Classification Methods for the Serological Status Based on Mixtures of Skew-Normal and Skew-t Distributions
Abstract
1. Introduction
2. Modeling Antibody Data: Skew-Normal and Skew-t Distributions
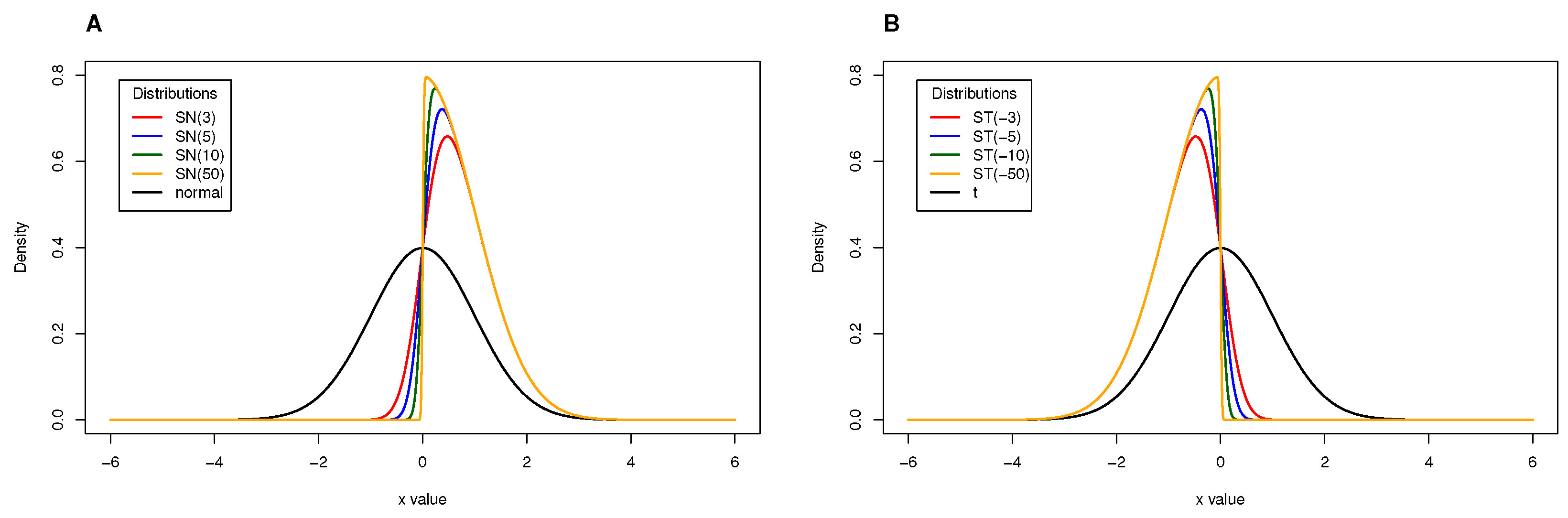
2.1. Skew-Normal Distribution
2.2. Skew-t Distribution
3. Finite Mixture Models to Describe Serological Data: Estimation of the Parameters
- E-step:The random variable takes the value 1 if the ith observation belongs to population k, and zero otherwise; thus, , with ; .In this step, one estimates the unobserved component membership, , i.e., the estimated probability that the ith observation comes from the kth population, , given the vector of the antibody levels, , and the current values for the unknown parameters:Afterwards, it estimates the probability of sampling from a seronegative or seropositive population, :
- M-step:In this step, one maximizes the weighted log-likelihood function (derived from Equation (5)), denoted by , with respect to :Therefore,
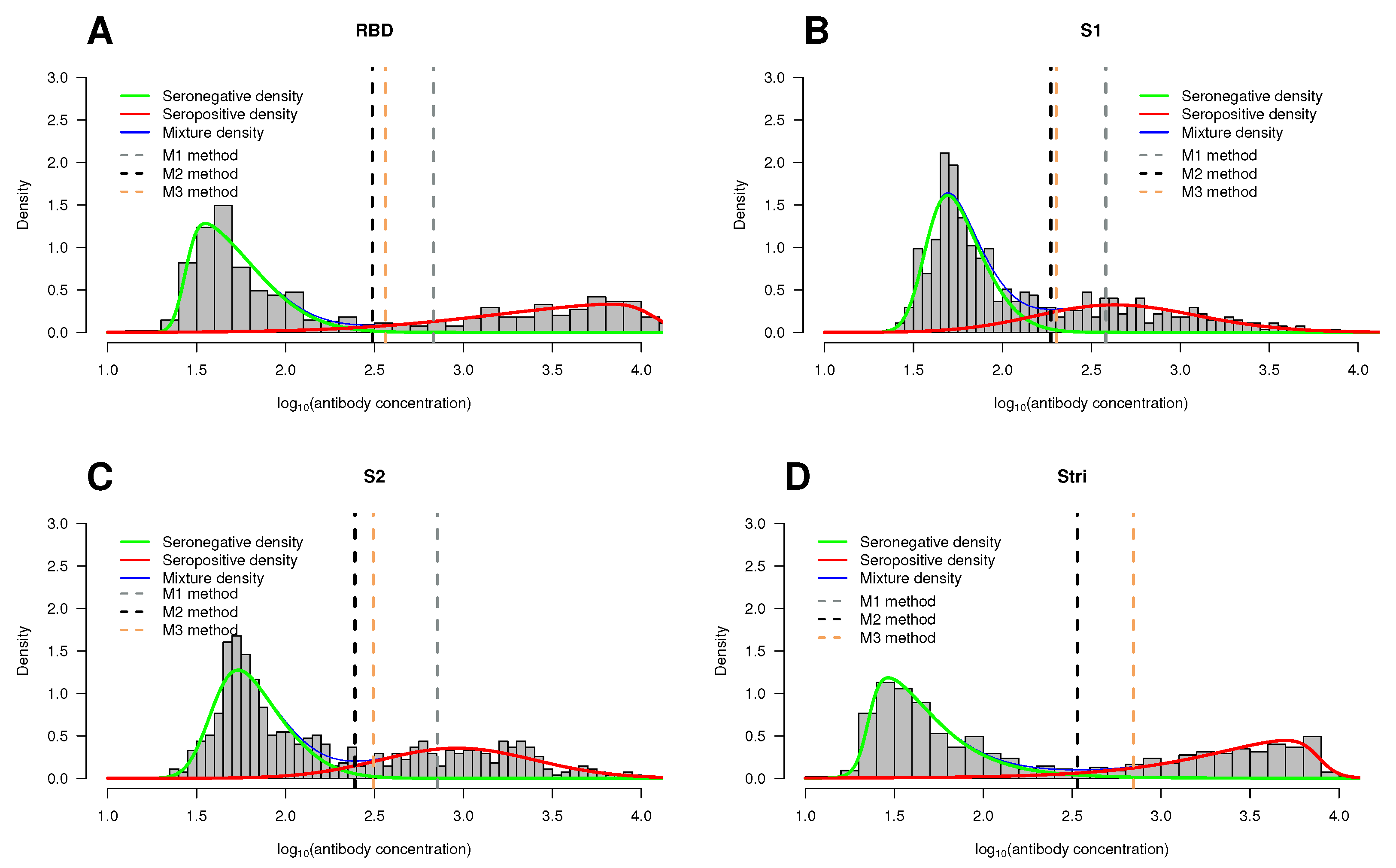
3.1. Definition of Seropositivity: Methods to Estimate the Cutoff Points in the Mixture Models
- -
- Method 1 (M1): It is based on the 99.9%-quantile associated with the estimated seronegative population. This method is the most popular in sero-epidemiology [32,33]. It is often called the rule because the 99.9%-quantile is given by the mean plus three times the standard deviation of a normally distributed seronegative population;
- -
- Method 2 (M2): It relies on the minimum of the density mixture functions. In the case of two latent populations, the cutoff corresponds to the absolute minimum. For three or more latent populations, the cutoff corresponds to the lowest relative minimum. This point can be calculated using Dekker’s algorithm [34]. It should be noted that the minimum of the mixing function is not expected to coincide with the point of intersection of the probability densities of each subpopulation;
- -
- Method 3 (M3): It imposes a threshold in the so-called conditional classification curves [32]. Under the assumption that all components but the first one referred to seropositive individuals, the conditional classification curve for the i-th individual given the antibody level is defined asIn turn, the classification curve of seronegative individuals is simply given byAfter calculating these curves, one can impose a minimum value for the classification of each individual. In this case, two cutoff values arise in the antibody distribution, one for the seronegative individuals and another for seropositive individuals. Mathematically, the classification rule is given as followswhere and are the cutoff values in the antibody distribution that ensure a minimum classification probability (say 90%). In practice, one can use the bisection method to calculate these cutoff values in practice, providing an initial interval where they might be located [32].
3.2. Software
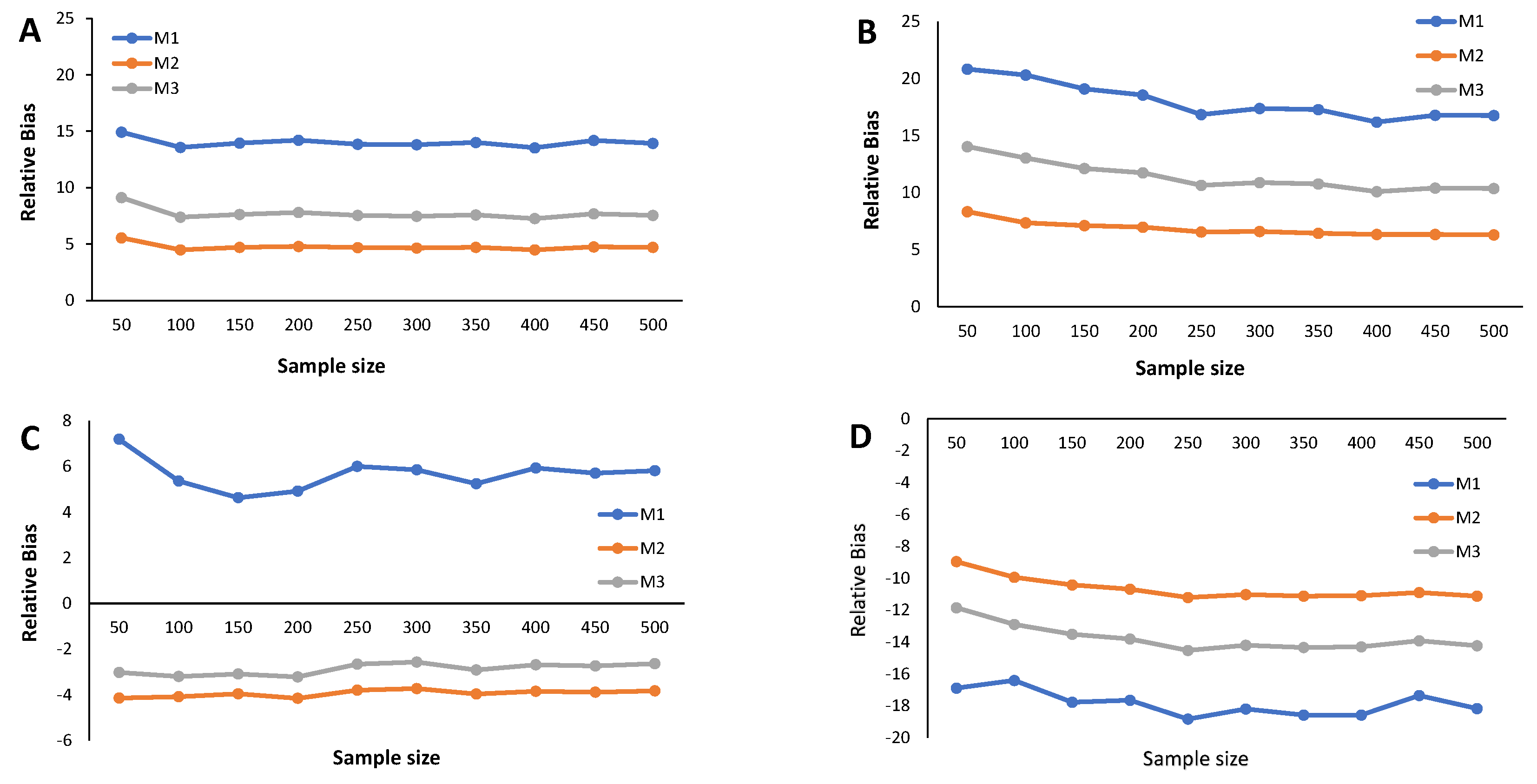
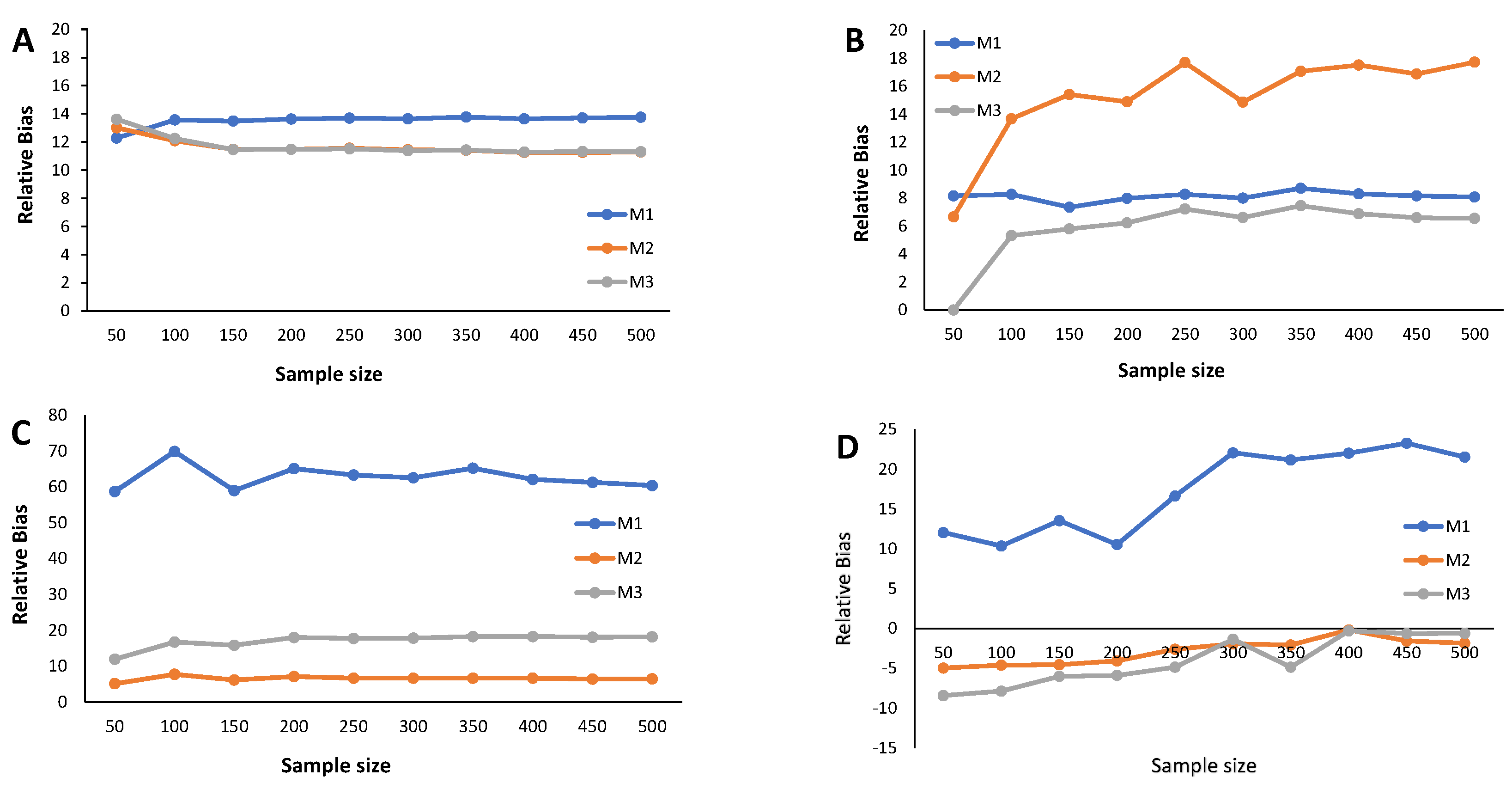
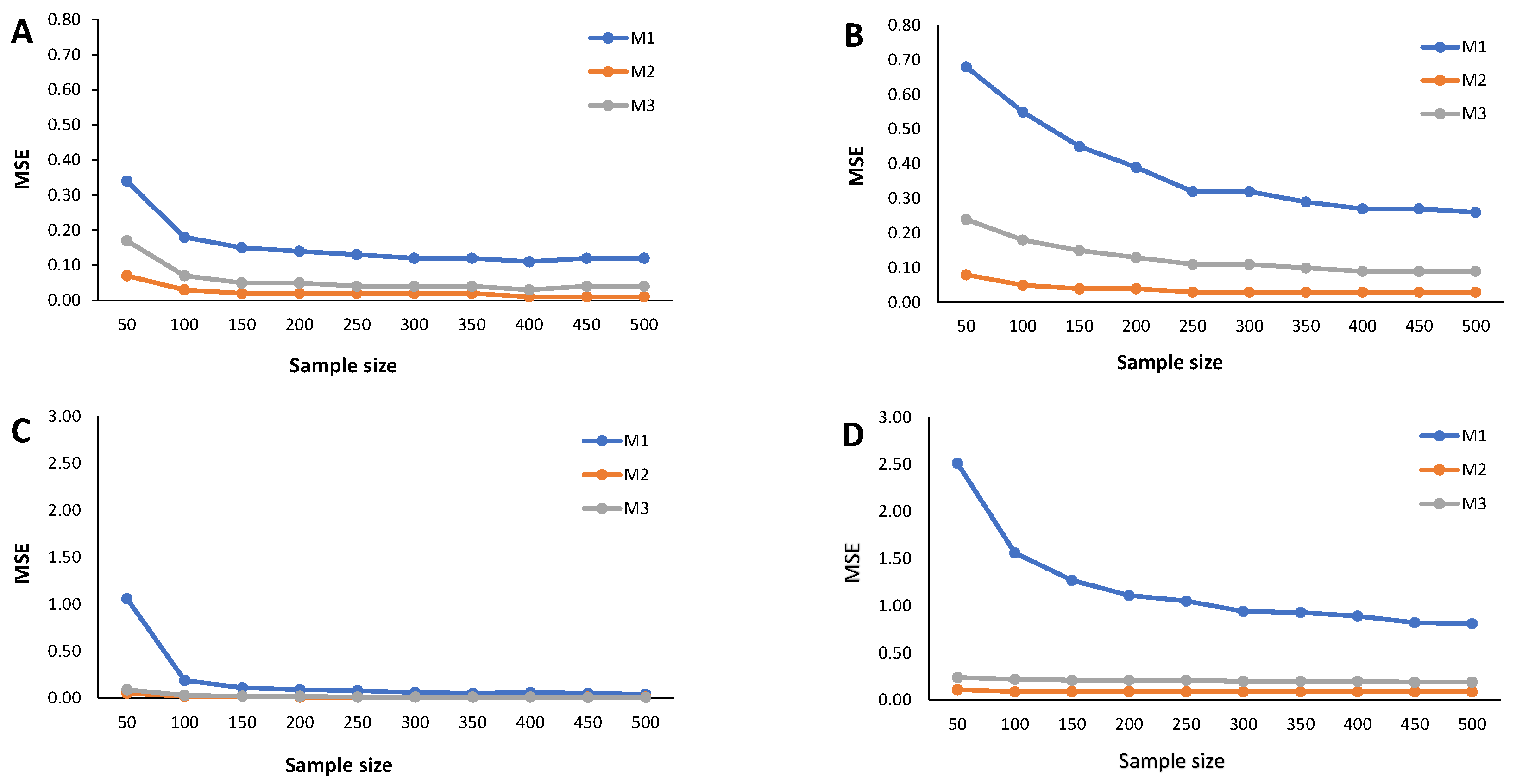
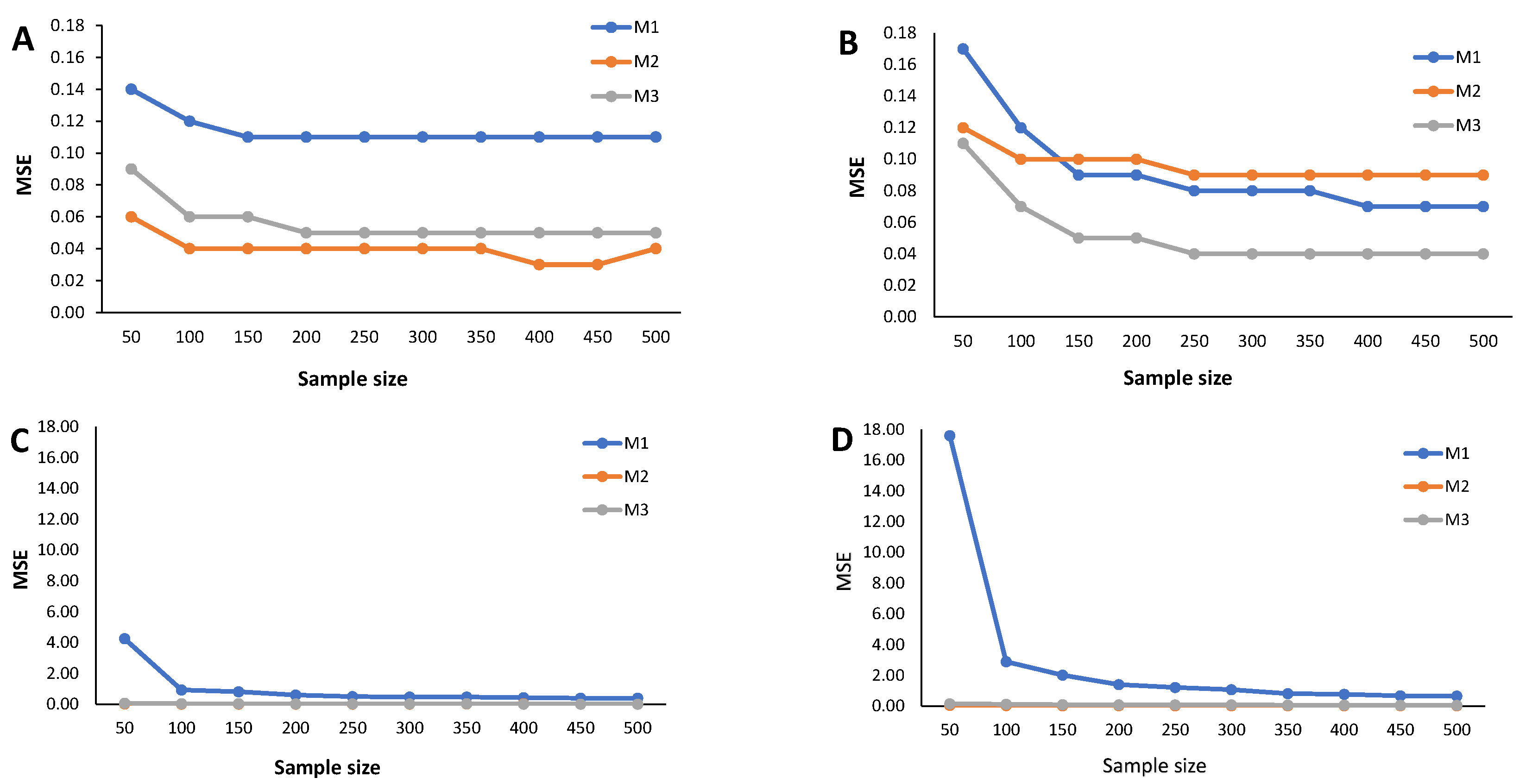
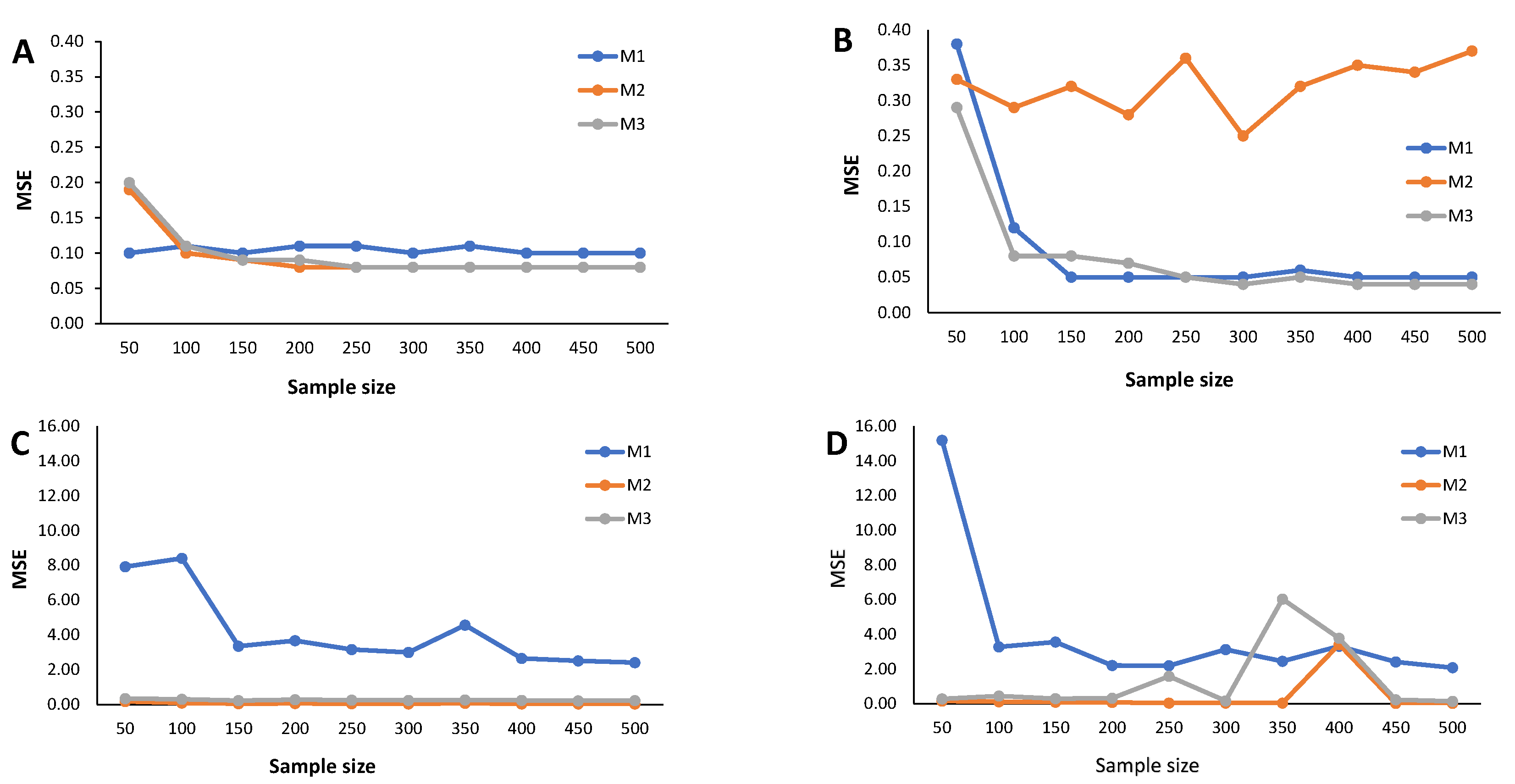
4. Simulation Study
- 1
- For to N (run N Monte Carlo simulations)
- S.1
- Simulate a sample with dimension n of antibodies concentration:
- Generate seronegative individuals using .
- The remaining individuals from the sample with dimension n are seropositive.
- Based on the theoretical model under consideration, generate a random sample of antibody concentration, with the sample size equal to n: the m observations of 1(S.1)i are drawn from the seronegative population, whereas the observations of 1(S.1)ii come from the seropositive population.
- S.2
- Fit a two-component mixture model to the simulated sample using the ECM algorithm described in Section 3.
- S.3
- Estimate the cutoff points based on the three methods under study, , where i denotes the ith simulated sample, represents the method under consideration, M1, M2, and M3, respectively.
- 2
- Store the estimated cutoff values in a matrix, , where the jth column contains the cutoff points’ sample with dimension N, for the jth method (), i.e., the N-dimensional column vector , and .
- 3
- Calculate the RB and the estimated MSE according to (9) for each cutoff points’ sample stored in the N-dimensional column vector
- 4
- Determine the empirical cumulative distribution function from the N-dimensional column vector , of the estimated cutoff points; then, construct a distribution-free approximate confidence interval for the true cutoff point from method , based on the percentile method [38].
5. Applications to SARS-CoV-2 Real Data
5.1. Patients’ Characteristics
5.2. Mixture Model Approach and Cutoff Points
6. Discussion and Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A. Bayesian Information Criteria (BIC), Sensitivity, Specificity, and Accuracy by Method for Each Antigen
| Method M1 | Method M2 | Method M3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antigen | Distribution | BIC | C | Sens (%) | Spec (%) | ACC (%) | C | Sens (%) | Spec (%) | ACC (%) | C | Sens (%) | Spec (%) | ACC (%) |
| RBD | Normal | 953.00 | 2.65 | 84.11 | 97.61 | 92.35 | 2.33 | 90.18 | 95.52 | 93.44 | 2.37 | 88.79 | 95.82 | 93.08 |
| Skew-Normal | 852.25 | 2.83 | 79.91 | 98.21 | 91.07 | 2.49 | 86.45 | 97.01 | 92.89 | 2.56 | 85.05 | 97.01 | 92.35 | |
| Student t | 959.60 | 4.16 | 0.09 | 100 | 61.38 | 2.34 | 90.18 | 95.52 | 93.44 | 2.38 | 88.79 | 96.42 | 93.44 | |
| Skew-t | 854.78 | 4.80 | — | 100.00 | — | 2.60 | 84.58 | 97.61 | 92.53 | 2.89 | 78.97 | 98.51 | 90.89 | |
| S1 | Normal | 561.81 | 2.43 | 63.08 | 97.91 | 84.34 | 2.13 | 81.31 | 95.52 | 89.98 | 2.12 | 82.71 | 95.52 | 90.53 |
| Skew-Normal | 561.63 | 2.58 | 50.93 | 98.81 | 80.15 | 2.27 | 71.03 | 97.01 | 86.89 | 2.30 | 69.63 | 97.31 | 86.52 | |
| Student t | 568.98 | 3.15 | 15.42 | 100.00 | 67.03 | 2.14 | 80.37 | 95.52 | 89.62 | 2.12 | 82.71 | 95.52 | 90.53 | |
| Skew-t | 568.27 | 3.27 | 10.28 | 100.00 | 65.03 | 2.27 | 71.03 | 97.01 | 86.89 | 2.31 | 69.16 | 97.31 | 86.34 | |
| S2 | Normal | 778.76 | 2.66 | 72.89 | 98.51 | 88.52 | 2.23 | 89.72 | 92.23 | 91.26 | 2.24 | 88.32 | 92.84 | 91.07 |
| Skew-Normal | 775.29 | 2.86 | 56.54 | 99.10 | 82.51 | 2.39 | 83.64 | 95.52 | 90.89 | 2.49 | 80.84 | 96.72 | 90.53 | |
| Student t | 785.73 | 3.51 | 9.35 | 100.00 | 64.66 | 2.24 | 88.32 | 92.84 | 91.07 | 2.25 | 87.38 | 93.13 | 90.89 | |
| Skew-t | 781.75 | 3.72 | 4.21 | 100.00 | 62.66 | 2.39 | 83.64 | 95.52 | 90.89 | 2.50 | 80.37 | 97.01 | 90.53 | |
| Normal | 1010.18 | 2.75 | 87.85 | 97.91 | 93.98 | 2.37 | 91.12 | 94.63 | 93.26 | 2.47 | 90.17 | 94.93 | 93.08 | |
| Skew-Normal | 916.15 | 2.98 | 79.44 | 99.40 | 91.62 | 2.46 | 90.19 | 94.93 | 93.08 | 2.58 | 89.25 | 96.12 | 93.44 | |
| Student t | 1016.84 | 4.34 | — | 100.00 | — | 2.39 | 90.65 | 94.63 | 93.08 | 2.48 | 89.72 | 95.22 | 93.08 | |
| Skew-t | 915.82 | 5.49 | — | 100.00 | — | 2.53 | 89.25 | 96.12 | 93.44 | 2.84 | 85.51 | 98.51 | 93.44 | |
Appendix B. Performance Measures for the Estimated Cutoff Point for Each Antigen
| Antigen | Cutoff | Sensitivity (%) | Specificity (%) | Accuracy (%) | AUC (CI 95%) |
|---|---|---|---|---|---|
| RBD | 2.15 | 94.39 | 94.33 | 94.35 | 98.50 (97.80, 99.30) |
| S1 | 2.07 | 86.92 | 93.73 | 91.07 | 96.10 (94.60, 97.60) |
| S2 | 2.33 | 86.92 | 94.63 | 91.62 | 94.90 (92.80, 97.00) |
| 2.81 | 86.92 | 98.51 | 93.98 | 98.30 (97.40, 99.20) |
Appendix C. Simulation Results
| Normal Distribution | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | cM1 | 95% CI (M1) | cM2 | 95% CI (M2) | cM3 | 95% CI (M3) | R.bias (M1) | MSE (M1) | R.bias (M2) | MSE (M2) | R.bias (M3) | MSE (M3) |
| = 0.3, , , | ||||||||||||
| 2.68 | (2.05–3.92) | 2.37 | (1.97–2.97) | 2.51 | (1.99–3.43) | 14.92 | 0.34 | 5.56 | 0.07 | 9.12 | 0.17 | |
| 2.64 | (2.19–3.34) | 2.34 | (2.08–2.65) | 2.47 | (2.10–2.93) | 13.57 | 0.18 | 4.49 | 0.03 | 7.39 | 0.07 | |
| 2.65 | (2.29–3.09) | 2.35 | (2.14–2.56) | 2.48 | (2.19–2.77) | 13.94 | 0.15 | 4.71 | 0.02 | 7.63 | 0.05 | |
| 2.66 | (2.35–3.03) | 2.35 | (2.18–2.52) | 2.48 | (2.23–2.73) | 14.20 | 0.14 | 4.79 | 0.02 | 7.81 | 0.05 | |
| 2.65 | (2.37–2.97) | 2.35 | (2.19–2.49) | 2.48 | (2.26–2.69) | 13.84 | 0.13 | 4.70 | 0.02 | 7.55 | 0.04 | |
| 2.65 | (2.39–2.95) | 2.35 | (2.12–2.49) | 2.47 | (2.29–2.69) | 13.81 | 0.12 | 4.65 | 0.02 | 7.46 | 0.04 | |
| 2.65 | (2.42–2.91) | 2.35 | (2.22–2.47) | 2.48 | (2.29–2.65) | 14.01 | 0.12 | 4.72 | 0.02 | 7.58 | 0.04 | |
| 2.64 | (2.43–2.88) | 2.34 | (2.23–2.46) | 2.47 | (2.30–2.63) | 13.54 | 0.11 | 4.49 | 0.01 | 7.26 | 0.03 | |
| 2.66 | (2.45–2.88) | 2.35 | (2.24–2.47) | 2.48 | (2.33–2.65) | 14.18 | 0.12 | 4.76 | 0.01 | 7.69 | 0.04 | |
| 2.65 | (2.48–2.88) | 2.35 | (2.25–2.45) | 2.48 | (2.34–2.64) | 13.92 | 0.12 | 4.71 | 0.01 | 7.54 | 0.04 | |
| = 0.6, , , | ||||||||||||
| 2.63 | (2.26–3.10) | 2.51 | (2.22–2.84) | 2.59 | (2.23–2.99) | 13.11 | 0.14 | 8.03 | 0.06 | 9.35 | 0.09 | |
| 2.64 | (2.35–2.94) | 2.50 | (2.30–2.72) | 2.58 | (2.32–2.86) | 13.23 | 0.12 | 7.66 | 0.04 | 8.94 | 0.06 | |
| 2.64 | (2.41–2.89) | 2.51 | (2.35–2.67) | 2.58 | (2.38–2.79) | 13.46 | 0.11 | 7.79 | 0.04 | 9.03 | 0.06 | |
| 2.65 | (2.47–2.83) | 2.51 | (2.38–2.64) | 2.58 | (2.41–2.75) | 13.68 | 0.11 | 7.87 | 0.04 | 9.11 | 0.05 | |
| 2.65 | (2.48–2.82) | 2.51 | (2.39–2.63) | 2.58 | (2.42–2.74) | 13.67 | 0.11 | 7.80 | 0.04 | 9.12 | 0.05 | |
| 2.65 | (2.50–2.81) | 2.51 | (2.40–2.61) | 2.59 | (2.44–2.72) | 13.72 | 0.11 | 7.88 | 0.04 | 9.18 | 0.05 | |
| 2.65 | (2.51–2.80) | 2.51 | (2.41–2.61) | 2.59 | (2.46–2.73) | 13.77 | 0.11 | 7.88 | 0.04 | 9.23 | 0.05 | |
| 2.65 | (2.53–2.78) | 2.51 | (2.42–2.59) | 2.58 | (2.47–2.71) | 13.65 | 0.11 | 7.75 | 0.03 | 9.06 | 0.05 | |
| 2.65 | (2.53–2.78) | 2.51 | (2.42–2.59) | 2.59 | (2.47–2.70) | 13.81 | 0.11 | 7.81 | 0.03 | 9.17 | 0.05 | |
| 2.65 | (2.54–2.76) | 2.51 | (2.43–2.59) | 2.59 | (2.48–2.69) | 13.89 | 0.11 | 7.89 | 0.04 | 9.23 | 0.05 | |
| = 0.9, , , | ||||||||||||
| 2.61 | (2.34–2.89) | 2.75 | (2.22–3.59) | 2.79 | (2.32–3.59) | 12.28 | 0.10 | 13.01 | 0.19 | 13.62 | 0.20 | |
| 2.64 | (2.45–2.82) | 2.72 | (2.53–2.99) | 2.76 | (2.50–3.03) | 13.56 | 0.11 | 12.09 | 0.10 | 12.24 | 0.11 | |
| 2.64 | (2.49–2.79) | 2.71 | (2.55–2.93) | 2.74 | (2.53–2.96) | 13.49 | 0.10 | 11.47 | 0.09 | 11.46 | 0.09 | |
| 2.65 | (2.51–2.78) | 2.71 | (2.57–2.89) | 2.74 | (2.55–2.91) | 13.63 | 0.11 | 11.47 | 0.08 | 11.48 | 0.09 | |
| 2.65 | (2.53–2.77) | 2.71 | (2.59–2.86) | 2.74 | (2.58–2.89) | 13.69 | 0.11 | 11.56 | 0.08 | 11.51 | 0.08 | |
| 2.65 | (2.54–2.76) | 2.71 | (2.60–2.83) | 2.74 | (2.59–2.87) | 13.64 | 0.10 | 11.45 | 0.08 | 11.38 | 0.08 | |
| 2.65 | (2.54–2.75) | 2.71 | (2.61–2.83) | 2.74 | (2.61–2.87) | 13.77 | 0.11 | 11.41 | 0.08 | 11.43 | 0.08 | |
| 2.65 | (2.55–2.74) | 2.70 | (2.61–2.81) | 2.73 | (2.61–2.85) | 13.64 | 0.10 | 11.25 | 0.08 | 11.28 | 0.08 | |
| 2.65 | (2.56–2.73) | 2.70 | (2.61–2.81) | 2.73 | (2.62–2.85) | 13.71 | 0.10 | 11.26 | 0.08 | 11.31 | 0.08 | |
| 2.65 | (2.57–2.73) | 2.70 | (2.62–2.80) | 2.73 | (2.63–2.84) | 13.75 | 0.10 | 11.29 | 0.08 | 11.30 | 0.08 | |
| Skew-Normal Distribution | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | cM1 |
95% CI (M1) | cM2 |
95% CI (M2) | cM3 |
95% CI (M3) |
R.bias (M1) |
MSE (M1) |
R.bias (M2) |
MSE (M2) |
R.bias (M3) |
MSE (M3) |
| = 0.3, , , | ||||||||||||
| 3.14 | (2.20–4.51) | 2.52 | (2.15–2.93) | 2.78 | (2.18–3.48) | 20.83 | 0.68 | 8.35 | 0.08 | 14.05 | 0.24 | |
| 3.13 | (2.29–4.28) | 2.50 | (2.23–2.76) | 2.76 | (2.24–3.33) | 20.32 | 0.55 | 7.37 | 0.05 | 13.04 | 0.18 | |
| 3.09 | (2.37–4.14) | 2.49 | (2.26–2.73) | 2.73 | (2.28–3.27) | 19.10 | 0.45 | 7.13 | 0.04 | 12.13 | 0.15 | |
| 3.08 | (2.39–3.94) | 2.49 | (2.28–2.67) | 2.72 | (2.30–3.16) | 18.56 | 0.39 | 6.98 | 0.04 | 11.74 | 0.13 | |
| 3.04 | (2.44–3.83) | 2.48 | (2.29–2.64) | 2.69 | (2.32–3.11) | 16.85 | 0.32 | 6.57 | 0.03 | 10.66 | 0.11 | |
| 3.05 | (2.47–3.81) | 2.48 | (2.29–2.64) | 2.70 | (2.36–3.12) | 17.39 | 0.32 | 6.62 | 0.03 | 10.89 | 0.11 | |
| 3.05 | (2.51–3.72) | 2.48 | (2.30–2.63) | 2.70 | (2.36–3.07) | 17.29 | 0.29 | 6.46 | 0.03 | 10.78 | 0.10 | |
| 3.02 | (2.51–3.72) | 2.48 | (2.32–2.62) | 2.68 | (2.37–3.04) | 16.19 | 0.27 | 6.35 | 0.03 | 10.10 | 0.09 | |
| 3.04 | (2.56–3.64) | 2.48 | (2.33–2.61) | 2.69 | (2.40–3.02) | 16.79 | 0.27 | 6.36 | 0.03 | 10.41 | 0.09 | |
| 3.04 | (2.56–3.58) | 2.48 | (2.34–2.60) | 2.69 | (2.39–2.99) | 16.77 | 0.26 | 6.31 | 0.03 | 10.37 | 0.09 | |
| = 0.6, , , | ||||||||||||
| 2.85 | (2.27–3.56) | 2.75 | (2.32–3.19) | 2.76 | (2.25–3.26) | 9.49 | 0.17 | 10.89 | 0.12 | 7.77 | 0.11 | |
| 2.85 | (2.39–3.37) | 2.74 | (2.39–3.08) | 2.74 | (2.38–3.09) | 9.67 | 0.12 | 10.38 | 0.10 | 7.32 | 0.07 | |
| 2.85 | (2.48–3.24) | 2.75 | (2.44–3.05) | 2.74 | (2.45–3.02) | 9.41 | 0.09 | 10.91 | 0.10 | 6.97 | 0.05 | |
| 2.85 | (2.51–3.23) | 2.75 | (2.45–3.01) | 2.73 | (2.46–3.00) | 9.47 | 0.09 | 10.78 | 0.10 | 6.92 | 0.05 | |
| 2.84 | (2.55–3.14) | 2.74 | (2.48–2.99) | 2.73 | (2.50–2.96) | 9.08 | 0.08 | 10.44 | 0.09 | 6.56 | 0.04 | |
| 2.84 | (2.58–3.12) | 2.75 | (2.49–2.98) | 2.73 | (2.52–2.93) | 9.22 | 0.08 | 10.75 | 0.09 | 6.61 | 0.04 | |
| 2.85 | (2.61–3.11) | 2.75 | (2.49–2.98) | 2.73 | (2.53–2.93) | 9.57 | 0.08 | 10.69 | 0.09 | 6.82 | 0.04 | |
| 2.84 | (2.60–3.08) | 2.74 | (2.49–2.97) | 2.72 | (2.53–2.90) | 9.02 | 0.07 | 10.49 | 0.09 | 6.37 | 0.04 | |
| 2.84 | (2.63–3.08) | 2.74 | (2.50–2.97) | 2.72 | (2.55–2.91) | 9.25 | 0.07 | 10.59 | 0.09 | 6.51 | 0.04 | |
| 2.85 | (2.65–3.06) | 2.75 | (2.51–2.96) | 2.73 | (2.56–2.89) | 9.39 | 0.07 | 10.85 | 0.09 | 6.60 | 0.04 | |
| = 0.9, , , | ||||||||||||
| 2.81 | (2.14–5.07) | 2.85 | (1.64–3.70) | 2.71 | (1.17–3.34) | 8.16 | 0.38 | 6.66 | 0.33 | 0.003 | 0.29 | |
| 2.82 | (2.48–3.14) | 3.04 | (2.36–3.69) | 2.86 | (2.49–3.21) | 8.27 | 0.12 | 13.68 | 0.29 | 5.32 | 0.08 | |
| 2.79 | (2.55–3.02) | 3.08 | (2.51–3.97) | 2.87 | (2.59–3.17) | 7.35 | 0.05 | 15.41 | 0.32 | 5.81 | 0.08 | |
| 2.81 | (2.62–2.99) | 3.07 | (2.59–3.90) | 2.88 | (2.64–3.12) | 7.98 | 0.05 | 14.89 | 0.28 | 6.24 | 0.07 | |
| 2.82 | (2.67–2.99) | 3.14 | (2.68–3.98) | 2.91 | (2.68–3.09) | 8.27 | 0.05 | 17.69 | 0.36 | 7.22 | 0.05 | |
| 2.81 | (2.66–2.96) | 3.07 | (2.69–3.56) | 2.89 | (2.73–3.07) | 8.01 | 0.05 | 14.87 | 0.25 | 6.62 | 0.04 | |
| 2.83 | (2.69–2.96) | 3.13 | (2.73–3.89) | 2.92 | (2.77–3.10) | 8.71 | 0.06 | 17.07 | 0.32 | 7.46 | 0.05 | |
| 2.82 | (2.67–2.95) | 3.14 | (2.72–3.96) | 2.90 | (2.73–3.06) | 8.31 | 0.05 | 17.52 | 0.35 | 6.88 | 0.04 | |
| 2.81 | (2.67–2.95) | 3.12 | (2.69–3.97) | 2.89 | (2.72–3.05) | 8.16 | 0.05 | 16.88 | 0.34 | 6.60 | 0.04 | |
| 2.81 | (2.67–2.94) | 3.14 | (2.71–4.00) | 2.89 | (2.74–3.05) | 8.08 | 0.05 | 17.72 | 0.37 | 6.55 | 0.04 | |
| Student t Distribution | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | cM1 |
95% CI (M1) | cM2 |
95% CI (M2) | cM3 |
95% CI (M3) |
R.bias (M1) |
MSE (M1) |
R.bias (M2) |
MSE (M2) |
R.bias (M3) |
MSE (M3) |
| = 0.3, , , | ||||||||||||
| 2.51 | (1.79–4.55) | 2.15 | (1.79–2.67) | 2.24 | (1.78–3.13) | 7.19 | 1.06 | −4.15 | 0.05 | −3.02 | 0.09 | |
| 2.47 | (1.98–3.69) | 2.16 | (1.95–2.36) | 2.24 | (1.95–2.54) | 5.36 | 0.19 | −4.09 | 0.02 | −3.20 | 0.03 | |
| 2.45 | (2.05–3.19) | 2.16 | (1.99–2.32) | 2.24 | (2.02–2.48) | 4.63 | 0.11 | −3.96 | 0.02 | −3.09 | 0.02 | |
| 2.46 | (2.08–3.14) | 2.15 | (2.02–2.28) | 2.24 | (2.04–2.44) | 4.92 | 0.09 | −4.16 | 0.01 | −3.22 | 0.02 | |
| 2.48 | (2.13–3.09) | 2.16 | (2.04–2.29) | 2.25 | (2.08–2.46) | 6.00 | 0.08 | −3.80 | 0.01 | −2.66 | 0.01 | |
| 2.48 | (2.17–2.97) | 2.16 | (2.05–2.28) | 2.25 | (2.09–2.42) | 5.86 | 0.06 | −3.73 | 0.01 | −2.57 | 0.01 | |
| 2.47 | (2.18–2.92) | 2.16 | (2.07–2.25) | 2.24 | (2.11–2.39) | 5.25 | 0.05 | −3.97 | 0.01 | −2.92 | 0.01 | |
| 2.48 | (2.18–2.95) | 2.16 | (2.07–2.26) | 2.25 | (2.11–2.39) | 5.93 | 0.06 | −3.85 | 0.01 | −2.69 | 0.009 | |
| 2.48 | (2.20–2.87) | 2.16 | (2.08–2.25) | 2.25 | (2.13–2.38) | 5.71 | 0.05 | −3.89 | 0.01 | −2.74 | 0.008 | |
| 2.48 | (2.23–2.85) | 2.16 | (2.08–2.24) | 2.25 | (2.13–2.28) | 5.82 | 0.04 | −3.83 | 0.01 | −2.64 | 0.007 | |
| = 0.6, , , | ||||||||||||
| 3.21 | (1.98–7.92) | 2.29 | (2.01–2.60) | 2.43 | (1.97–2.95) | 36.95 | 4.25 | −1.41 | 0.02 | 2.26 | 0.07 | |
| 2.91 | (2.10–5.01) | 2.30 | (2.11–2.49) | 2.44 | (2.08–2.79) | 24.15 | 0.92 | −1.35 | 0.01 | 2.47 | 0.04 | |
| 2.93 | (2.13–4.49) | 2.30 | (2.12–2.46) | 2.45 | (2.10–2.73) | 25.03 | 0.81 | −1.31 | 0.008 | 2.86 | 0.03 | |
| 2.92 | (2.22–4.15) | 2.31 | (2.17–2.44) | 2.46 | (2.20–2.70) | 24.72 | 0.59 | −1.05 | 0.005 | 3.39 | 0.02 | |
| 2.90 | (2.29–3.87) | 2.31 | (2.18–2.43) | 2.46 | (2.23–2.68) | 23.88 | 0.49 | −1.03 | 0.004 | 3.47 | 0.02 | |
| 2.90 | (2.30–3.89) | 2.31 | (2.20–2.43) | 2.46 | (2.26–2.68) | 23.79 | 0.47 | −1.02 | 0.004 | 3.49 | 0.02 | |
| 2.90 | (2.38–3.78) | 2.31 | (2.21–2.41) | 2.47 | (2.28–2.65) | 23.83 | 0.46 | −1.12 | 0.02 | 3.53 | 0.03 | |
| 2.89 | (2.39–3.73) | 2.31 | (2.22–2.39) | 2.47 | (2.29–2.64) | 23.73 | 0.43 | −1.04 | 0.03 | 3.54 | 0.01 | |
| 2.88 | (2.39–3.68) | 2.31 | (2.22–2.39) | 2.46 | (2.29–2.62) | 22.88 | 0.39 | −1.04 | 0.002 | 3.52 | 0.01 | |
| 2.89 | (2.42–3.63) | 2.31 | (2.23–2.39) | 2.47 | (2.32–2.62) | 23.57 | 0.39 | −0.97 | 0.002 | 3.69 | 0.01 | |
| = 0.9, , , | ||||||||||||
| 3.72 | (2.09–11.21) | 2.57 | (1.92–3.27) | 2.77 | (1.90–3.61) | 58.71 | 7.92 | 5.10 | 0.18 | 11.94 | 0.34 | |
| 3.98 | (2.11–8.04) | 2.63 | (2.20–3.12) | 2.89 | (2.16–3.61) | 69.90 | 8.40 | 7.72 | 0.09 | 16.72 | 0.31 | |
| 3.73 | (2.21–6.92) | 2.59 | (2.29–2.94) | 2.87 | (2.22–3.37) | 58.98 | 3.35 | 6.13 | 0.05 | 15.87 | 0.23 | |
| 3.87 | (2.35–7.11) | 2.61 | (2.37–2.94) | 2.92 | (2.37–3.39) | 65.12 | 3.67 | 7.09 | 0.06 | 18.01 | 0.28 | |
| 3.83 | (2.39–6.06) | 2.60 | (2.38–2.86) | 2.91 | (2.38–3.34) | 63.32 | 3.16 | 6.66 | 0.05 | 17.74 | 0.25 | |
| 3.81 | (2.45–5.85) | 2.60 | (2.41–2.85) | 2.92 | (2.39–3.29) | 62.54 | 2.99 | 6.67 | 0.04 | 17.82 | 0.24 | |
| 3.87 | (2.62–5.71) | 2.60 | (2.45–2.82) | 2.93 | (2.59–3.27) | 65.22 | 4.56 | 6.65 | 0.07 | 18.26 | 0.27 | |
| 3.79 | (2.69–5.49) | 2.60 | (2.44–2.80) | 2.93 | (2.58–3.27) | 62.09 | 2.65 | 6.66 | 0.04 | 18.29 | 0.24 | |
| 3.78 | (2.75–5.29) | 2.59 | (2.46–2.77) | 2.92 | (2.62–3.20) | 61.28 | 2.51 | 6.43 | 0.03 | 18.06 | 0.22 | |
| 3.76 | (2.81–5.22) | 2.59 | (2.47–2.77) | 2.93 | (2.67–3.23) | 60.43 | 2.41 | 6.46 | 0.03 | 18.21 | 0.23 | |
| Skew-t Distribution | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | cM1 |
95% CI (M1) | cM2 |
95% CI (M2) | cM3 |
95% CI (M3) |
R.bias (M1) |
MSE (M1) |
R.bias (M2) |
MSE (M2) |
R.bias (M3) |
MSE (M3) |
| = 0.3, , , | ||||||||||||
| 3.08 | (1.93–5.14) | 2.37 | (1.93–2.93) | 2.55 | (1.93–3.36) | −16.90 | 2.51 | −8.96 | 0.11 | −11.85 | 0.24 | |
| 3.09 | (2.11–5.81) | 2.34 | (2.04–2.68) | 2.52 | (2.07–3.14) | −16.41 | 1.56 | −9.94 | 0.09 | −12.89 | 0.22 | |
| 3.05 | (2.16–5.73) | 2.33 | (2.09–2.60) | 2.51 | (2.12–3.05) | −17.78 | 1.27 | −10.42 | 0.09 | −13.51 | 0.21 | |
| 3.05 | (2.19–5.49) | 2.32 | (2.09–2.57) | 2.49 | (2.13–2.98) | −17.65 | 1.11 | −10.69 | 0.09 | −13.81 | 0.21 | |
| 3.01 | (2.24–5.15) | 2.31 | (2.12–2.55) | 2.48 | (2.16–2.95) | −18.84 | 1.05 | −11.21 | 0.09 | −14.53 | 0.21 | |
| 3.03 | (2.28–5.12) | 2.32 | (2.13–2.53) | 2.49 | (2.18–2.91) | −18.21 | 0.94 | −11.02 | 0.09 | −14.21 | 0.20 | |
| 3.02 | (2.28–4.93) | 2.31 | (2.14–2.52) | 2.48 | (2.18–2.89) | −18.58 | 0.93 | −11.12 | 0.09 | −14.34 | 0.20 | |
| 3.02 | (2.30–4.75) | 2.31 | (2.14–2.51) | 2.48 | (2.19–2.86) | −18.58 | 0.89 | −11.10 | 0.09 | −14.30 | 0.20 | |
| 3.06 | (2.34–4.86) | 2.32 | (2.17–2.50) | 2.49 | (2.22–2.86) | −17.36 | 0.82 | −10.9 | 0.09 | −13.93 | 0.19 | |
| 3.03 | (2.37–4.83) | 2.31 | (2.17–2.49) | 2.49 | (2.24–2.85) | −18.18 | 0.81 | −11.12 | 0.09 | −14.23 | 0.19 | |
| = 0.6, , , | ||||||||||||
| 3.84 | (2.09–9.23) | 2.49 | (2.10–2.93) | 2.69 | (2.10–3.36) | 3.57 | 17.61 | −3.46 | 0.06 | −6.28 | 0.15 | |
| 3.52 | (2.17–8.77) | 2.47 | (2.16–2.77) | 2.67 | (2.17–3.23) | −4.89 | 2.89 | −4.39 | 0.04 | −7.05 | 0.12 | |
| 3.49 | (2.23–7.43) | 2.47 | (2.21–2.73) | 2.68 | (2.24–3.18) | −5.88 | 2.02 | −4.24 | 0.03 | −6.72 | 0.10 | |
| 3.43 | (2.31–6.72) | 2.47 | (2.26–2.70) | 2.68 | (2.29–3.14) | −7.52 | 1.41 | −4.17 | 0.02 | −6.60 | 0.08 | |
| 3.41 | (2.31–6.31) | 2.47 | (2.26–2.66) | 2.68 | (2.29–3.07) | −7.99 | 1.21 | −4.09 | 0.02 | −6.49 | 0.08 | |
| 3.44 | (2.37–6.06) | 2.47 | (2.29–2.66) | 2.69 | (2.34–3.08) | −7.14 | 1.07 | −4.97 | 0.03 | −7.16 | 0.08 | |
| 3.43 | (2.39–5.78) | 2.48 | (2.30–2.64) | 2.69 | (2.35–3.03) | −7.45 | 0.82 | −4.95 | 0.02 | −7.10 | 0.07 | |
| 3.45 | (2.41–5.58) | 2.48 | (2.31–2.64) | 2.71 | (2.36–3.04) | −6.91 | 0.77 | −4.64 | 0.02 | −6.62 | 0.07 | |
| 3.44 | (2.47–5.55) | 2.48 | (2.33–2.63) | 2.71 | (2.41–3.03) | −7.08 | 0.67 | −4.61 | 0.02 | −6.53 | 0.06 | |
| 3.42 | (2.47–5.38) | 2.48 | (2.32–2.63) | 2.70 | (2.39–3.01) | −7.68 | 0.66 | −4.79 | 0.02 | −6.83 | 0.06 | |
| = 0.9, , , | ||||||||||||
| 4.15 | (2.16–10.27) | 2.74 | (2.06–3.38) | 2.96 | (2.09–3.64) | 12.05 | 15.18 | −4.97 | 0.15 | −8.42 | 0.28 | |
| 4.08 | (2.25–9.52) | 2.75 | (1.98–3.33) | 2.98 | (2.09–3.69) | 10.36 | 3.27 | −4.61 | 0.11 | −7.88 | 0.45 | |
| 4.21 | (2.25–10.14) | 2.75 | (2.21–3.24) | 3.04 | (2.16–3.73) | 13.53 | 3.55 | −4.56 | 0.09 | −6.02 | 0.29 | |
| 4.09 | (2.35–8.18) | 2.76 | (2.36–3.25) | 3.04 | (2.32–3.72) | 10.53 | 2.20 | −4.06 | 0.07 | −5.89 | 0.32 | |
| 4.32 | (2.39–8.52) | 2.81 | (2.35–3.13) | 3.07 | (2.38–3.68) | 16.64 | 2.19 | −2.61 | 0.04 | −4.86 | 1.58 | |
| 4.52 | (2.38–8.19) | 2.82 | (2.42–3.17) | 3.19 | (2.41–3.68) | 22.05 | 3.12 | −1.94 | 0.04 | −1.36 | 0.16 | |
| 4.49 | (2.46–7.49) | 2.82 | (2.45–3.10) | 3.07 | (2.37–3.62) | 21.15 | 2.45 | −2.06 | 0.03 | −4.87 | 6.03 | |
| 4.52 | (2.53–8.22) | 2.88 | (2.46–3.08) | 3.22 | (2.45–3.65) | 22.01 | 3.31 | −0.18 | 3.42 | −0.29 | 3.77 | |
| 4.57 | (2.64–8.01) | 2.84 | (2.52–3.06) | 3.21 | (2.54–3.62) | 23.27 | 2.41 | −1.56 | 0.02 | −0.64 | 0.23 | |
| 4.50 | (2.56–7.14) | 2.83 | (2.48–3.06) | 3.21 | (2.48–3.61) | 21.54 | 2.07 | −1.85 | 0.02 | −0.62 | 0.14 | |
References
- Dávila, V.H.L.; Cabral, C.R.B.; Zeller, C.B. Finite Mixture of Skewed Distributions; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Lin, T.I.; Lee, J.C.; Yen, S.Y. Finite mixture modelling using the Skew-Normal distribution. Stat. Sin. 2007, 17, 909–927. [Google Scholar]
- Govaert, G.; Nadif, M. Clustering with block mixture models. Pattern Recognit. 2003, 36, 463–473. [Google Scholar] [CrossRef]
- Melnykov, V.; Wang, Y. Conditional mixture modeling and model-based clustering. Pattern Recognit. 2023, 133, 108994. [Google Scholar] [CrossRef]
- De Nicola, G.; Sischka, B.; Kauermann, G. Mixture models and networks: The stochastic blockmodel. Stat. Model. 2022, 22, 67–94. [Google Scholar] [CrossRef]
- Wine, Y.; Horton, A.P.; Ippolito, G.C.; Georgiou, G. Serology in the 21st Century: The Molecular-Level Analysis of the Serum Antibody Repertoire. Curr. Opin. Immunol. 2015, 35, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Rosado, J.; Pelleau, S.; Cockram, C.; Merkling, S.H.; Nekkab, N.; Demeret, C.; Meola, A.; Kerneis, S.; Terrier, B.; Fafi-Kremer, S.; et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: An antibody-based diagnostic and machine learning study. Lancet Microbe 2020, 2, E60–E69. [Google Scholar] [CrossRef] [PubMed]
- Domingues, T.; Mouriño, H.; Sepúlveda, N. Analysis of antibody data using Finite Mixture Models based on Scale Mixtures of Skew-Normal distributions. medRxiv 2021. [Google Scholar] [CrossRef]
- Parker, R.A.; Erdman, D.D.; Anderson, L.J. Use of mixture models in determining laboratory criterion for identification of seropositive individuals: Application to parvovirus B19 serology. J. Virol. Methods 1990, 27, 135–144. [Google Scholar] [CrossRef]
- Kafatos, G.; Andrews, N.J.; McConway, K.J.; Maple, P.A.; Brown, K.; Farrington, C.P. Is it appropriate to use fixed assay cut-offs for estimating seroprevalence? Epidemiol. Infect. 2016, 144, 887–895. [Google Scholar] [CrossRef][Green Version]
- Ridge, S.E.; Vizard, A.L. Determination of the optimal cutoff value for a serological assay: An example using the Johne’s Absorbed EIA. J. Clin. Microbiol. 1993, 31, 1256–1261. [Google Scholar] [CrossRef]
- Maple, P.A.C.; Simms, I.; Kafatos, G.; Solomou, M.; Fenton, K. Application of a noninvasive oral fluid test for detection of treponemal IgG in a predominantly HIV-infected population. Eur. J. Clin. Microbiol. Infect. Dis. 2006, 25, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Tong, D.D.; Buxser, S.; Vidmar, T.J. Application of a mixture model for determining the cutoff threshold for activity in high-throughput screening. Comput. Stat. Data Anal. 2007, 51, 4002–4012. [Google Scholar] [CrossRef]
- Baughman, A.L.; Bisgard, K.M.; Lynn, F.; Meade, B.D. Mixture model analysis for establishing a diagnostic cut-off point for pertussis antibody levels. Stat. Med. 2006, 25, 2994–3010. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Prata, S.; Domingues, T.D.; Leal, R.O.; Nunes, T.; Tavares, L.; Almeida, V.; Sepúlveda, N.; Gil, S. Detection and modeling of anti-Leptospira IgG prevalence in cats from Lisbon area and its correlation to retroviral infections, lifestyle, clinical and hematologic changes. Vet. Anim. Sci. 2020, 10, 100144. [Google Scholar] [CrossRef] [PubMed]
- Domingues, T.D.; Mouriño, H.; Sepúlveda, N. A statistical analysis of serological data from the UK myalgic encephalomyelitis/chronic fatigue syndrome biobank. AIP Conf. Proc. 2020, 2293, 420099. [Google Scholar]
- Hasibi, M.; Jafari, M.S.; Mortazavi, H.; Asadollahi, M.; Djavid, G.E. Determination of the accuracy and optimal cut-off point for ELISA test in diagnosis of human brucellosis in Iran. Acta Medica Iran. 2013, 51, 687–692. [Google Scholar]
- Rota, M.M.; Antolini, L. Finding the optimal cut-point for Gaussian and Gamma distributed biomarkers. Comput. Stat. Data Anal. 2014, 69, 1–14. [Google Scholar] [CrossRef]
- Habibzadeh, F.; Habibzadeh, P.; Yadollahie, M. On determining the most appropriate test cut-off value: The case of tests with continuous results. Biochem. Medica 2016, 26, 297–307. [Google Scholar] [CrossRef]
- Blacksell, S.; Lim, C.; Tanganuchitcharnchai, A.; Jintaworn, S.; Kantipong, P.; Richards, A.L.; Paris, D.H.; Limmathurotsakul, D.; Day, N. Optimal cutoff and accuracy of an IgM enzyme-linked immunosorbent assay for diagnosis of acute scrub typhus in northern Thailand: An alternative reference method to the IgM immunofluorescence assay. J. Clin. Microbiol. 2016, 54, 1472–1478. [Google Scholar] [CrossRef]
- Perkins, N.J.; Schisterman, E.F. The inconsistency of “optimal” cut-points using two ROC based criteria. Am. J. Epidemiol. 2006, 163, 670–675. [Google Scholar] [CrossRef]
- Unal, I. Defining an optimal cut-point value in ROC analysis: An alternative approach. Comput. Math. Methods Med. 2017, 2017, 3762651. [Google Scholar] [CrossRef] [PubMed]
- Migchelsen, S.J.; Martin, D.L.; Southisombath, K.; Turyaguma, P.; Heggen, A.; Rubangakene, P.P.; Joof, H.; Makalo, P.; Cooley, G.; Gwyn, S.; et al. Defining Seropositivity Thresholds for Use in Trachoma Elimination Studies. PLoS Neglected Trop. Dis. 2017, 11, e0005230. [Google Scholar] [CrossRef] [PubMed]
- Gay, N.J. Analysis of serological surveys using mixture models: Application to a survey of parvovirus B19. Stat. Med. 1996, 15, 1567–1573. [Google Scholar] [CrossRef]
- Azzalini, A. The Skew-Normal and Related Families; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Basso, R.M.; Lachos, V.H.; Cabral, C.R.B.; Gosh, P. Robust mixture modelling based on scale mixtures of skew-normal distributions. Comput. Stat. Data Anal. 2010, 54, 2926–2941. [Google Scholar] [CrossRef]
- Domingues, T.; Mouriño, H.; Sepúlveda, N. Analysis of antibody data using Skew-Normal and Skew-t mixture models. REVSTAT-Stat. J. (Fourthcoming) 2022. Available online: https://revstat.ine.pt/index.php/REVSTAT/article/view/455 (accessed on 24 November 2023).
- Dempster, A.P.; Rubin, D.B. Maximum likelihood estimation from incomplete data via the EM algorithm. J. R. Stat. Soc. 1977, 39, 1–38. [Google Scholar]
- Meng, X.L.; Rubin, D.B. Maximum likelihood estimation via the ECM algorithm: A general framework. Biometrika 1993, 80, 267–278. [Google Scholar] [CrossRef]
- Liu, C.; Rubin, D.B. The ECME algorithm: A simple extension of EM and ECM with faster monotone convergence. Biometrika 1994, 81, 633–648. [Google Scholar] [CrossRef]
- McLachlan, G.J.; Krishnan, T. The EM Algorithm and Extensions; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Sepúlveda, N.; Stresman, G.; White, M.T.; Drakeley, C.J. Current Mathematical Models for Analyzing Anti-Malarial Antibody Data with an Eye to Malaria Elimination and Eradication. J. Immunol. Res. 2015, 10, 738030. [Google Scholar] [CrossRef]
- Saraswati, K.; Phanichkrivalkosil, M.; Day, N.; Blacksell, S.D. The validity of diagnostic cut-offs for commercial and in-house scrub typhus IgM and IgG ELISAs: A review of the evidence. PLoS Neglected Trop. Dis. 2019, 13, e0007158. [Google Scholar] [CrossRef]
- Brent, R.P. Algorithms for Minimization Without Derivatives; Prentice-Hall: Hoboken, NJ, USA, 1973; pp. 73–76. [Google Scholar]
- Prates, M.O.; Lachos, V.H.; Cabral, C. Fitting finite mixture of scale mixture of skew-normal distributions. J. Stat. Softw. 2013, 54, 1–20. [Google Scholar] [CrossRef]
- Wolodzko, T. Additional Univariate and Multivariate Distributions. R CRAN. 2020. Available online: https://github.com/twolodzko/extraDistr (accessed on 24 November 2023).
- Azzalini, A. The Skew-Normal and Related Distributions Such as the Skew-t. R CRAN. 2020. Available online: http://azzalini.stat.unipd.it/SN/ (accessed on 24 November 2023).
- Meeker, W.Q.; Han, G.J.; Escobar, L.A. Statistical Intervals: A Guide for Practitioners and Researchers; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Stringhini, S.; Wisniak, A.; Piumatti, G.; Azman, A.; Lauer, S.; Baysson, H.; Ridder, D.; Petrovic, D.; Schrempft, S.; Marcus, K.; et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 2020, 396, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Larremore, D.; Fosdick, B.; Bubar, K.; Zhang, S.; Kissler, S.; Metcalf, C.; Buckee, C.; Grad, Y. Estimating SARS-CoV-2 seroprevalence and epidemiological parameters with uncertainty from serological surveys. Elife 2021, 10, e64206. [Google Scholar] [CrossRef] [PubMed]
- López-Ratón, M.; Rodríguez-Álvarez, M.X.; Cadarso-Suárez, C.; Gude-Sampedro, F. OptimalCutpoints: An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. J. Stat. Softw. 2014, 61, 1–36. [Google Scholar] [CrossRef]



| Seronegative Population | Seropositive Population | |||||||
|---|---|---|---|---|---|---|---|---|
| Distribution | ||||||||
| Normal | 1.72 | 0.30 | 0.00 | — | 3.35 | 0.60 | 0.00 | — |
| Skew-Normal | 1.41 | 0.40 | 5.77 | — | 4.09 | 0.90 | −9.12 | — |
| Student-t | 1.65 | 0.10 | 0.00 | 3.00 | 3.35 | 0.60 | 0.00 | 3.00 |
| Skew-t | 1.46 | 0.20 | 3.64 | 2.91 | 4.08 | 0.90 | −7.93 | 18.07 |
| Distribution | M1 | M2 | M3 | Distribution | M1 | M2 | M3 | ||
|---|---|---|---|---|---|---|---|---|---|
| Mixture of Normals | 0.3 | 2.33 | 2.24 | 2.30 | Mixture of Student-t | 0.3 | 2.34 | 2.25 | 2.31 |
| 0.6 | 2.33 | 2.33 | 2.37 | 0.6 | 2.34 | 2.33 | 2.38 | ||
| 0.9 | 2.33 | 2.43 | 2.46 | 0.9 | 2.34 | 2.44 | 2.48 | ||
| Mixture of Skew-Normals | 0.3 | 2.60 | 2.33 | 2.44 | Mixture of Skew-t | 0.3 | 3.71 | 2.38 | 2.64 |
| 0.6 | 2.60 | 2.48 | 2.56 | 0.6 | 3.71 | 2.58 | 2.87 | ||
| 0.9 | 2.60 | 2.67 | 2.71 | 0.9 | 3.71 | 2.88 | 3.23 |
| Seronegative Population | Seropositive Population | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Antigen | Distribution | v | v | ||||||
| RBD | Skew-Normal | 1.435 | 0.125 | 6.318 | — | 4.077 | 0.767 | −7.634 | — |
| S1 | Skew-Normal | 1.569 | 0.062 | 2.687 | — | 2.339 | 0.321 | 1.062 | — |
| S2 | Skew-Normal | 1.583 | 0.096 | 2.804 | — | 2.817 | 0.212 | 0.450 | — |
| Skew-t | 1.352 | 0.121 | 5.751 | 4.873 | 3.885 | 0.367 | −6.482 | 4.873 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dias-Domingues, T.; Mouriño, H.; Sepúlveda, N. Classification Methods for the Serological Status Based on Mixtures of Skew-Normal and Skew-t Distributions. Mathematics 2024, 12, 217. https://doi.org/10.3390/math12020217
Dias-Domingues T, Mouriño H, Sepúlveda N. Classification Methods for the Serological Status Based on Mixtures of Skew-Normal and Skew-t Distributions. Mathematics. 2024; 12(2):217. https://doi.org/10.3390/math12020217
Chicago/Turabian StyleDias-Domingues, Tiago, Helena Mouriño, and Nuno Sepúlveda. 2024. "Classification Methods for the Serological Status Based on Mixtures of Skew-Normal and Skew-t Distributions" Mathematics 12, no. 2: 217. https://doi.org/10.3390/math12020217
APA StyleDias-Domingues, T., Mouriño, H., & Sepúlveda, N. (2024). Classification Methods for the Serological Status Based on Mixtures of Skew-Normal and Skew-t Distributions. Mathematics, 12(2), 217. https://doi.org/10.3390/math12020217






