In this section, the proposed EGBO is evaluated in two experiments; the first is global optimization, and the second is feature selection. All results of the proposed EGBO are compared to six algorithms: GBO, particle swarm optimization (PSO) [
21], genetic algorithms (GA) [
22], differential evolution (DE) [
23], dragonfly algorithm (DA) [
24], and moth-flame optimization (MFO) [
25]. These algorithms are selected because they have shown stability and good results in the literature. They have different behaviors in exploring the search space; for instance, the PSO uses the particle’s position and velocity, whereas the GA uses three phases: selection, crossover, and mutation.
The parameters of all algorithms were set as mentioned in their original paper, whereas the global parameters were as follows: the search agents’ numbers = 25 and the maximum iteration number = 100. The number of the fitness function evaluation is set to 2500 as a stop condition, and each algorithm is applied with 30 independent runs.
4.1. First Experiment: Solving Global Optimization Problem
In this section, the proposed EGBO method is assessed using CEC2019 [
26] benchmark functions to solve global optimization functions and the results are compared to some well-known algorithms. This experiment aims to evaluate the proposed EGBO in solving different types of test functions.
The comparison is performed using a set of performance measures: the mean (Equation (
29)), min, max, and standard deviations (Equation (
30)) of the fitness functions, and the computation times for all algorithms. All experimental results are listed in
Table 1,
Table 2,
Table 3,
Table 4 and
Table 5.
where
f is the produced fitness value and
denotes the mean value of
f.
N indicates the size of the sample.
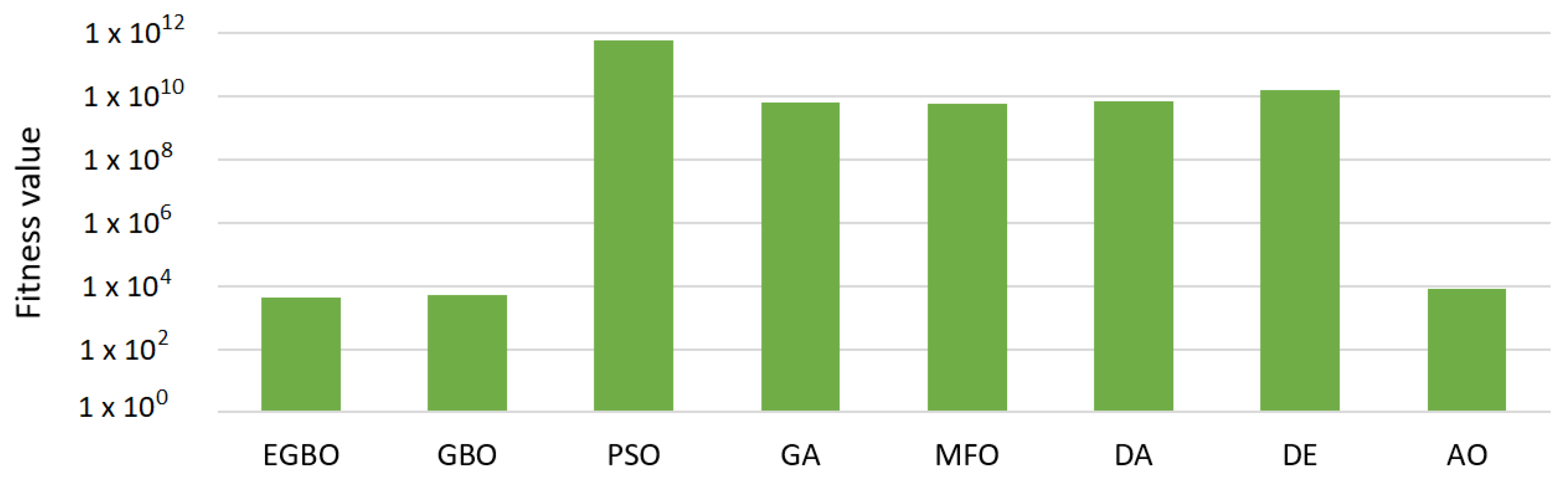
Table 1 tabulates the results of the mean of the fitness function measure. In that table, the proposed EGBO outperformed the other methods in six out of ten functions (i.e., F1–F5, F9); therefore, it was ranked first. The GA was ranked second by obtaining the best values in two functions, F7 and F8. The GBO came in the third rank with the best values in three functions, F1–F3, followed by the MFO. The rest of the algorithms were ordered: the AO, DE, DA, and PSO, respectively.
Figure 1 illustrates the average fitness function values for all methods over all functions.
The results of the standard deviation measure are reported in
Table 2 to show the stability of each method. In this measure, the smallest value indicates good stability behavior within the independent runs. The results show that the proposed EGBO showed the best stability in 70% of the functions compared to the other methods followed by the DE and GBO. The GA, AO, and MFO came in the fourth, fifth, and sixth ranks.
The results of the max measure are listed in
Table 3. In this table, the worst value of each algorithm for each function is recorded. As seen in the table, the worst values of the proposed EGBO were better than the compared methods in six out of ten functions (i.e., F1–F3, F5, and F8–F9). The GA was ranked second by obtaining the best values in both F6 and F10. The MFO and GBO came in the third and fourth ranks, followed by the DE and AO.
Furthermore, the results of the min measure are presented in
Table 4. In this measure, the best values so far obtained by each algorithm are recorded. As seen in the results, the proposed EGBO obtained the best values in four out of ten functions (i.e., F1, F2, F5, and F9) and obtained good results in the rest of the functions. The GA ranked second by obtaining the minimum values in F4 and F8 functions. The GBO came in third, followed by the MFO, DA, D, PSO, and AO.
The computation time is also considered in
Table 5. In this measure, the MFO algorithm was the fastest among all methods, followed by the PSO and AO. The EGBO consumed an acceptable computation time in each function and was ranked fourth, followed by GBO, GA, and DE. The slower algorithm was the DA; it recorded the longest computation time in each function.
Figure 2 illustrates the average computation times for all methods over all functions.
Moreover,
Figure 3 illustrates the convergence curves for all methods during the optimization process. From this figure, it can be seen that the proposed EGBO can effectively maintain the populations to converge toward the optimal value as shown in F1, F4, F5, and F9. The PSO, GBO, and MFO also showed second-best convergence. In contrast, the DA algorithm showed the worst convergence in most functions.
Furthermore,
Table 6 records the Wilcoxon rank-sum test as a statistical test to check if there is a statistical difference between the EGBO and the compared algorithms at a
p-value < 0.05. The results, as recorded in
Table 6, show significant differences between the proposed EGBO and the compared algorithms, especially with MFO, DA, DE, and AO, which indicates the effectiveness of the EGBO in solving global optimization problems.
In light of the above results, the proposed EGBO method can effectively solve global optimization problems and provide promising results compared to the other methods. Therefore, in the next section, it will be evaluated in solving different feature selection problems.
4.2. Second Experiment: Solving Feature Selection Problem
In this part, the proposed EGBO method is assessed in solving different feature selection problems using twelve well-known feature selection datasets from [
27];
Table 7 lists their descriptions.
The performance of the proposed method is evaluated by five measures, namely fitness value (Equation (
31)), accuracy (Acc) as in Equation (
32), standard deviation (Std) as in Equation (
30), number of selected features, and the computation time for each algorithm. The results are recorded in
Table 8,
Table 9,
Table 10,
Table 11,
Table 12 and
Table 13.
where
denotes the error value of the classification process (kNN is used in this paper as a classifier). The second term defines the selected feature number.
is used to balance the number of the selected features and the classification error.
c and
C are the current and ’all-feature’ numbers, respectively.
where the number of positive classes correctly classified is (
), whereas the number of negative classes correctly classified is (
). Both
and
are the numbers of positive and negative classes incorrectly classified, respectively.
Table 8 shows the results of the fitness function values for all algorithms over all datasets. These results indicate that the proposed EGBO obtained the best fitness value among the compared algorithms and performed well in all datasets. The DE obtained the second-best results in 75% of the datasets. The PSO came in third, followed by GBO, GA, DA, AO, and MFO.
Figure 4 illustrates the average of this measure over all datasets, which shows that the EGBO obtained the smallest average among all methods.
The stability results of all algorithms are listed in
Table 9. As in that table, the proposed EGBO showed the most stable algorithm in 7 out of 12 datasets: glass, tic-tac-toe, waveform, clean1data, SPECT, Zoo, and heart. The GA and DE ranked second and third for stability, respectively, followed by PSO, DA, and GBO.
As mentioned above, this experiment aims to decrease each dataset’s features by deleting the redundant descriptors and keeping the best ones. Therefore,
Table 10 reports the number of selected features in each dataset. From
Table 10, we can see that the EGBO chose the smallest number of features in 9 out of 12 datasets and showed good performance in the remaining datasets. The GBO was ranked second, followed by MFO, AO, DE, PSO, and GA. In this regard, the small number of features is sometimes the best; therefore, we used the classification accuracy measure to evaluate the obtained feature by each algorithm.
Consequently, the classification accuracy measure was used to evaluate the proposed method’s ability to classify the samples of each dataset correctly. The results of this measure were recorded in
Table 11. This table shows that the EGBO ranked first; it outperformed the other methods and obtained high-accuracy results in all datasets. The DE came in the second rank, whereas the PSO obtained the third, followed by the GBO, GA, DA, and AO. In contrast, the MFO recorded the worst accuracy measures in most datasets.
The computation times for all methods were also measured.
Table 12 records the time consumed by each algorithm. By analyzing the results, we can note that all algorithms consumed similar times to some extent. In detail, the EGBO was the fastest in 59% of the datasets, followed by PSO, MFO, DE, GBO, and DA, respectively, whereas the GA and AO recorded the longest computation time.
Furthermore, an example of the convergence curves for all methods is illustrated in
Figure 5. In this figure, a sample of the datasets is presented, which shows that the proposed EGBO method converged to the optimal values faster than the compared algorithms, which indicates the good convergence behaviors of the EGBO when solving feature selection problems.
In addition,
Table 13 shows the Wilcoxon rank-sum test as a statistical test to check if there is a statistical difference between the proposed method and the other algorithms at a
p-value < 0.05. From this table, we can see that there are significant differences between the proposed methods and the other algorithms in most datasets, indicating the EGBO’s effectiveness in solving different feature selection problems.
From the previous results, the proposed EGBO method outperformed the compared algorithms in most cases in solving global optimization problems and selecting the most relative features. These promising results can be attributed to a few reasons, e.g., the local escaping operator of the GBO was extended to include the expanded and narrowed exploration behavior, which added more flexibility and reliability to the traditional GBO algorithm. This extension increased the ability of the GBO to explore more areas in the search space, reflecting on effectively reaching the optimal point. On the other hand, although the EGBO obtained good results in most cases, it failed to show the best results in the computation time measure, namely in the global optimization experiment. This defect can be due to the traditional GBO consuming a relatively longer time than the compared algorithm when performing an optimization task. Therefore, this defect can be studied in future work. Generally, the exploration behaviors used in the EGBO increased the performance of the traditional algorithm in terms of performance measures.