1. Introduction
Random graphs are widely used to model complex systems such as social networks, biological networks, and the internet. The degree distribution is an important characteristic of a network, as it provides information about the connectivity of nodes in the network [
1], and its shape determines many network phenomena, such as robustness [
2,
3,
4] or spreading processes [
5,
6,
7]. Inhomogeneous random graphs are a type of random graph where the nodes are not equally likely to be connected. Instead, the probability of two nodes being connected depends on their attributes or characteristics. Example applications of inhomogeneous random graphs are social network analysis [
8,
9], modelling biological networks [
10], or modelling transportation networks [
11].
In the literature of network science and random graphs, inhomogeneous random graphs are not a well-defined random graph model, but they are a family of random graph models, where the nodes have varying degrees of connectivity. One example for inhomogeneous random graphs is the stochastic block model [
8,
9]. A stochastic block model (SBM) is defined by a
partition of the vertex set and a
symmetric
P edge probability matrix. For any two vertices
and
, the draw probability of the
undirected edge is
. Therefore, for any
vertex pair, the
probability that the nodes
u and
v are connected is directly determined by the model parameters (the
P matrix), and the links are drawn independently if the
P matrix is given. A second example is the generalized random graph [
12,
13]. In case of a generalized random graph (GRG), the inhomogeneity is introduced into the model using vertex weights. For any
i node of the network, there is given a
vertex weight, and the probability that a edge is drawn between the nodes
i and
j is equal to
where
is the total weight of all vertices. The consequence of this definition is that vertices with high weight are more likely to have many neighbours than vertices with small weights, and vertices with extremely high weights could act as hubs observed in many real-world networks. Furthermore, if the
parameters are deterministic and given, the edge probabilities can be computed with (
1), and the edges are independent. A third example for inhomogeneous random graphs is the biased static edge voting model [
14]. We use this model in our numerical tests in
Section 4, and it is briefly described in
Section 4.2. Similarly to the SBM and GRG models, if the model parameters are given, the
link probability of any
node pair can be directly computed (see Equation (
66)), and the edges are drawn independently.
For all these example inhomogeneous random graph models, the common property is that the edge probabilities can be directly computed from the model definition, and the edges are drawn independently. We define the inhomogeneous random graph (IRG) model via these properties (as it is defined in [
13] (Section 6.7)). The inhomogeneous random graph is a random graph model on the vertex set
, where the
draw probability of any
edge is given, and the edges are drawn independently. The IRG model can be considered as the natural generalization of the Erdős–Rényi (ER) random graph [
15], where each link of the graph is drawn independently with a fixed
p probability. If we set all the edge probabilities of the IRG model to a fixed
p value, then we obtain an ER random graph. It is well known that the degree distribution of the ER model is close to the Poisson distribution [
15]. When we observe the degree sequence of real-world networks, we often see that their empirical degree distribution has a fat tail [
13]. Therefore, the ER random graph cannot be used to model real-world networks.
If the parameters are deterministic and given, the SBM, GRG, and the static edge voting model can be represented by an IRG. A further example for such a model is the Chung–Lu random graph [
16]. However, not all inhomogeneous random graph models can be expressed as IRG. For example, the Norros–Reittu model [
17] is a random multigraph model, while IRG is a model of a simple graph. A second example is the GRG model with random weights. Using random weights in GRG breaks the independence of the edges. A third example is the Barabási–Albert (BA) model [
18]. The BA model is a dynamic network growth model, and for this case, we cannot derive the edge probabilities.
In this paper, we discuss a novel algorithm what allow us to compute the exact degree distribution of the IRG model and an approximation method to estimate the IRG degree distribution. The hardness of computing the degree distribution of the IRG model comes from the fact that each edge candidate of the network may have different draw probabilities; therefore, the degree distribution of any node is Poisson binomial (PB) [
19,
20]. The algorithm that we have developed to compute the degree distribution of the IRG model is based on the DFT-CF method invented by Yili Hong [
19], and the approximation method uses the Poisson, binomial, and the Gaussian distributions to approximate the PB distribution. The proposed algorithms can be used to compute or approximate the degree distribution of any random graph model that can be represented by an IRG.
The structure of the remaining part of this paper is as follows:
Section 2 contains the mathematical preliminaries of our study. In
Section 2.1, we introduce the necessary notations and definitions. In
Section 2.2, we briefly discuss the DFT-CF algorithm for computing the Poisson binomial distribution.
Section 2.3 contains selected results about the approximation of the Poisson binomial distribution. In
Section 3, we discuss the proposed algorithms to compute or approximate the degree distribution of the IRG model. In
Section 3.1, we formally define the problem that we aim to solve. In
Section 3.2, we present an exact algorithm to compute the degree distribution of the inhomogeneous random graph, and in
Section 3.3, we discuss an approximation method to estimate this distribution. In the first part of
Section 3.3, we outline the general scheme of the approximation method; then, we provide an upper bound of the approximation error for the special cases, when the approximator distribution is Poisson (
Section 3.3.1), binomial (
Section 3.3.2), and Gaussian (
Section 3.3.3). The results of the numerical experiments are provided in
Section 4. The study is concluded with the discussion in
Section 5.
The contribution of the authors are a novel algorithm to compute the exact degree distribution of the IRG model utilizing the DFT-CF method (
Section 3.2) and the analysis of the estimation method for this distribution (
Section 3.3). The idea of the approximation scheme is simple and not new: we group the similar nodes into clusters and apply the same approximator distribution within a cluster. Our contribution here is the analysis of the approximation error in the specific cases when the approximator distribution is Poisson (
Section 3.3.1), binomial (
Section 3.3.2), and Gaussian (
Section 3.3.3).
4. Numerical Experiments
We demonstrate the developed methods with numerical experiments. In
Section 4.1 we compute the degree distribution of the ER random graph using Algorithm 2. In
Section 4.3 and
Section 4.4, we experimentally test the precision of the approximation method. In
Section 4.3, we observe how the approximation error changes as the network size changes. For this test, we use two IRG types: in the first type, the network has a block structure, and within one block, the degree distribution of the nodes is the same. In the second type, there is no such a structure; for any different
a and
b nodes,
and
follow different distributions. We group the
a and
b nodes to the same
M cluster only if
and
have the same distribution; therefore, for the second IRG type, every group contains only a single node. In
Section 4.4, we fix an IRG with the second type: for any different
a and
b nodes,
and
have different distributions. For this experiment, we create partitions
of the
V nodes, where
S denotes the common cluster size in the
partition. Therefore, for a fixed
partition and any
node cluster, the distribution of
and
are different if
and
. We observe how approximation precision changes as the cluster size changes. We use the biased static edge voting model [
14] to generate the test IRGs with appropriate structures. Hence, in
Section 4.2, we briefly discuss the biased static edge voting model.
4.1. The ER Test
In the special case when all the
edge probabilities of an
are equal to the same
p value, we obtain the ER random graph with the parameter
p. We know that the degree distribution of the ER random graph model with a fixed
n node number and
p link probability is binomial [
15] with parameters
and
p. Therefore, the degree distribution of the ER random graph with parameters
n and
p can be expressed as:
where
is a uniform random variable on the
node set. Let us denote the IRG model on the node set
by
, where all the
edge probabilities are set to the same
p probability. It is clear that
and
denote the same random graph model; therefore, we expect that if we compute the degree distribution of
using Algorithm 2, we obtain exactly the degree distribution of the
model given in (
62). We experimentally tested this statement setting the
n network size to be
and computed the degree distribution of
using Algorithm 2 for each
. Let us denote the degree distribution of
computed by Algorithm 2 with
, and calculate the total variation distance between the degree distributions
and
:
The magnitude of the total variation distance values for each p = 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 parameter value was , which means that we can consider the distributions and to be the same.
4.2. The Biased Static Edge Voting Model
We used the biased static edge voting model [
14] to generate the appropriate IRG parameterizations for the experiments in
Section 4.3 and
Section 4.4. The model with
N nodes is defined by the parameter set
and a single positive real value
. For any
, the parameter
controls the local behaviour of node
a, while
is a model-level control parameter. We can group the nodes based on their
parameter values: nodes
a and
b belong to the same
S group if and only if
. This naturally defines a partition of the nodes. We suppose that the
parameters are in the set
}, and we index a cluster with the common parameter of the nodes within the cluster:
. Denote as
a random variable that represents the vote of the
a node for the
edge candidate. We assume that for all
and
,
follows the same probability distribution. For any pair of different nodes
a and
b, the probability that there will be an edge between the nodes
a and
b depends on the incoming votes, and it is given by
, where
s is the edge probability function. The biased edge voting model specifies this definition in the following way: for any
and
nodes, the
random variable is Bernoulli distributed with a parameter of
, i.e.,
. The edge probability function is given as:
where
is the control parameter of the model, and
The
probability that the
link is drawn is given by the formula [
14]:
where:
The model is defined by the parameters
and
, or equivalently by the
number and the partition
of the nodes, where for each
,
(in some cases,
can be empty). Given these parameters, links are drawn independently, and the probability that the edge between nodes
a and
b is drawn is given by (
66). We use this model to generate different IRG parametrizations by applying Equation (
66). It is clear that different parametrizations of the voting model lead to different
models. As we have seen, parametrization means to fix the value of the control parameter
and the sequence
. To generate the
sequence, we used two methods: range and lognormal. Range is defined as the first
N non-negative integer:
. We denote the lognormal sequence generator by
, where
N is the length of the sequence, and
and
are the parameters of the lognormal distribution. A positive random variable
X is log-normally distributed with parameters
and
if
is normally distributed with mean
and standard deviation
. For the lognormal sequence generator algorithm, we suppose that the rate of nodes with parameter
k is approximately
, where
is the cumulative distribution function of the lognormal distribution with parameters
and
. Therefore, the number of nodes with parameter
k is approximately
. The algorithm is given in Algorithm 5. Its input parameter
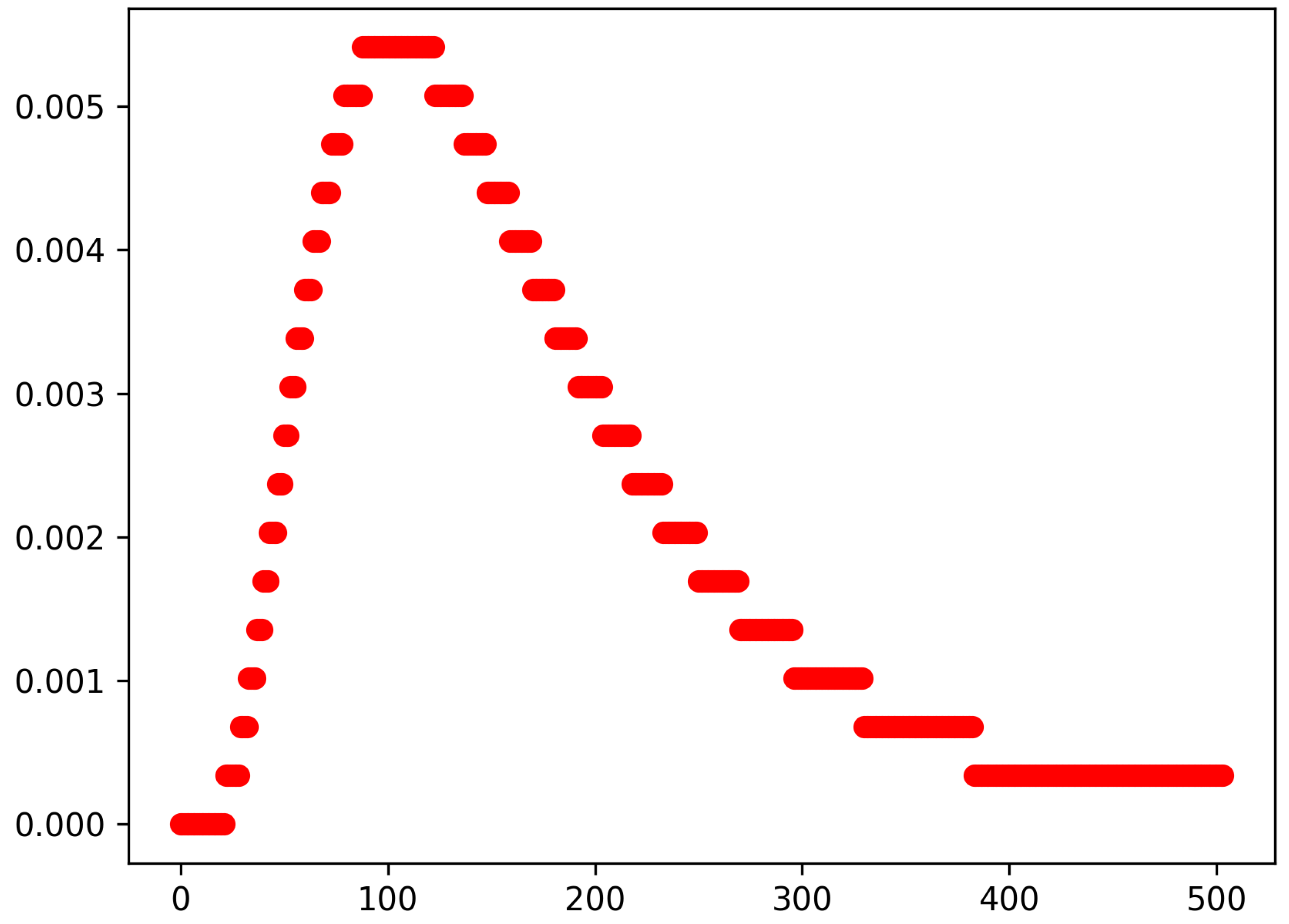
can be any cumulative distribution function. We plotted the empirical density function of the sequence
in
Figure 1. The
parameter controls the global behaviour of the model. In the rest of this paper, we fix the value of
to 2.0.
| Algorithm 5 parameter_sequence_generator(node_nr, cdf) |
parameter_sequence = empty list
not_finished_nodes = node_nr
max_param = node_nr − 1
normalizer = cdf(max_param + 0.5)
for param = 0 to max_param do
m = param − 0.5
M = param + 0.5
p = (cdf(M) − cdf(m))/normalizer
nr = min(round(), not_finished_nodes)
Add param to the degree_parameter list nr times
not_finished_nodes = not_finished_nodes − nr
if not_finished_nodes ≤ 0 then
Break
end if
end for
return parameter_sequence |
4.3. The Effect of Network Size on Approximation Accuracy
In this test, we experimentally observe the effect of network size on approximation accuracy. For a sequence of networks with increasing network size, we compare the degree distributions returned by the approximation method (Algorithm 4) to the exact degree distributions computed by Algorithm 2. We use the total variation distance for comparison. When the approximator uses the Poisson or the binomial distributions, we can directly use the total variation distance; however, when the Gaussian distribution is used for approximation, we use the following discretization to obtain a discrete probability distribution: if
X is a continuous random variable with
cdf, then its discretized version
has a pdf:
To create the test IRG parametrizations, we used the biased static edge voting model using lognormal and range parametrization methods. For both cases, the network sizes are 50, 100, 300, 500, 1000, 1500, 2000, 2500, and 3000.
We denote the biased static edge voting model with parametrization
and
by
, and the IRG generated from
using (
66) by
. It is clear that in
, for all different nodes
a and
b,
. Similarly, we denote the static edge voting model with parametrization
by
, and the IRG generated from
using (
66) by
. The values of
and
for the
parametrization for each
n can be found in
Table 1. Because of the construction,
has a block structure. It contains clusters, and within a cluster, the nodes have the same degree distribution. In
Table 1, we collected basic statistics about the clusters for the used
parametrizations.
Let us denote the exact degree distribution of
computed with Algorithm 2 by
, where
T can be
R or
. Similarly, we denote the approximated degree distribution of
computed with Algorithm 4 by
, where
T can be
R or
and
G stands for the used approximation distribution:
P (Poisson),
B (binomial), or
G (Gaussian). For example, we denote the approximated degree distribution of
using the Poisson distribution by
. We calculated the total variation distance between the approximated and the exact degree distribution:
The calculated
values are collected in
Table 2 and
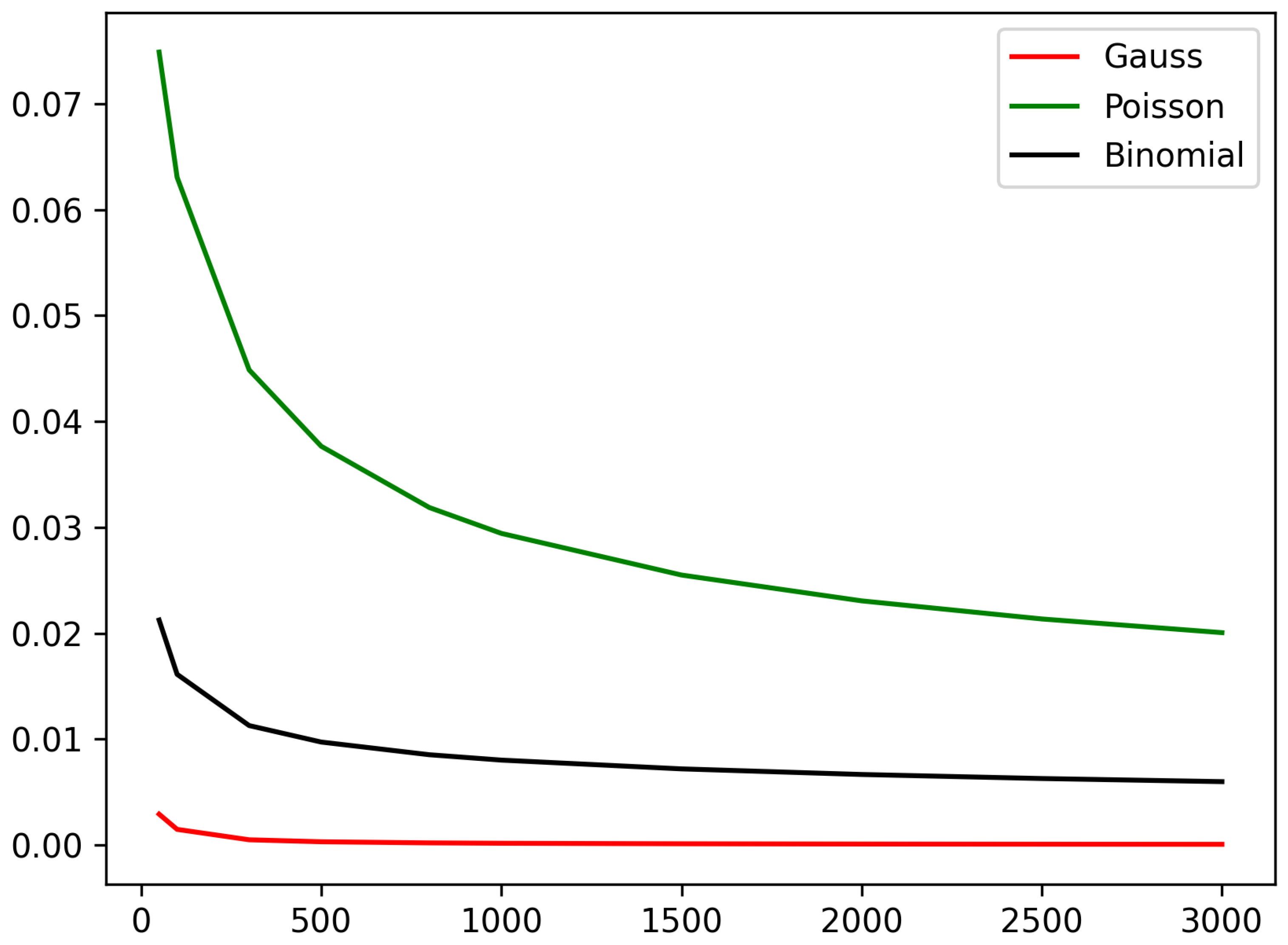
Table 3. We also plotted the total variation distances in the function of the network size in
Figure 2 and
Figure 3. We can observe that in the case of the
parametrization, the approximation error monotonically decreases with the network size, and we achieve the best approximation using the Gaussian approximation. At the
parametrization, we can observe an initial fluctuation in the approximation error, and after this, there is a monotone decreasing trend in the approximation error in function of the network size. In this case, we obtain the smallest approximation error when we use the binomial distribution.
4.4. The Effect of Cluster Size on Approximation Accuracy
In this experiment, we test how the accuracy of the approximation method depends on the cluster size.
denotes the IRG model generated from the biased edge voting model with parametrization
using (
66). This IRG is “very inhomogeneous” in the sense that all edge probabilities are different; therefore, the degree distribution of each node is different. We compute the exact degree distribution of
using Algorithm 2 and denote the result by
. We plotted
in
Figure 4.
We denote the number of nodes by
n (
in the current setting) and identify the nodes by their parameter in the static edge voting model used to generate the
. This means that for any
node, the
parameter of the node in the generator biased edge voting model was
a. Let us fix a cluster size
and suppose that
n is divisible by
S. We define a
partition of
as:
The degree distributions of all nodes within a cluster of partition
are different. We tested the approximation method described in
Section 3.3, where the node clusters are given by
, and the
S cluster size is in
= {1500, 1000, 750, 600, 500, 300, 200, 100, 75, 60, 50, 30, 20, 10, 5, and 1}. For example, if the
S cluster size is 1000, then we have 3 node clusters:
. We computed the approximated degree distribution of
using Algorithm 4 and denoted the result distributions by
, where
S denotes the cluster size and
D represents the type of distribution used in the approximation, which can be
P (Poisson),
B (binomial), or
G (Gaussian). For all
S cluster sizes from
, we calculated the total variation distance between the exact degree distribution (plotted in
Figure 4) and the approximated degree distributions:
The results are collected in
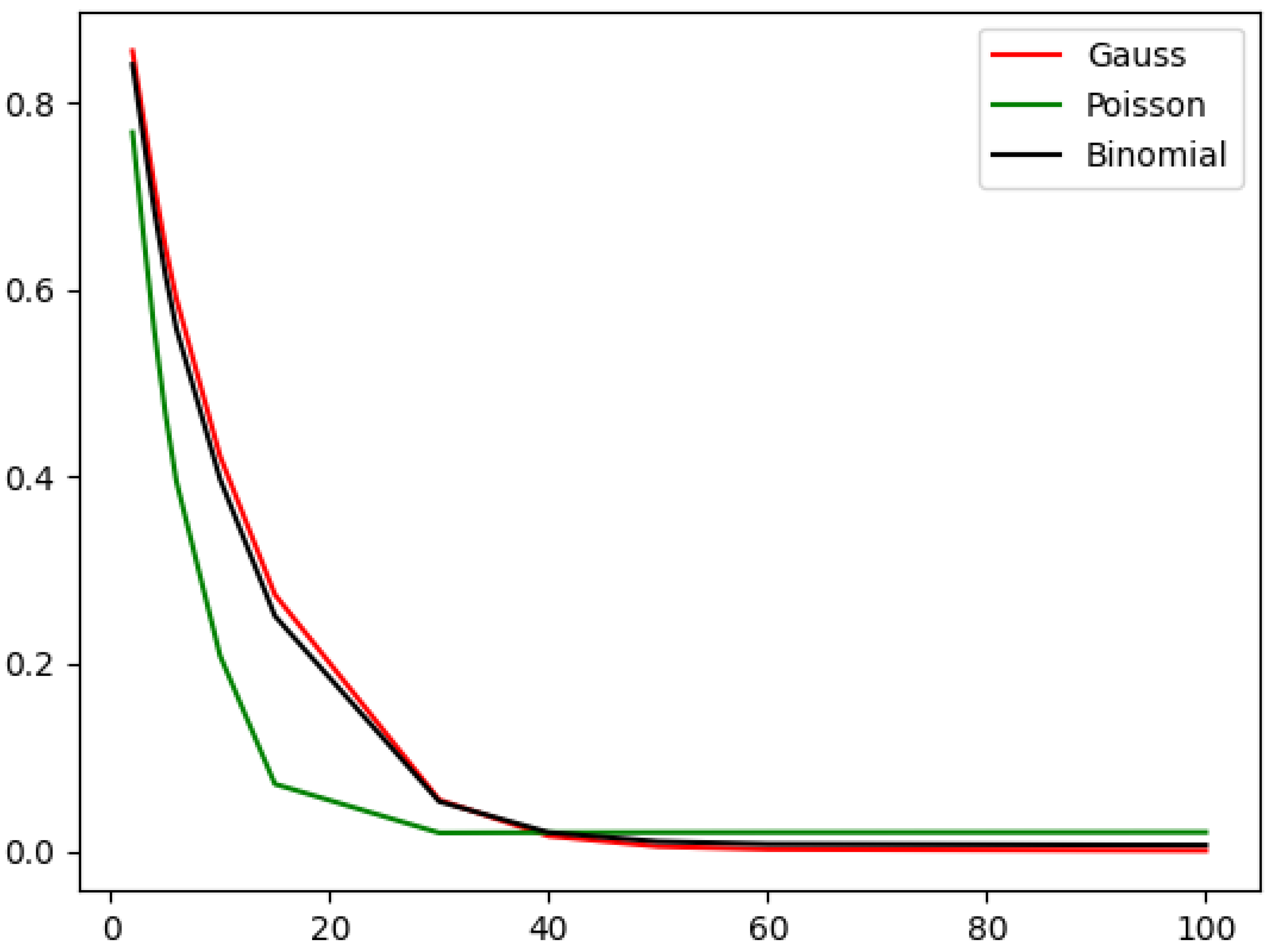
Table 4 and plotted in
Figure 5. We can observe that the approximation error decreases as the cluster shrinks (or the number of clusters increases). If the cluster size is huge, then the Poisson approximation gives the smallest approximation error, while if the clusters are small, the Gaussian approximation gives the best results, although in the case of small clusters, the difference between the approximators is small.
5. Discussion
Inhomogeneous random graph is a random graph model where the links of the graph are drawn independently and the link probabilities can be different. It can be seen as the generalization of the Erdős–Rényi random graph [
15], where the edges are drawn independently, but the probability that any different two nodes are linked is a fixed
p value. The degree distribution of a deterministic or random graph with
n nodes is defined as the probability that the degree of a uniformly chosen node equals to
k for all
. The degree distribution has a central role in network science, not only because it is needed to compute several other network properties [
1], but also because the shape of the degree distribution mostly determines the outcome of many important network processes, such as the spread of viruses [
5], diffusion of innovations [
6,
7], or attacks against critical infrastructure [
2,
3,
4]. The degree distribution of many real-world networks have fat tails; therefore, the ER random graph model is not suitable to model real world networks, because its degree distribution is binomial [
15]. Therefore, many alternative random graph models have been proposed to be able to model real world networks, such as the stochastic block model [
8], generalized random graphs [
12], Chung–Lu random graphs [
16], the Norros–Reittu model [
17], and the static edge voting model [
14] (
Section 4.2), which we used to generate the appropriate IRG parametrisations to test our algorithms in
Section 4. Using different parametrizations of the IRG model, one can achieve random network models with very different degree distributions. IRG is interesting not only as the generalization of the ER random graph but also as a tool to analyse other random graph models, such as the stochastic block model, generalized random graphs, or the static edge voting model.
In this paper, we focused on the calculation of the degree distribution of the IRG model. In
Section 3.2, we discussed an algorithm to compute the exact degree distribution utilizing the DFT-CF method [
19] developed by Yili Hong. The proposed algorithm (Algorithm 2) is highly parallelizable since the sub-step given in Algorithm 3 can be called independently. Furthermore, if the IRG model has a block structure, Algorithm 2 can take advantage of it. In
Section 3.3, we presented a method to approximate the degree distribution of the IRG model. There are several reasons why one would apply approximation even if an exact computational method is available. One reason is that approximation is computationally cheaper than the exact method. A second reason is that the approximation method may also be used in the case when the IRG is not fully defined. At the beginning of
Section 3.3, we discussed the general scheme of the proposed approximation method, which is presented in Algorithm 4. The idea of the approximation method is simple: we group the nodes of the network according to their statistical behaviour. For each node group, we approximate the common behaviour of the nodes within the group, and finally, we aggregate these group approximations. As a result, we obtain a mixture model, which we use as an approximation of the exact degree distribution. Similarly to the exact algorithm, it can be implemented effectively in a multi-thread environment. In Algorithm 4, we did not specify how to approximate the common behaviour of a node group, because it can be done in many ways, but in the subsequent subsections, we analysed three possible ways: using the Poisson distribution (
Section 3.3.1), the binomial distribution (
Section 3.3.2), and the Gaussian distribution (
Section 3.3.3). Furthermore, we derived an upper bound for the approximation error for all three cases: Equation (
48) for the Poisson and the binomial approximations, and Equation (
60) for the Gaussian approximation.
Determining which distribution to use for optimal results is a natural question. Unfortunately, we do not have a clear answer to this. During the numerical experiments in
Section 4, we found that the structure of the IRG and the granularity of the clustering influences which distribution will lead to the most accurate approximation. In
Section 4.3, we tested the approximation method on IRG models having a block structure (see
Figure 2 and
Table 2), where within each block, the nodes obey the same degree distribution. In this case, we could observe that using the binomial distribution gave the most accurate results. However, in the case where the degree distribution of each node was different and we did not apply grouping, using the Gaussian distribution gave the best results (see
Figure 3 and
Table 3). In
Section 4.4, we tested the effect of the clustering granularity on the approximation precision. We fixed an IRG model where the degree distribution of each node is different and applied the approximation with clustering, where the cluster size was different for each test case. We found that for larger cluster sizes, the usage of the Poisson distribution gave the most accurate estimate, and for smaller cluster sizes, the Gauss distribution gave the best results (see
Figure 5 and
Table 4).
The approximation method can be extended or improved in several ways. In this study, we analysed the usage of Poisson, binomial and Gaussian distributions. However, there are other distributions that we could use in a similar way. One obvious possibility is using the PB distribution itself. Another candidate for this is the translated Poisson distribution [
20,
33] or the Pólya approximation of the PB distribution [
34]. Another direction can be the optimal selection of the group approximation method. We have seen in
Section 4.3 and
Section 4.4 and in the previous paragraph that the structure of the IRG and the granularity of the clustering influences which distribution will lead to the most accurate approximation. It is an open question if we can implement a selection method to find the optimal approximator distribution.