Identifying the Informational/Signal Dimension in Principal Component Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Definition of Signal and Noise
2.2. Stopping Rules
2.2.1. Kaiser–Guttman
2.2.2. Broken Stick
2.2.3. Information Dimension
2.2.4. Rencher Bartlett-Kind Test
2.2.5. Eigenvalues p-Value
2.2.6. Pseudo-F Ratio
2.2.7. RV Coefficient
2.2.8. Random Average under Permutation
2.2.9. Bootstrap and Parallel Permutation
- Apply PCA to the original matrix , saving the resulting units’ coordinates in a matrix .
- Extract from the units set (the pseudo sampling universe in bootstrap re-sampling) a bootstrap sample of size , e.g., a data table , and submit it to PCA, obtaining a matrix of coordinates .
- Due to the nature of the bootstrap sampling (extraction of units with replacement, and thus possible repetition), the units in are a subset of . Thus, from , a matrix must be extracted, with the same units and in the same order of .
- For each dimension k of PCA, after a Procrustes adjustment [49] and using the first k principal components, compute the Pearson correlation between the coordinates of the kth principal components of and , respectively; the higher is this correlation, the better is the agreement between bootstrap and reference ordination and the more stable is its representation through the k-dimensional sample.
- Generate a matrix by randomly permuting the data within each variable of and repeat Steps 1–4 for this new matrix.
- Compare the correlation with the correlation obtained at Step 5.
- Repeat Steps 1–6 B times to get a p-value as the proportion of permutations for which resulted .
- Starting with the least ordination axis, and iterating towards the first one, a p-value suggests that this dimension is significantly more stable than the one found for the same dimension in the PCA of a random dataset. Thus, it is interpreted as signal.
- Once the kth dimension is deemed to carry signal, the test may stop and all larger dimensions are also taken into account, irrespective of their corresponding probabilities. Otherwise, the kth ordination dimension is considered noise, because it is both unstable and indistinguishable from an ordination of random data, and the probability of the next th axis is examined. See Pillar [19] for further details.
- As the comparison between real and bootstrapped data is performed considering the correlation among units’ coordinates, the method works with any kind of multidimensional scaling (including non-metric one) applied to any kind of data and resemblance measure. In addition, other measures of agreement, such as rank correlation, may be used instead.
- The bootstrap procedure to build the empirical probability of the obtained correlation may be repeated for the increasing size s of the bootstrapped sample, up to the table size n. This way the method is able to evaluate the sufficiency of the sample size, as it results by the stability of probabilities across increasing bootstrap sample sizes (as in [50]). Indeed, while increasing the sample size, should the probabilities associated to the kth dimension keep stable and larger than , this ordination axis is truly noise. Should they be decreasing, but still larger than even with a bootstrap sample of size n, this may be interpreted as the need of an even larger sample size to ensure that the ordination axis under examination is confirmed as either signal or noise.
2.3. The Simulation
2.3.1. Data Generation
- The eigenvalues of the simulated data are specified and normalized to sum up to their number.
- A data table is built with random numbers extracted from a specified distribution, representing a large number of units in rows and a specified number of variables in columns.
- The PCA of the data table is computed.
- The data table is reconstructed through Equation (1), substituting the eigenvalues issued by the PCA with the predefined ones.
- Steps 3 and 4 are iterated. This way the data table’s eigenvalues converge to the predefined ones, ensuring that the sum of the absolute differences between specified and resulting eigenvalues through iteration converges to zero. The procedure stops when the sum results less than a threshold, here fixed at
- A random sample with the specified number of units is extracted.
- Noise is added to the sample. It is a normally distributed random variable with zero mean and specified variance. The larger is the variance, the larger is the noise added to the defined eigenstructure. This way, to population zero-eigenvalues may correspond non-zero ones in the samples.
2.3.2. The Experiment
- A population with 1000 units and 9 variables was generated with the procedure described in Section 2.3.1 with the specified eigenvalues.
- A sample with 30 units was randomly extracted from the population.
- To each simulated observation noise was added, extracted by a normally distributed random variable with zero mean and fixed variance.
- PCA was computed on the sample and the eigenvalues were sorted according to decreasing size.
- The ten methods were applied. For the distribution-free rules, each obtained statistics was compared to a number B of statistics obtained by applying the same method with the observations randomly permuted within each original variable. Then, a p-value for was calculated by the proportion of permutations for which resulted . If the p-value was smaller or equal than , it was considered significant and its corresponding dimension accounted for signal. Since and 1000 tests were applied in the following step, we set permutations, considering that in each distribution-free test this was enough to know whether .
- Steps (1)–(5) were repeated 1000 times for each of the 11 eigenstructures and each of nine fixed noise variances, ranging from 0 (no-noise) to 0.08 with step 0.01. For each repetition, a check was done whether the method correctly identified the expected dimension of the signal and treated the following as noise. This gave the proportion of correct answer for both power and type-I error, respectively.
- For each method, mean power and type-I error resulted for 99 combinations of 11 eigenstructures and 9 noise levels.
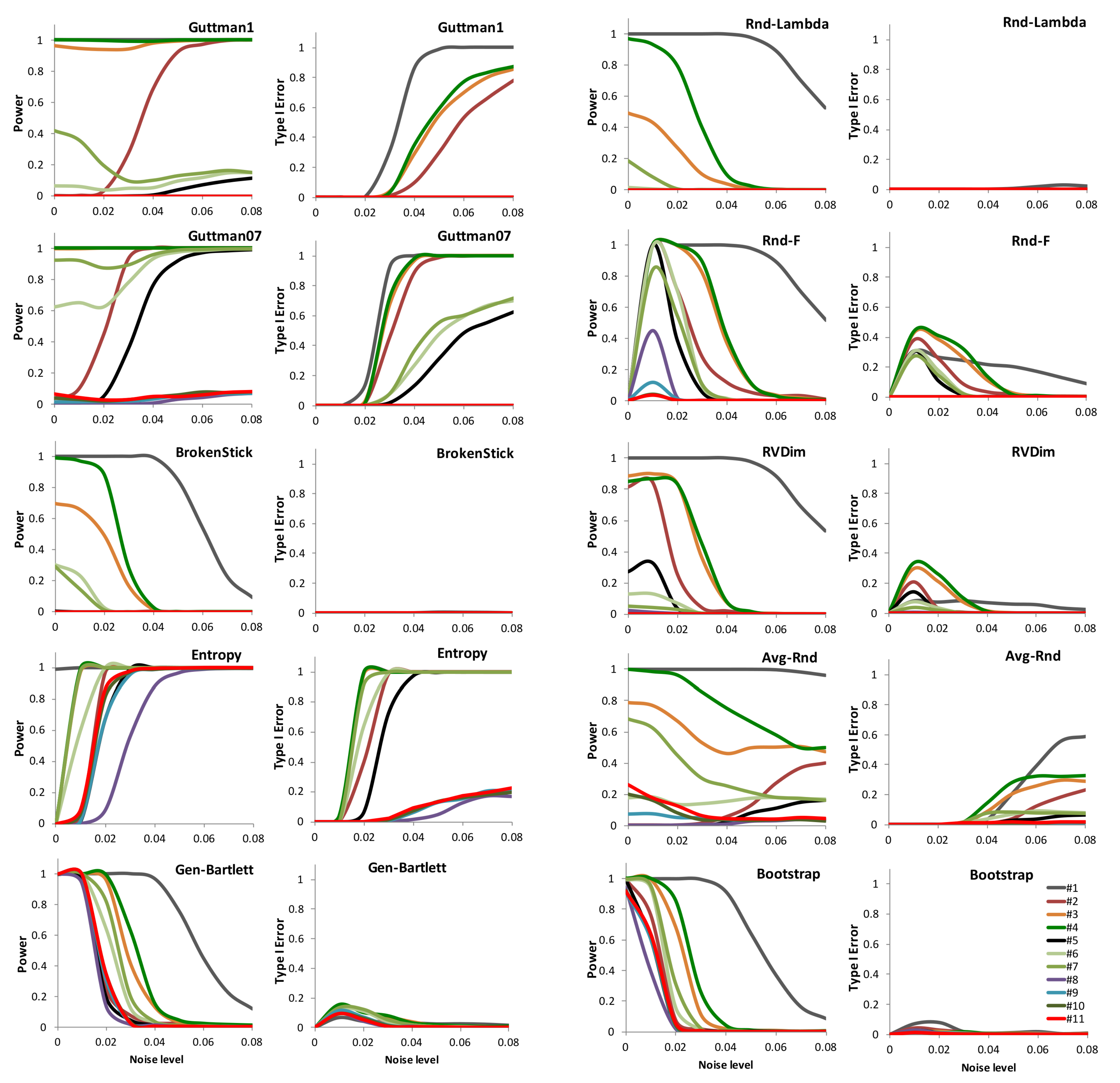
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gnanadesikan, R.; Kettenring, J. Robust estimates, residuals, and outlier detection with multiresponse data. Biometrics 1972, 28, 81–124. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis; Springer: Berlin, Germany, 2002. [Google Scholar]
- Rencher, A.C. Methods of Multivariate Analysis; Wiley Interscience: New York, NY, USA, 2002. [Google Scholar]
- Lebart, L.; Piron, M.; Morineau, A. Statistique Exploratoire Multidimensionnelle—Visualisation et Inférence en Fouilles de Données; Dunod: Paris, France, 2016. [Google Scholar]
- Guttman, L. Some necessary conditions for common-factor analysis. Psychometrika 1954, 19, 149–161. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Discarding Variables in a Principal Component Analysis. I: Artificial Data. Appl. Stat. 1972, 21, 160–173. [Google Scholar] [CrossRef]
- Cattell, R.B. The scree test for the number of factors. Multivar. Behav. Res. 1966, 1, 245–276. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.A. Stopping Rules in Principal Components Analysis: A Comparison of Heuristical and Statistical Approaches. Ecology 1993, 74, 2204–2214. [Google Scholar] [CrossRef]
- Peres-Neto, P.R.; Jackson, D.A.; Somers, K.M. How many principal components? stopping rules for determining the number of non-trivial axes revisited. Comput. Stat. Data Anal. 2005, 49, 974–997. [Google Scholar] [CrossRef]
- Frontier, S. Étude de la décroissance des valeurs propres dans une analyse en composantes principales: Comparaison avec le modèle du bâton brisé. J. Exp. Mar. Biol. Ecol. 1976, 25, 67–75. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, NY, USA, 1998. [Google Scholar]
- Caron, P.O. A Monte Carlo examination of the broken-stick distribution to identify components to retain in principal component analysis. J. Stat. Comput. Simul. 2016, 86, 2405–2410. [Google Scholar] [CrossRef]
- Bartlett, M.S. A note on the multiplying factors for various χ 2 approximations. J. R. Stat. Soc. Ser. B Math. 1954, 16, 296–298. [Google Scholar]
- Wold, S. Cross-validatory estimation of the number of components in factor and principal components models. Technometrics 1978, 20, 397–405. [Google Scholar] [CrossRef]
- Eastment, H.; Krzanowski, W. Cross-validatory choice of the number of components from a principal component analysis. Technometrics 1982, 24, 73–77. [Google Scholar] [CrossRef]
- Minka, T.P. Automatic choice of dimensionality for PCA. In Proceedings of the 13th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 3–8 December 2001; pp. 598–604. [Google Scholar]
- Auer, P.; Gervini, D. Choosing principal components: A new graphical method based on Bayesian model selection. Commun. Stat. Simul. Comput. 2008, 37, 962–977. [Google Scholar] [CrossRef]
- Wang, M.; Kornblau, S.M.; Coombes, K.R. Decomposing the Apoptosis Pathway into Biologically Interpretable Principal Components. Cancer Inform. 2017, 17. [Google Scholar] [CrossRef]
- Pillar, V.D. The bootstrapped ordination re-examined. J. Veg. Sci. 1999, 10, 895–902. [Google Scholar] [CrossRef]
- Vieira, V.M. Permutation tests to estimate significances on Principal Components Analysis. Comput. Ecol. Softw. 2012, 2, 103–123. [Google Scholar]
- Camiz, S.; Pillar, V.D. Comparison of Single and Complete Linkage Clustering with the Hierarchical Factor Classification of Variables. Community Ecol. 2007, 8, 25–30. [Google Scholar] [CrossRef]
- Feoli, E.; Zuccarello, V. Fuzzy Sets and Eigenanalysis in Community Studies: Classification and Ordination are “Two Faces of the Same Coin”. Community Ecol. 2013, 14, 164–171. [Google Scholar] [CrossRef]
- Jolliffe, I.T. A note on the use of principal components in regression. J. R. Stat. Soc. Ser. C Appl. Stat. 1982, 31, 300–303. [Google Scholar] [CrossRef]
- Céréghino, R.; Pillar, V.; Srivastava, D.; de Omena, P.M.; MacDonald, A.A.M.; Barberis, I.M.; Corbara, B.; Guzman, L.M.; Leroy, C.; Bautista, F.O.; et al. Constraints on the Functional Trait Space of Aquatic Invertebrates in Bromeliads. Funct. Ecol. 2018, 32, 2435–2447. [Google Scholar] [CrossRef]
- Ferré, L. Selection of components in principal component analysis: A comparison of methods. Comput. Stat. Data Anal. 1995, 19, 669–682. [Google Scholar] [CrossRef]
- Dray, S. On the number of principal components: A test of dimensionality based on measurements of similarity between matrices. Comput. Stat. Data Anal. 2008, 52, 2228–2237. [Google Scholar] [CrossRef]
- Karr, J.; Martin, T. Random number and principal components: Further searches for the unicorn. In The Use of Multivariate Statistics in Wildlife Habitat; Capen, D., Ed.; United Forest Service: Washington, DC, USA, 1981; pp. 20–24. [Google Scholar]
- Gauch, H.G.J. Reduction by Eigenvector Ordinations. Ecology 1982, 63, 1643–1649. [Google Scholar] [CrossRef]
- Jackson, D.A.; Somers, K.M.; Harvey, H.H. Null models and fish communities: Evidence of nonrandom patterns. Am. Nat. 1992, 139, 930–951. [Google Scholar] [CrossRef]
- Abdi, H. Singular Value Decomposition (SVD) and Generalized Singular Value Decomposition (GSVD). In Encyclopedia of Measurement and Statistics; Salkind, N., Ed.; Sage: Thousand Oaks, CA, USA, 2007. [Google Scholar]
- Eckart, C.; Young, G. The approximation of one matrix by another of lower rank. Psychometrika 1936, 1, 211–218. [Google Scholar] [CrossRef]
- Basilevsky, A. Statistical Factor Analysis and Related Methods: Theory and Applications; Wiley-Blackwell: New York, NY, USA, 1994. [Google Scholar]
- Malinvaud, E. Data analysis in applied socio-economic statistics with special consideration of correspondence analysis. In Proceedings of the Academy of Marketing Science (AMS) Annual Conference, Bal Harbour, FL, USA, 27–30 May 1987. [Google Scholar]
- Ben Ammou, S.; Saporta, G. On the connection between the distribution of eigenvalues in multiple correspondence analysis and log-linear models. Revstat Stat. J. 2003, 1, 42–79. [Google Scholar]
- Wishart, J. The Generalised Product Moment Distribution in Samples from a Normal Multivariate Population. Biometrika 1928, 20, 32–52. [Google Scholar] [CrossRef]
- Anderson, T. Asymptotic Theory for Principal Component Analysis. Ann. Math. Stat. 1963, 34, 122–148. [Google Scholar] [CrossRef]
- Jackson, J.E. A User’s Guide to Principal Components; John Wiley & Sons: New York, NY, USA, 1991. [Google Scholar]
- Efron, B. Bootstrap methods: Another look at jackknife. Ann. Stat. 1979, 7, 1–26. [Google Scholar] [CrossRef]
- Manly, B.F. Randomization, Bootstrap and Monte Carlo Methods in Biology; Texts in Statistical Science; Chapman & Hall/CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Efron, B.; Tibshirani, R. An Introduction to the Bootstrap; Chapman and Hall: New York, NY, USA, 1993. [Google Scholar]
- Barton, D.; David, F. Some notes on ordered random intervals. J. R. Stat. Soc. Ser. B Methodol. 1956, 18, 79–94. [Google Scholar]
- Cangelosi, R.; Goriely, A. Component retention in principal component analysis with application to cDNA microarray data. Biol. Direct 2007, 2, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Jost, L. Entropy and diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Ter Braak, C.J. CANOCO—A FORTRAN Program for Canonical Community Ordination by [Partial][Detrended][Canonical] Correspondence Analysis, Principal Components Analysis and Redundancy Analysis (Version 2.1); Technical Report; Agricultural Mathematic Group: Wageningen, The Netherlands, 1988. [Google Scholar]
- Ter Braak, C.J. CANOCO Version 3.1, Update Notes; Technical Report; Agricultural Mathematics Group: Wageningen, The Netherlands, 1990. [Google Scholar]
- Escoufier, Y. Le Traitement des Variables Vectorielles. Biometrics 1973, 29, 751–760. [Google Scholar] [CrossRef]
- Robert, P.; Escoufier, Y. A Unifying Tool for Linear Multivariate Statistical Methods: The RV-Coefficient. Appl. Stat. 1976, 25, 257–265. [Google Scholar] [CrossRef]
- Josse, J.; Pagès, J.; Husson, F. Testing the significance of the RV coefficient. Comput. Stat. Data Anal. 2008, 53, 82–91. [Google Scholar] [CrossRef]
- Schönemann, P.H.; Carroll, R.M. Fitting one matrix to another under choice of a central dilation and a rigid motion. Psychometrika 1970, 35, 245–255. [Google Scholar] [CrossRef]
- Pillar, V.D. Sampling sufficiency in ecological surveys. Abstr. Bot. 1998, 22, 37–48. [Google Scholar]
- Stapleton, J. Linear Statistical Models; Wiley: New York, NY, USA, 1995. [Google Scholar]
- Camacho, J.; Ferrer, A. Cross-validation in PCA models with the element-wise k-fold (ekf) algorithm: Theoretical aspects. J. Chemom. 2012, 26, 361–373. [Google Scholar] [CrossRef]
- Camacho, J.; Ferrer, A. Cross-validation in PCA models with the element-wise k-fold (ekf) algorithm: Practical aspects. Chemom. Intell. Lab. Syst. 2014, 131, 37–50. [Google Scholar] [CrossRef]

| Eigen | Specified Eigenvalues (Proportions) | TrueDim | Ratio | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1.0 |
| #2 | 0.810 | 0.140 | 0.050 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 3 | 16.1 |
| #3 | 0.545 | 0.273 | 0.182 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 3 | 3.0 |
| #4 | 0.416 | 0.315 | 0.268 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 3 | 1.6 |
| #5 | 0.568 | 0.201 | 0.109 | 0.071 | 0.051 | 0.000 | 0.000 | 0.000 | 0.000 | 5 | 11.2 |
| #6 | 0.359 | 0.221 | 0.167 | 0.136 | 0.117 | 0.000 | 0.000 | 0.000 | 0.000 | 5 | 3.1 |
| #7 | 0.251 | 0.211 | 0.191 | 0.178 | 0.168 | 0.000 | 0.000 | 0.000 | 0.000 | 5 | 1.5 |
| #8 | 0.386 | 0.193 | 0.129 | 0.096 | 0.077 | 0.064 | 0.055 | 0.000 | 0.000 | 7 | 7.0 |
| #9 | 0.225 | 0.170 | 0.145 | 0.129 | 0.118 | 0.110 | 0.103 | 0.000 | 0.000 | 7 | 2.2 |
| #10 | 0.181 | 0.157 | 0.145 | 0.137 | 0.131 | 0.126 | 0.122 | 0.000 | 0.000 | 7 | 1.5 |
| #11 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.000 | 0.000 | 7 | 1.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camiz, S.; Pillar, V.D. Identifying the Informational/Signal Dimension in Principal Component Analysis. Mathematics 2018, 6, 269. https://doi.org/10.3390/math6110269
Camiz S, Pillar VD. Identifying the Informational/Signal Dimension in Principal Component Analysis. Mathematics. 2018; 6(11):269. https://doi.org/10.3390/math6110269
Chicago/Turabian StyleCamiz, Sergio, and Valério D. Pillar. 2018. "Identifying the Informational/Signal Dimension in Principal Component Analysis" Mathematics 6, no. 11: 269. https://doi.org/10.3390/math6110269
APA StyleCamiz, S., & Pillar, V. D. (2018). Identifying the Informational/Signal Dimension in Principal Component Analysis. Mathematics, 6(11), 269. https://doi.org/10.3390/math6110269





