E.; Lin, A.; Nejatbakhsh, A.; Varol, E.; Sun, R.; Mena, G.E.; Samuel, A.D.; Paninski, L.; Venkatachalam, V.; Hobert, O. NeuroPAL: A multicolor atlas for whole-brain neuronal identification in C. elegans. Cell 2021, 184, 272–288. [Google Scholar] [CrossRef] [PubMed]
- Altun, Z.; Hall, D. Muscle System, Somatic Muscle. WormAtlas. 2009. Available online: https://www.wormatlas.org/hermaphrodite/musclesomatic/MusSomaticframeset.html (accessed on 22 April 2023).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 1–12. [Google Scholar]









| Parameter | Unit | Initialization | Constraint | Number |
|---|---|---|---|---|
| second (s) | 1 | 469 | ||
| millivolt (mV) | - | 469 | ||
| millivolt (mV) | 1 | 3324 2 | ||
| - | 3 | 4869 | ||
| - | 3 | 1433 | ||
| - | - | 1223 | ||
| 4 | - | 1 | 3 | 3 |
| Controller | PID Controller | C. elegans Neural Network | ||||
|---|---|---|---|---|---|---|
| Initial Position (mm) | Crawling Distance (mm) | Chemotaxis Index | Initial Concentration | Crawling Distance (mm) | Chemotaxis Index | Initial Concentration |
| Random | ||||||
| 0.61 | 0.61 | |||||
| 0.14 | 0.14 | |||||
| 0.01 | 0.01 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Yu, Y.; Xue, X. A Connectome-Based Digital Twin Caenorhabditis elegans Capable of Intelligent Sensorimotor Behavior. Mathematics 2023, 11, 2442. https://doi.org/10.3390/math11112442
Chen Z, Yu Y, Xue X. A Connectome-Based Digital Twin Caenorhabditis elegans Capable of Intelligent Sensorimotor Behavior. Mathematics. 2023; 11(11):2442. https://doi.org/10.3390/math11112442
Chicago/Turabian StyleChen, Zhongyu, Yuguo Yu, and Xiangyang Xue. 2023. "A Connectome-Based Digital Twin Caenorhabditis elegans Capable of Intelligent Sensorimotor Behavior" Mathematics 11, no. 11: 2442. https://doi.org/10.3390/math11112442
APA StyleChen, Z., Yu, Y., & Xue, X. (2023). A Connectome-Based Digital Twin Caenorhabditis elegans Capable of Intelligent Sensorimotor Behavior. Mathematics, 11(11), 2442. https://doi.org/10.3390/math11112442







