A Fuzzy-Based Fast Feature Selection Using Divide and Conquer Technique in Huge Dimension Dataset
Abstract
:1. Introduction
2. Feature Selection Methods
2.1. Filter Methods
2.1.1. Mutual Information
2.1.2. Correlation
2.1.3. Chi-Squared
2.2. Wrapper Method
2.3. Embedded Method
3. Related Work
3.1. Minimum Redundancy Maximal Relevance Criteria (mRMR)
3.2. Least Angle Regression (LARS)
- A slight adjustment to LARS implements LASSO and calculates all possible LASSO estimates for a given problem.
- Another variation of LARS efficiently executes forward stagewise linear regression.
- A rough estimate of the degrees of freedom of a LARS estimate is available, allowing for a calculated prediction error estimate based on C. This enables a deliberate choice among the possible LARS estimates.
3.3. Hilbert–Schmidt Independence Criterion LASSO (HSIC-LASSO)
3.4. Conditional Covariance Minimization (CCM)
3.5. Binary Coyote Optimization Algorithm (BCOA)
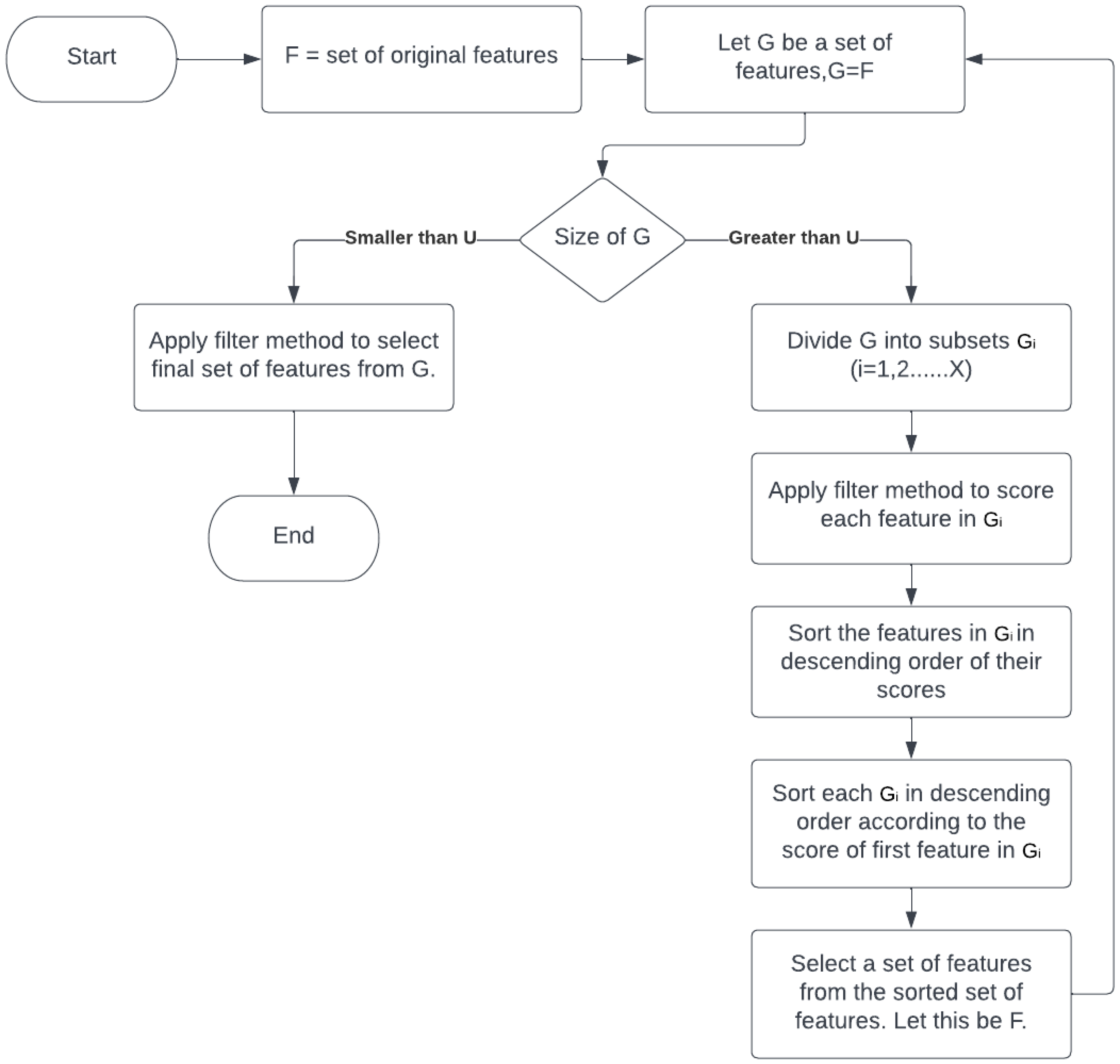
4. Proposed Method
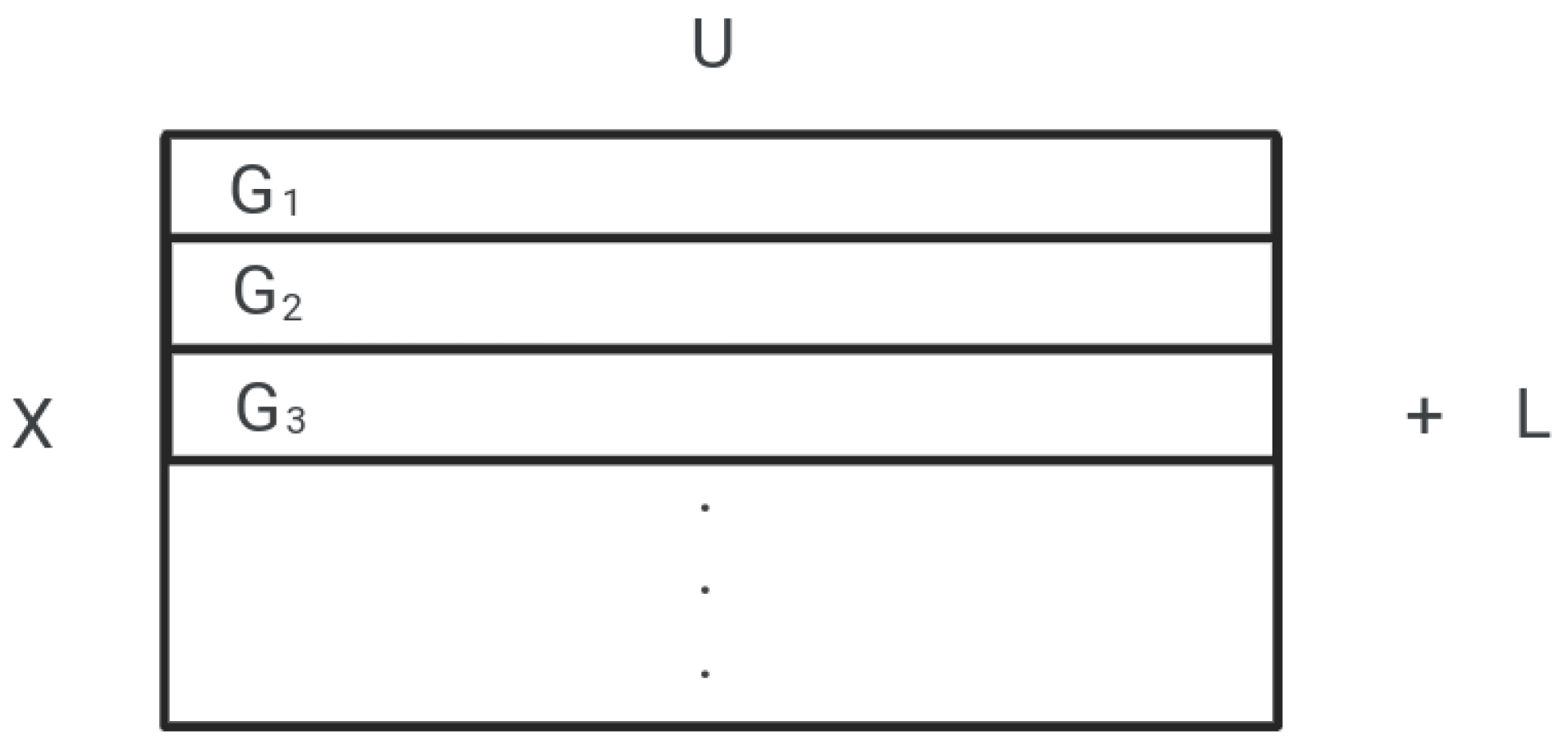
| Let, U = number of features; efficiently selected by feature selection method from a group of features. U will be the number of features in each subset, any feature selection method would have to select a maximum of U features from each subset. To divide the features into equal subsets, Let, G = some set of features. Initially, we will take the original set of features, G = F. Let, X = number of subsets, L = set of features left after the division of G into equal subsets, G = subset of G, i = 1, 2, 3, ⋯ , X; Then, sizeof(G) = X × U + sizeof(L) sizeof() represents the size of a set in single dimension. Given the values of G and U, we can find the value of X and L by, X = , sizeof(L) = sizeof(G) - X × U |
| top n features of G, or top n-1 features of G + top feature of G, or top n-2 features of G + top 2 features of G, or top n-2 features of G + top feature of G + top feature of G, or top n-3 features of G + top 3 features of G, or top n-3 features of G + top 2 features of G + top feature of G, . . . Keeping this in mind, the new subset of features(G) will be selected as, G’ = {}, initially it will be an empty set, For every subset of features G, i = 1, 2, ⋯, X Let, , G = G + top y feature of G The previous set of features G will be updated as, G = G + L |
| Algorithm 1 Pseudocode of the proposed approach. |
Require:
Main Function: main_fun(features,upper_limit,mod,k)
|
5. Experimental Results
5.1. Dataset Used
5.2. Classifier Used
| ’n estimators’: np.arange(50,200,30) ’max features’: np.arange(0.1, 1, 0.1) ’max depth’: [3, 5, 7, 9, 50, 100] ’max samples’: [0.3, 0.5, 0.8, 1] |
- n_estimators: The quantity of trees in the forest.
- max_features: The number of features considered to find the optimal split.
- max_depth: The highest level of the tree.
- max_samples: The number of samples taken from X to train each base estimator.
- np.arange: Produces evenly spaced values within a specified range.
| ’c’ :[0.01, 1, 5, 10, 100] ’kernel’:(’linear’, ’poly’, ’rbf’, ’sigmoid’) ’gamma’ :(’scale’, ’auto’) |
- c: A regularization constant.
- kernel: Determines the type of kernel used in the algorithm.
- gamma: The kernel coefficient for ‘rbf’, ‘poly’, and ‘sigmoid’.
5.3. Result and Discussion
6. Application of the Proposed Work
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chandrashekar, G.; Sahin, F. A survey on feature selection methods. Comput. Electr. Eng. 2014, 40, 16–28. [Google Scholar] [CrossRef]
- Altman, N.; Krzywinski, M. The curse(s) of dimensionality. Nat. Methods 2018, 15, 399–400. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Baştanlar, Y.; Özuysal, M. Introduction to machine learning. In miRNomics: MicroRNA Biology and Computational Analysis; Humana Press: Totowa, NJ, USA, 2014; pp. 105–128. [Google Scholar]
- Law, M.H.; Figueiredo, M.A.; Jain, A.K. Simultaneous feature selection and clustering using mixture models. IEEE Trans. Pattern Anal. Mach. Intell. 2004, 26, 1154–1166. [Google Scholar] [CrossRef] [PubMed]
- Kuncheva, L.I.; Matthews, C.E.; Arnaiz-González, Á.; Rodríguez, J.J. Feature selection from high-dimensional data with very low sample size: A cautionary tale. arXiv 2020, arXiv:2008.12025. [Google Scholar]
- Kohavi, R.; John, G.H. Wrappers for feature subset selection. Artif. Intell. 1997, 97, 273–324. [Google Scholar] [CrossRef]
- Blum, A.L.; Langley, P. Selection of relevant features and examples in machine learning. Artif. Intell. 1997, 97, 245–271. [Google Scholar] [CrossRef]
- Jović, A.; Brkić, K.; Bogunović, N. A review of feature selection methods with applications. In Proceedings of the 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 25–29 May 2015; pp. 1200–1205. [Google Scholar]
- Liu, H.; Sun, J.; Liu, L.; Zhang, H. Feature selection with dynamic mutual information. Pattern Recognit. 2009, 42, 1330–1339. [Google Scholar] [CrossRef]
- Battiti, R. Using mutual information for selecting features in supervised neural net learning. IEEE Trans. Neural Netw. 1994, 5, 537–550. [Google Scholar] [CrossRef]
- Wah, Y.B.; Ibrahim, N.; Hamid, H.A.; Abdul-Rahman, S.; Fong, S. Feature Selection Methods: Case of Filter and Wrapper Approaches for Maximising Classification Accuracy. Pertanika J. Sci. Technol. 2018, 26, 329–340. [Google Scholar]
- El Aboudi, N.; Benhlima, L. Review on wrapper feature selection approaches. In Proceedings of the 2016 International Conference on Engineering & MIS (ICEMIS), Agadir, Morocco, 22–24 September 2016; pp. 1–5. [Google Scholar]
- Bolón-Canedo, V.; Sánchez-Marono, N.; Alonso-Betanzos, A.; Benítez, J.M.; Herrera, F. A review of microarray datasets and applied feature selection methods. Inf. Sci. 2014, 282, 111–135. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, M.; Liu, Q. An embedded feature selection method for imbalanced data classification. IEEE/CAA J. Autom. Sin. 2019, 6, 703–715. [Google Scholar] [CrossRef]
- Liu, H.; Motoda, H. Feature Extraction, Construction and Selection: A Data Mining Perspective; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1998; Volume 453. [Google Scholar]
- Aziz, R.; Verma, C.; Srivastava, N. A fuzzy based feature selection from independent component subspace for machine learning classification of microarray data. Genom. Data 2016, 8, 4–15. [Google Scholar] [CrossRef]
- Yasmin, G.; Das, A.K.; Nayak, J.; Pelusi, D.; Ding, W. Graph based feature selection investigating boundary region of rough set for language identification. Expert Syst. Appl. 2020, 158, 113575. [Google Scholar] [CrossRef]
- Reimann, M.; Doerner, K.; Hartl, R.F. D-ants: Savings based ants divide and conquer the vehicle routing problem. Comput. Oper. Res. 2004, 31, 563–591. [Google Scholar] [CrossRef]
- Song, Q.; Ni, J.; Wang, G. A fast clustering-based feature subset selection algorithm for high-dimensional data. IEEE Trans. Knowl. Data Eng. 2011, 25, 1–14. [Google Scholar] [CrossRef]
- Zhao, Q.; Meng, D.; Xu, Z. A recursive divide-and-conquer approach for sparse principal component analysis. arXiv 2012, arXiv:1211.7219. [Google Scholar]
- Sun, Y.; Todorovic, S.; Goodison, S. Local-learning-based feature selection for high-dimensional data analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 32, 1610–1626. [Google Scholar]
- Armanfard, N.; Reilly, J.P.; Komeili, M. Local feature selection for data classification. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 1217–1227. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. (Methodol.) 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Chen, X.; Yuan, G.; Nie, F.; Huang, J.Z. Semi-supervised Feature Selection via Rescaled Linear Regression. In Proceedings of the IJCAI, Melbourne, Australia, 19–25 August 2017; Volume 2017, pp. 1525–1531. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; Hastie, T.; Johnstone, I.; Tibshirani, R. Least angle regression. Ann. Stat. 2004, 32, 407–499. [Google Scholar] [CrossRef]
- Yamada, M.; Jitkrittum, W.; Sigal, L.; Xing, E.P.; Sugiyama, M. High-dimensional feature selection by feature-wise kernelized lasso. Neural Comput. 2014, 26, 185–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Stern, M.; Wainwright, M.J.; Jordan, M.I. Kernel feature selection via conditional covariance minimization. Adv. Neural Inf. Process. Syst. 2017, 30, 6949–6958. [Google Scholar]
- Pierezan, J.; Coelho, L.D.S. Coyote optimization algorithm: A new metaheuristic for global optimization problems. In Proceedings of the 2018 IEEE congress on evolutionary computation (CEC), Brisbane, Australia, 10–15 June 2018; pp. 1–8. [Google Scholar]
- Rösler, U. On the analysis of stochastic divide and conquer algorithms. Algorithmica 2001, 29, 238–261. [Google Scholar] [CrossRef]
- Smith, D.R. The design of divide and conquer algorithms. Sci. Comput. Program. 1985, 5, 37–58. [Google Scholar] [CrossRef]
- Guyon, I.; Gunn, S.; Nikravesh, M. Feature Extraction: Studies in Fuzziness and Soft Computing; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Luukka, P. Feature selection using fuzzy entropy measures with similarity classifier. Expert Syst. Appl. 2011, 38, 4600–4607. [Google Scholar] [CrossRef]
- Parkash, O.; Gandhi, C. Applications of trigonometric measures of fuzzy entropy to geometry. Int. J. Math. Comput. Sci 2010, 6, 76–79. [Google Scholar]
- Yu, L.; Liu, H. Feature selection for high-dimensional data: A fast correlation-based filter solution. In Proceedings of the 20th International Conference on Machine Learning (ICML-03), Washington, DC, USA, 21–24 August 2003; pp. 856–863. [Google Scholar]
- Yu, K.; Ding, W.; Wu, X. LOFS: A library of online streaming feature selection. Knowl.-Based Syst. 2016, 113, 1–3. [Google Scholar] [CrossRef]
- De Souza, R.C.T.; de Macedo, C.A.; dos Santos Coelho, L.; Pierezan, J.; Mariani, V.C. Binary coyote optimization algorithm for feature selection. Pattern Recognit. 2020, 107, 107470. [Google Scholar] [CrossRef]
- Available online: https://github.com/git-arihant/feature-importance (accessed on 25 December 2022).
- Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 25 December 2022).
- Available online: https://github.com/jranaraki/NCBIdataPrep (accessed on 25 December 2022).
- Afshar, M.; Usefi, H. High-dimensional feature selection for genomic datasets. Knowl.-Based Syst. 2020, 206, 106370. [Google Scholar] [CrossRef]
- Sheykhmousa, M.; Mahdianpari, M.; Ghanbari, H.; Mohammadimanesh, F.; Ghamisi, P.; Homayouni, S. Support vector machine versus random forest for remote sensing image classification: A meta-analysis and systematic review. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2020, 13, 6308–6325. [Google Scholar] [CrossRef]
- Yuanyuan, S.; Yongming, W.; Lili, G.; Zhongsong, M.; Shan, J. The comparison of optimizing SVM by GA and grid search. In Proceedings of the 2017 13th IEEE International Conference on Electronic Measurement & Instruments (ICEMI), Yangzhou, China, 20–22 October 2017; pp. 354–360. [Google Scholar]
- Sumathi, B. Grid search tuning of hyperparameters in random forest classifier for customer feedback sentiment prediction. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 173–178. [Google Scholar]


| Datasets | Samples | Original Features | Cleaned Features | Labels |
|---|---|---|---|---|
| GDS-1615 | 127 | 22,200 | 13,600 | Three |
| GDS-2546 | 167 | 12,600 | 9500 | Four |
| GDS-968 | 171 | 12,600 | 9100 | Four |
| GDS-2545 | 171 | 12,600 | 9300 | Four |
| GDS-3929 | 183 | 24,500 | 19,300 | Two |
| GDS-1962 | 180 | 54,600 | 29,100 | Four |
| GDS-531 | 173 | 12,600 | 9300 | Two |
| GDS-2547 | 164 | 12,600 | 9300 | Four |
| Datasets | Features Selected | RF | SVM | Time Taken (sec) |
|---|---|---|---|---|
| GDS1615 | 20 | 86.31 | 83.15 | 41.29 |
| 30 | 88.42 | 86.31 | 41.2 | |
| 40 | 86.31 | 82.1 | 43.25 | |
| 50 | 87.36 | 88.42 | 43.87 | |
| 60 | 86.31 | 88.42 | 44.97 | |
| GDS968 | 20 | 76.52 | 77.41 | 35.9 |
| 30 | 74.89 | 75.84 | 36.43 | |
| 40 | 73.35 | 74.21 | 37.26 | |
| 50 | 76.55 | 78.8 | 38.4 | |
| 60 | 78.06 | 81.26 | 38.82 | |
| GDS531 | 20 | 84.49 | 78.27 | 29.44 |
| 30 | 85.29 | 79.84 | 22.32 | |
| 40 | 86.86 | 86.06 | 22.52 | |
| 50 | 86.09 | 83.75 | 22.9 | |
| 60 | 86.83 | 83.75 | 23.75 | |
| GDS2545 | 20 | 72.67 | 68.76 | 37.98 |
| 30 | 78.09 | 71.04 | 38.49 | |
| 40 | 75.01 | 73.29 | 38.9 | |
| 50 | 70.15 | 69.53 | 39.84 | |
| 60 | 75.01 | 71.81 | 40.82 | |
| GDS1962 | 20 | 79.25 | 72.59 | 129.13 |
| 30 | 78.51 | 73.33 | 116.72 | |
| 40 | 80 | 77.77 | 129.6 | |
| 50 | 79.25 | 76.29 | 130.06 | |
| 60 | 79.25 | 76.29 | 130.82 | |
| GDS3929 | 20 | 74.44 | 69.33 | 46.38 |
| 30 | 72.93 | 69.41 | 45.37 | |
| 40 | 76.66 | 67.88 | 45.91 | |
| 50 | 74.36 | 67.91 | 46.37 | |
| 60 | 75.26 | 67.88 | 48.54 | |
| GDS2546 | 20 | 95.2 | 84 | 37.87 |
| 30 | 95.2 | 85.59 | 38.01 | |
| 40 | 96 | 80.8 | 38.66 | |
| 50 | 95.2 | 82.4 | 36.88 | |
| 60 | 95.2 | 80 | 38.39 | |
| GDS2547 | 20 | 69.23 | 67.59 | 37.77 |
| 30 | 70.03 | 68.39 | 38.2 | |
| 40 | 70.06 | 67.56 | 39.1 | |
| 50 | 70.83 | 68.39 | 38.07 | |
| 60 | 70.86 | 65.93 | 38.79 |
| Classifier | Datasets | mRMR | LARS | HSIC | Fast | Group | CCM | BCOA | Proposed |
|---|---|---|---|---|---|---|---|---|---|
| LASSO | OSFS | SAOLA | CCM | BCOA | Work | ||||
| SVM | GDS1615 | 87.37(40) | 91.67(26) | 91.35(18) | 84.31(17) | 83.13(12) | 80.82(29) | 84.9(33) | 88.42(50) |
| GDS968 | 80.87(39) | 83.73(38) | - | 72.41(19) | 70.53(14) | 78.82(34) | 76.19(32) | 81.26(60) | |
| GDS531 | 69.78(30) | 79.96(27) | 67.93(4) | 77.43(26) | 77.7(11) | 80.82(30) | 74.17(32) | 86.06(40) | |
| GDS2545 | 75.9(34) | 79.02(33) | 76.4(33) | 74.95(18) | 75.55(12) | 70.82(30) | 75.4(29) | 73.29(40) | |
| GDS1962 | 65.12(39) | 76.56(32) | 76.81(31) | 65.15(24) | 66.59(10) | 66.82(40) | 66.89(35) | 77.77(40) | |
| GDS3929 | 73.57(41) | 83.78(41) | - | 83.11(40) | 76.97(21) | 75.82(39) | 72.12(41) | 69.41(30) | |
| GDS2546 | 74.13(33) | 83.51(32) | 77.69(27) | 81.25(26) | 80.88(17) | 73.82(35) | 72.98(32) | 85.59(30) | |
| GDS2547 | 67.31(39) | 73.88(32) | 71.16(12) | 73.13(23) | 76.85(24) | 66.82(28) | 67.35(26) | 68.39(30) | |
| RF | GDS1615 | 81.96(32) | 88.24(20) | 92.88(22) | 82.34(15) | 82.26(13) | 79.55(31) | 81.08(30) | 88.42(30) |
| GDS968 | 79.44(44) | 79.77(42) | - | 72.84(19) | 71.28(18) | 77.53(41) | 76.42(40) | 78.06(60) | |
| GDS531 | 63.69(23) | 71.44(20) | 67.82(4) | 75.48(14) | 74.67(16) | 77.36(23) | 73.92(21) | 86.86(40) | |
| GDS2545 | 79.31(31) | 75.81(33) | 80.64(33) | 74.16(14) | 76.05(12) | 74.82(34) | 75.63(33) | 78.09(30) | |
| GDS1962 | 72.37(29) | 72.41(30) | 78.45(42) | 69.88(21) | 63.28(13) | 69.17(32) | 67.95(30) | 80(40) | |
| GDS3929 | 71.94(29) | 73.44(28) | - | 70.49(28) | 71.56(15) | 67.5(28) | 65.13(24) | 76.66(40) | |
| GDS2546 | 70.53(36) | 75.86(34) | 83.09(45) | 77.04(25) | 78.46(18) | 72.9(36) | 75.28(31) | 96(40) | |
| GDS2547 | 68.44(22) | 71.68(24) | 81.67(32) | 75.85(30) | 77.1(20) | 69.7(25) | 71.28(24) | 70.86(60) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanwar, A.; Alghamdi, W.; Alahmadi, M.D.; Singh, H.; Rana, P.S. A Fuzzy-Based Fast Feature Selection Using Divide and Conquer Technique in Huge Dimension Dataset. Mathematics 2023, 11, 920. https://doi.org/10.3390/math11040920
Tanwar A, Alghamdi W, Alahmadi MD, Singh H, Rana PS. A Fuzzy-Based Fast Feature Selection Using Divide and Conquer Technique in Huge Dimension Dataset. Mathematics. 2023; 11(4):920. https://doi.org/10.3390/math11040920
Chicago/Turabian StyleTanwar, Arihant, Wajdi Alghamdi, Mohammad D. Alahmadi, Harpreet Singh, and Prashant Singh Rana. 2023. "A Fuzzy-Based Fast Feature Selection Using Divide and Conquer Technique in Huge Dimension Dataset" Mathematics 11, no. 4: 920. https://doi.org/10.3390/math11040920
APA StyleTanwar, A., Alghamdi, W., Alahmadi, M. D., Singh, H., & Rana, P. S. (2023). A Fuzzy-Based Fast Feature Selection Using Divide and Conquer Technique in Huge Dimension Dataset. Mathematics, 11(4), 920. https://doi.org/10.3390/math11040920







