Graph Convolutional Network Design for Node Classification Accuracy Improvement
Abstract
:1. Introduction
- We propose a GCN layer for improving the classification accuracy of nodes in graph data.
- The proposed approach is tested with seven different datasets and is compared with related previous studies.
2. Graph Architecture
2.1. Graph Notation
2.2. Graph Convolutional Networks
3. Proposed Model
| Algorithm 1 Graph Convolution Layer |
| Input , Output: l, 1: 2: 3: 4: 5: 6: 7: End. |
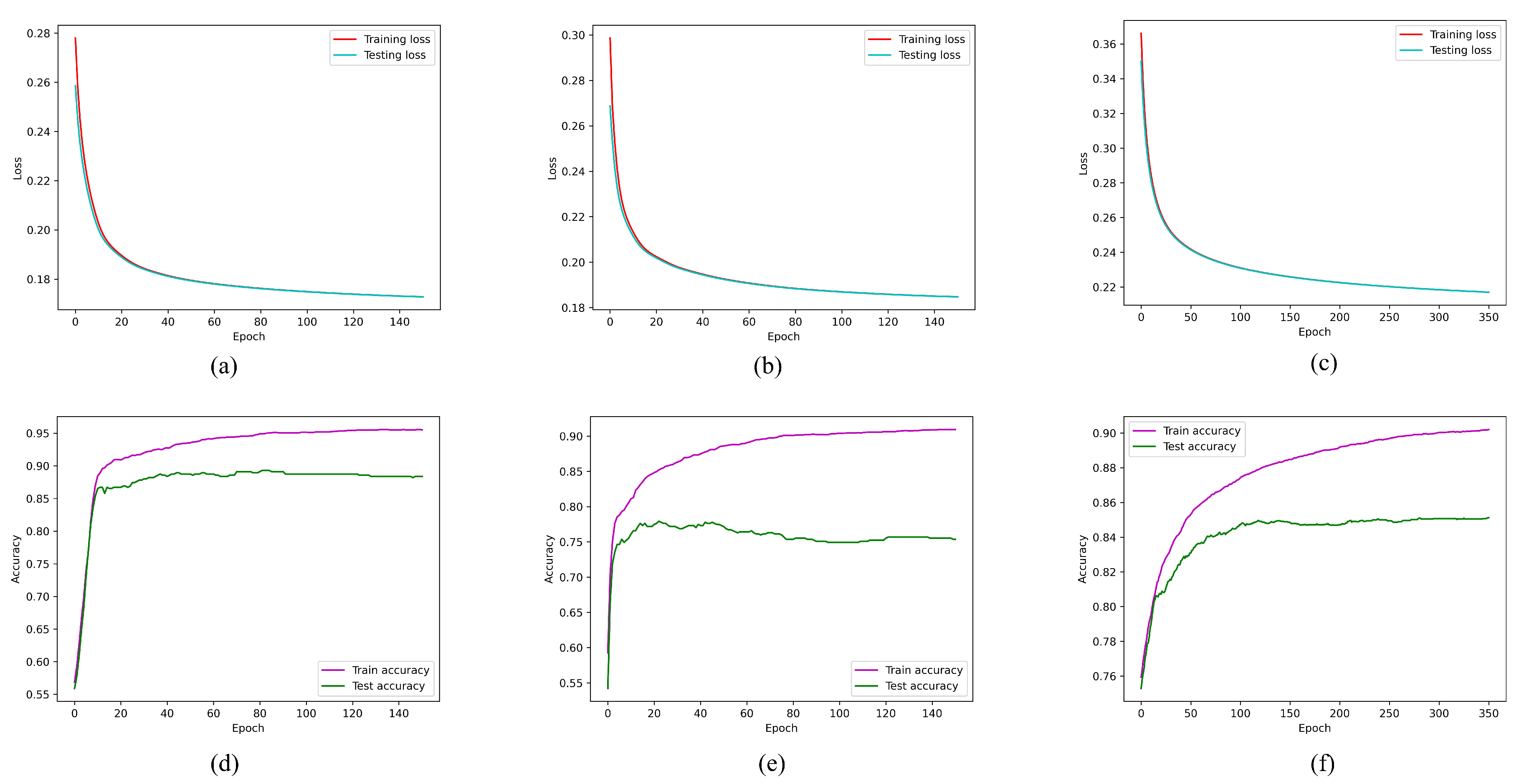
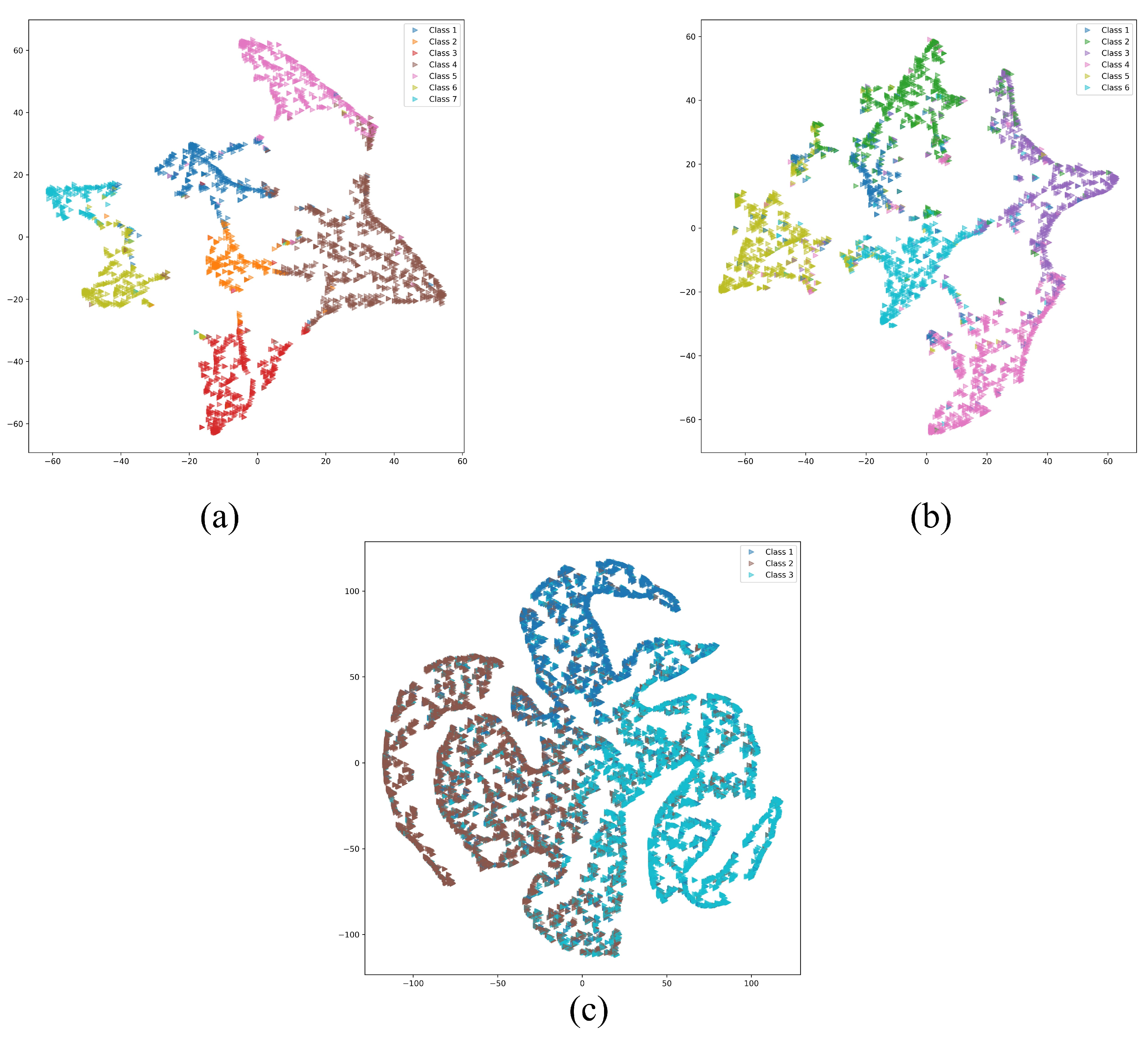
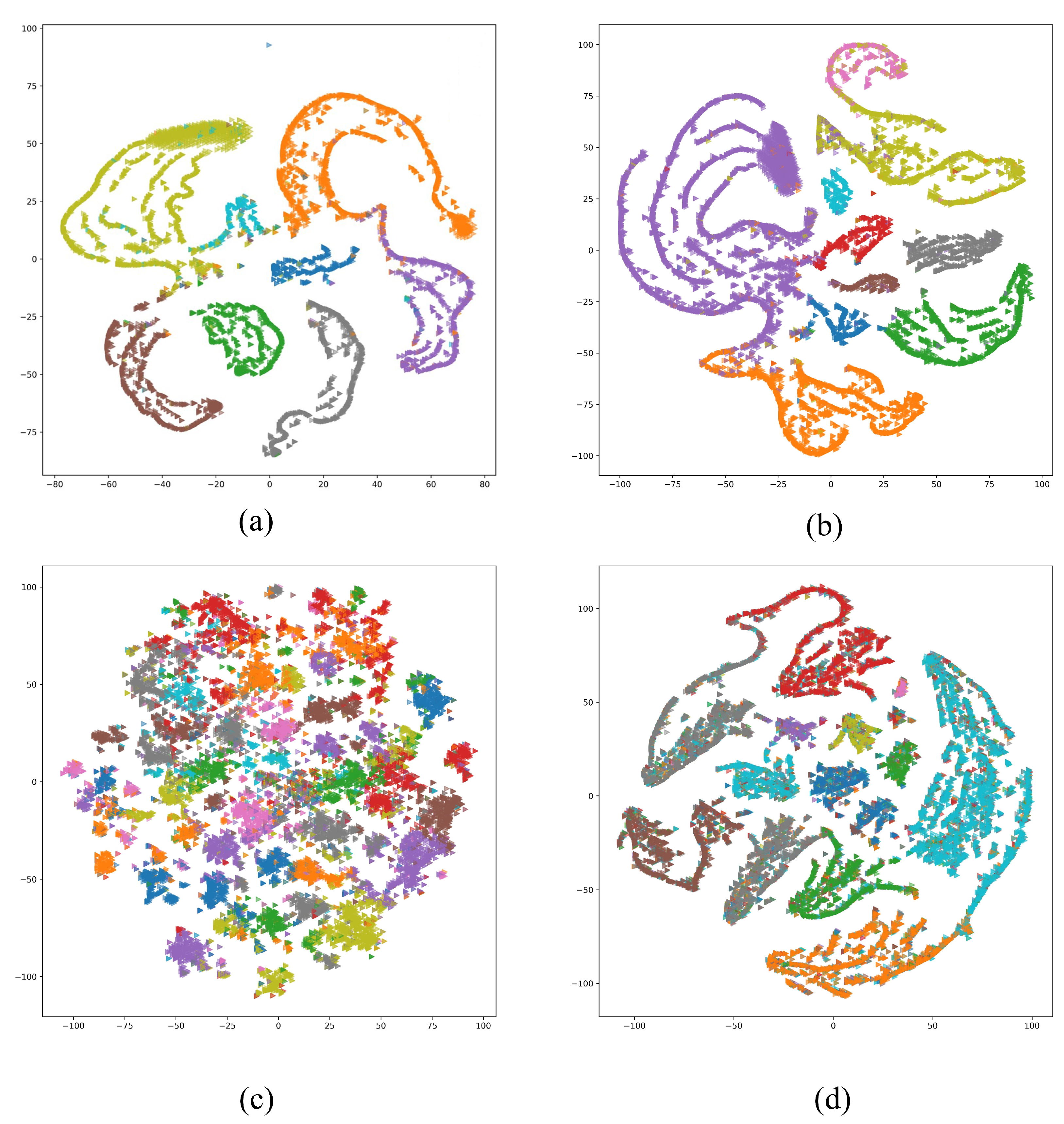
4. Results
| Algorithm 2 Proposed model training and evaluation steps. |
| 1: Input: Train data feature (), labels(y), Adjacency matrix (, Loss function, optimizer(Adam), coefficient value (q), max-episode. 2: Output: Optimized model. 3: A learnable parameters list is created with size of . 4: Adam optimizer function is initiated. for episode is not max-episode do 5: Get parameters state. 6: Using Algorithm 1 calculate the loss for the current parameters. 7: Update the value of parameters using the optimizer function. end for 8: Save the model with the minimized loss value. 9: Return the model. 10: Run inference for the input feature and find node class. 11: End |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.; Chien, J.; Yong, J.; Kuang, R. Network-based machine learning and graph theory algorithms for precision oncology. NPJ Precis. Oncol. 2017, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Pan, S.; Chen, F.; Long, G.; Zhang, C.; Philip, S.Y. A comprehensive survey on graph neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4–24. [Google Scholar] [CrossRef] [PubMed]
- Girvan, M.; Newman, M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Rosvall, M.; Bergstrom, C.T. Maps of random walks on complex networks reveal community structure. Proc. Natl. Acad. Sci. USA 2008, 105, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Li, B.; Pi, D. Learning deep neural networks for node classification. Expert Syst. Appl. 2019, 137, 324–334. [Google Scholar] [CrossRef]
- Ying, R.; He, R.; Chen, K.; Eksombatchai, P.; Hamilton, W.L.; Leskovec, J. Graph convolutional neural networks for web-scale recommender systems. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 974–983. [Google Scholar]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Hamilton, W.; Ying, Z.; Leskovec, J. Inductive representation learning on large graphs. Adv. Neural Inf. Process. Syst. 2017, 30, 1025–1035. [Google Scholar]
- Chen, J.; Ma, T.; Xiao, C. Fastgcn: Fast learning with graph convolutional networks via importance sampling. arXiv 2018, arXiv:1801.10247. [Google Scholar]
- Dabhi, S.; Parmar, M. Nodenet: A graph regularised neural network for node classification. arXiv 2020, arXiv:2006.09022. [Google Scholar]
- Prakash, S.K.A.; Tucker, C.S. Node classification using kernel propagation in graph neural networks. Expert Syst. Appl. 2021, 174, 114655. [Google Scholar] [CrossRef]
- Maurya, S.K.; Liu, X.; Murata, T. Simplifying approach to node classification in Graph Neural Networks. J. Comput. Sci. 2022, 62, 101695. [Google Scholar] [CrossRef]
- Wang, T.; Jin, D.; Wang, R.; He, D.; Huang, Y. Powerful graph convolutional networks with adaptive propagation mechanism for homophily and heterophily. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual conference, 22 Feburary–1 March 2022; Volume 36, pp. 4210–4218. [Google Scholar]
- Zhang, S.; Tong, H.; Xu, J.; Maciejewski, R. Graph convolutional networks: A comprehensive review. Comput. Soc. Netw. 2019, 6, 1–23. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, S.; Gilvary, C.; Elemento, O.; Zhou, J.; Wang, F. Graph convolutional networks for computational drug development and discovery. Briefings Bioinform. 2020, 21, 919–935. [Google Scholar] [CrossRef] [PubMed]
- Abu-El-Haija, S.; Kapoor, A.; Perozzi, B.; Lee, J. N-gcn: Multi-scale graph convolution for semi-supervised node classification. In Proceedings of the 35th Uncertainty in Artificial Intelligence Conference, Tel Aviv, Israel, 22–25 July 2019; pp. 841–851. [Google Scholar]
- Fout, A.; Byrd, J.; Shariat, B.; Ben-Hur, A. Protein interface prediction using graph convolutional networks. Adv. Neural Inf. Process. Syst. 2017, 30, 6533–6542. [Google Scholar]
- Pei, H.; Wei, B.; Chang, K.C.C.; Lei, Y.; Yang, B. Geom-gcn: Geometric graph convolutional networks. arXiv 2020, arXiv:2002.05287. [Google Scholar]
- Dong, S.; Zhou, W. Improved influential nodes identification in complex networks. J. Intell. Fuzzy Syst. 2021, 41, 6263–6271. [Google Scholar] [CrossRef]
- Liao, R.; Zhao, Z.; Urtasun, R.; Zemel, R.S. Lanczosnet: Multi-scale deep graph convolutional networks. arXiv 2019, arXiv:1901.01484. [Google Scholar]
- Hong, D.; Gao, L.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph convolutional networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2020, 59, 5966–5978. [Google Scholar] [CrossRef]
- Chiang, W.L.; Liu, X.; Si, S.; Li, Y.; Bengio, S.; Hsieh, C.J. Cluster-gcn: An efficient algorithm for training deep and large graph convolutional networks. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 257–266. [Google Scholar]
- Xu, K.; Hu, W.; Leskovec, J.; Jegelka, S. How powerful are graph neural networks? arXiv 2018, arXiv:1810.00826. [Google Scholar]
- Sen, P.; Namata, G.; Bilgic, M.; Getoor, L.; Galligher, B.; Eliassi-Rad, T. Collective classification in network data. AI Mag. 2008, 29, 93. [Google Scholar] [CrossRef]
- Rossi, R.; Ahmed, N. The network data repository with interactive graph analytics and visualization. In Proceedings of the AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 29. [Google Scholar]
- Namata, G.; London, B.; Getoor, L.; Huang, B.; Edu, U. Query-driven active surveying for collective classification. In Proceedings of the 10th International Workshop on Mining and Learning with Graphs, Edinburgh, Scotland, 1 July 2012; Volume 8, p. 1. [Google Scholar]
- McAuley, J.; Targett, C.; Shi, Q.; Van Den Hengel, A. Image-based recommendations on styles and substitutes. In Proceedings of the 38th International ACM SIGIR Conference on Research and Development in Information Retrieval, Santiago, Chile, 9–13 August 2015; pp. 43–52. [Google Scholar]
- Bojchevski, A.; Günnemann, S. Deep gaussian embedding of graphs: Unsupervised inductive learning via ranking. arXiv 2017, arXiv:1707.03815. [Google Scholar]
- Velickovic, P.; Fedus, W.; Hamilton, W.L.; Liò, P.; Bengio, Y.; Hjelm, R.D. Deep graph infomax. ICLR (Poster) 2019, 2, 4. [Google Scholar]
- Xu, K.; Li, C.; Tian, Y.; Sonobe, T.; Kawarabayashi, K.I.; Jegelka, S. Representation learning on graphs with jumping knowledge networks. In Proceedings of the International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; pp. 5453–5462. [Google Scholar]
- Gasteiger, J.; Bojchevski, A.; Günnemann, S. Predict then propagate: Graph neural networks meet personalized pagerank. arXiv 2018, arXiv:1810.05997. [Google Scholar]
- Gasteiger, J.; Weißenberger, S.; Günnemann, S. Diffusion improves graph learning. Adv. Neural Inf. Process. Syst. 2019, 32, 1336–13378. [Google Scholar]
- Vashishth, S.; Yadav, P.; Bhandari, M.; Talukdar, P. Confidence-based graph convolutional networks for semi-supervised learning. In Proceedings of the 22nd International Conference on Artificial Intelligence and Statistics, Naha, Okinawa, Japan, 16–18 April 2019; pp. 1792–1801. [Google Scholar]
- Zhu, S.; Pan, S.; Zhou, C.; Wu, J.; Cao, Y.; Wang, B. Graph geometry interaction learning. Adv. Neural Inf. Process. Syst. 2020, 33, 7548–7558. [Google Scholar]
- Zheng, R.; Chen, W.; Feng, G. Semi-supervised node classification via adaptive graph smoothing networks. Pattern Recognit. 2022, 124, 108492. [Google Scholar] [CrossRef]
- Chapelle, O.; Scholkopf, B.; Zien, A. Semi-supervised learning (chapelle, o. et al., eds.; 2006)[book reviews]. IEEE Trans. Neural Netw. 2009, 20, 542. [Google Scholar] [CrossRef]
- Veličković, P.; Fedus, W.; Hamilton, W.L.; Liò, P.; Bengio, Y.; Hjelm, R.D. Deep graph infomax. arXiv 2018, arXiv:1809.10341. [Google Scholar]
- Li, Q.; Han, Z.; Wu, X.M. Deeper insights into graph convolutional networks for semi-supervised learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Orleans, LA, USA, 2–7 February 2018; Volume 32. [Google Scholar]
- Velickovic, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. Stat 2017, 1050, 10–48550. [Google Scholar]
- Xu, H.; Jiang, B.; Huang, L.; Tang, J.; Zhang, S. Multi-head collaborative learning for graph neural networks. Neurocomputing 2022, 499, 47–53. [Google Scholar] [CrossRef]



| Parameter | Symbol |
|---|---|
| Graph | |
| Set of node or vertices | |
| Set of edges | |
| Adjacency matrix | |
| Feature matrix | |
| Signal vector | |
| GNN output at the lth layer | |
| Weight matrix | |
| Identity matrix | |
| Activation matrix | |
| Activation function | |
| Sigmoid function | |
| Output of final layer | Y |
| Model depth | K |
| Dataset | Nodes | Edges | Features | Labels | Label Rate | Edge Density |
|---|---|---|---|---|---|---|
| Cora [26] | 2708 | 5429 | 1433 | 7 | 0.0563 | 0.0004 |
| Citeseer [27] | 3327 | 4732 | 3703 | 6 | 0.0569 | 0.0004 |
| Pubmed [28] | 19,717 | 44,338 | 500 | 3 | 0.0030 | 0.0001 |
| Amazon photo [29] | 7487 | 119,043 | 745 | 8 | 0.0214 | 0.0011 |
| Amazon computers [29] | 13,752 | 491,722 | 767 | 10 | 0.0149 | 0.0007 |
| Cora Full [30] | 19,793 | 65,311 | 8710 | 70 | 0.0745 | 0.0001 |
| Coauthor CS [29] | 18,333 | 163,788 | 6805 | 15 | 0.0149 | 0.0007 |
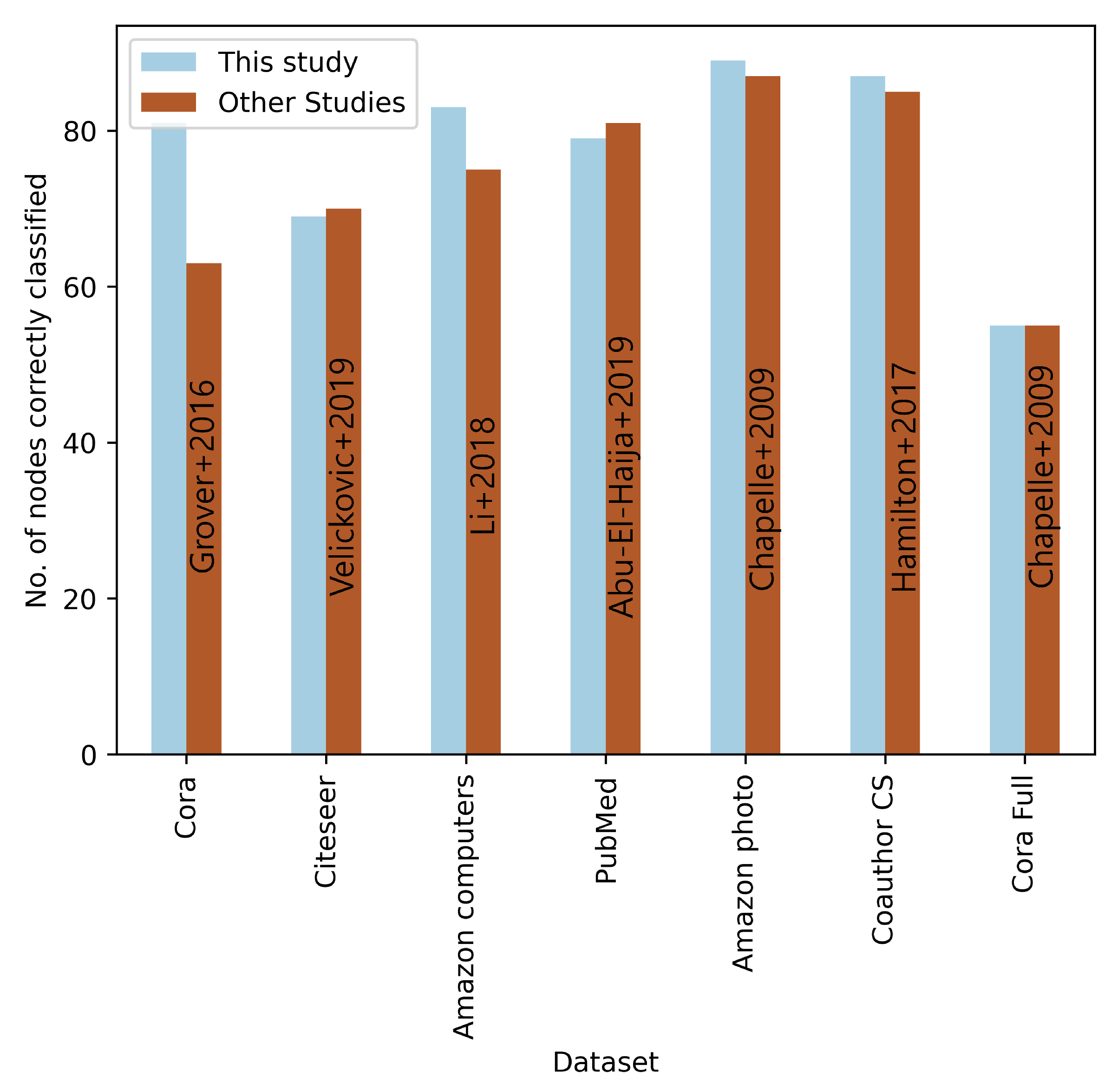
| Ref. | Method | Classification Data | Cora | Citeseer | Pubmed |
|---|---|---|---|---|---|
| [9] | Node2vec | % | % | % | |
| [31] | GAT | % | % | % | |
| [31] | GAT | % | % | % | |
| [32] | JK NET | % | % | % | |
| [33] | APPNP | % | % | % | |
| [34] | APPNP | % | % | % | |
| [35] | ConfGNN | % | % | % | |
| [18] | NGCN | % | % | % | |
| [36] | GIL | % | % | ||
| [37] | AGSN | % | % | ||
| This study | GCN | % | % |
| Ref. | Method | Classification Data | Amazon Photo | Amazon Computers | Cora Full | Coauthor CS |
|---|---|---|---|---|---|---|
| [38] | LabelProp NL | % | % | % | % | |
| [10] | GraphSAGE | , K, | % | % | % | % |
| [39] | DGI | % | % | % | % | |
| [40] | Co-training | % | % | % | % | |
| [5] | GCN | % | % | % | % | |
| [41] | GAT | % | % | % | % | |
| [42] | MCL-GCNs | % | % | |||
| This study | GCN | % | % |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sejan, M.A.S.; Rahman, M.H.; Aziz, M.A.; Baik, J.-I.; You, Y.-H.; Song, H.-K. Graph Convolutional Network Design for Node Classification Accuracy Improvement. Mathematics 2023, 11, 3680. https://doi.org/10.3390/math11173680
Sejan MAS, Rahman MH, Aziz MA, Baik J-I, You Y-H, Song H-K. Graph Convolutional Network Design for Node Classification Accuracy Improvement. Mathematics. 2023; 11(17):3680. https://doi.org/10.3390/math11173680
Chicago/Turabian StyleSejan, Mohammad Abrar Shakil, Md Habibur Rahman, Md Abdul Aziz, Jung-In Baik, Young-Hwan You, and Hyoung-Kyu Song. 2023. "Graph Convolutional Network Design for Node Classification Accuracy Improvement" Mathematics 11, no. 17: 3680. https://doi.org/10.3390/math11173680






