Estimation of Unknown Parameters of Truncated Normal Distribution under Adaptive Progressive Type II Censoring Scheme
Abstract
:1. Introduction
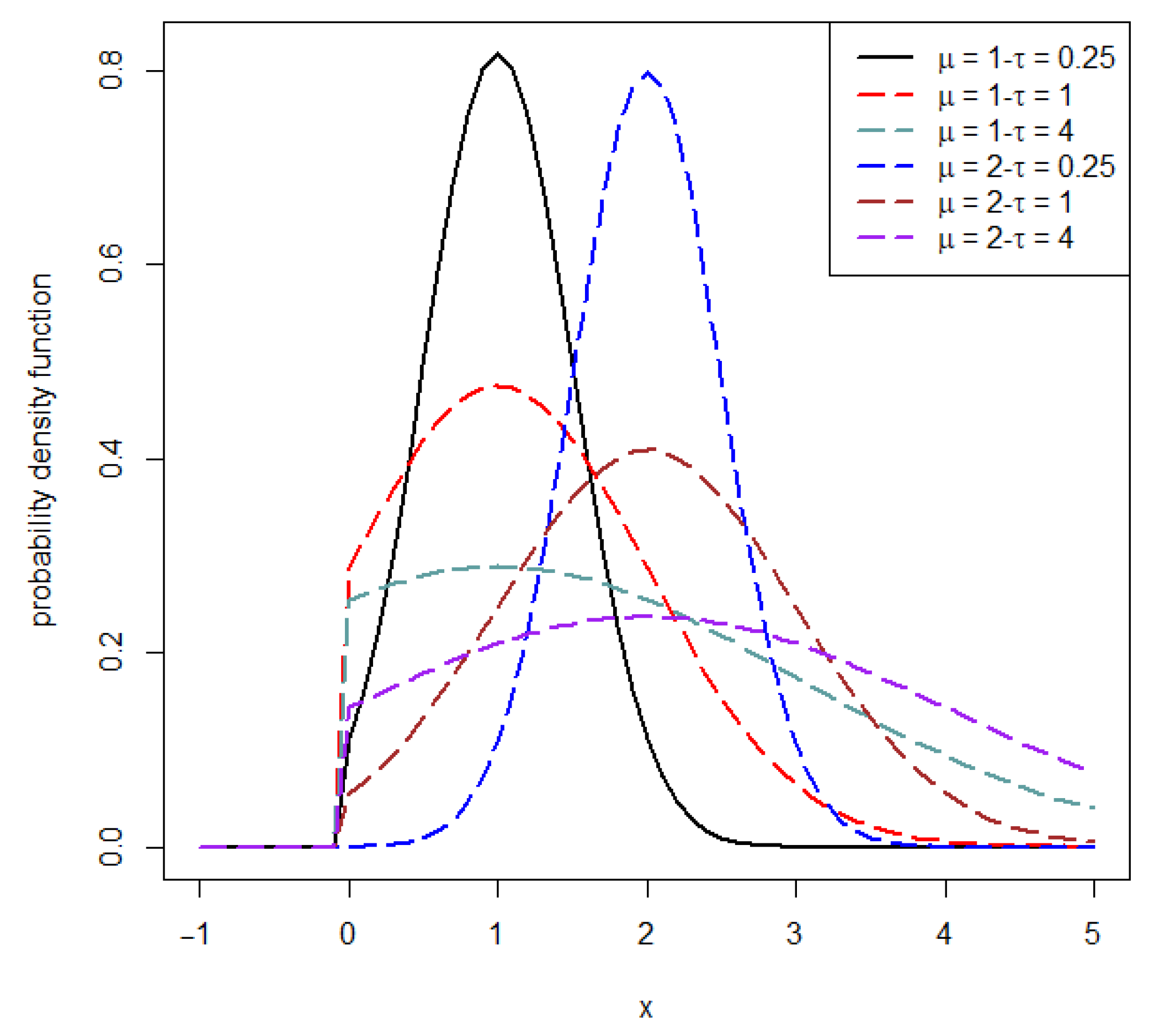
1.1. Truncated Normal Distribution
1.2. Adaptive Progressive Type-II Censored Scheme
2. Maximum Likelihood Estimation
2.1. Point Estimation
2.2. Asymptotic Confidence Interval
2.2.1. Observed Fisher Information Matrix
2.2.2. Expected Fisher Information Matrix
3. Bayesian Estimation
3.1. Three Loss Functions
3.1.1. Square Error Loss Function (SELF)
3.1.2. Linex Loss Function (LLF)
3.1.3. General Entropy Loss Function (GELF)
3.2. Lindley Approximation
- (1)
- Estimation under the square error loss function.Bayes estimate is written asSimilarly, Bayes estimate is given as
- (2)
- Estimation under Linex loss function.In this case u becomes an exponential function about unknown parameter. Then the Bayes estimate is written asSimilarly, Bayes estimate is given as
- (3)
- Estimation under general entropy loss function.In this situation, u changes to a power function then the Bayes estimate is obtained asSimilarly, Bayes estimate is given as -4.6cm0cm
3.3. Importance Sampling
- According to Function (18), all the hyperparameters are positive. Then c and are known. So .
- According to Cauchy–Schwarz inequality, we can obtain , andthen is divided to three parts.
- (a)
- (b)
- (c)
- Because all hyperparameters and integer are positive, and . Thus .
To sum up, all three parts are positive, so . Then belongs to Inverse Gamma distribution naturally.
- Prefix the number of samples .
- Generate from .
- Generate from .
- Repeat step 2 and 3 to produce a series of samples (), (), ⋯, ().
4. Bootstrap Confidence Interval
- step 1 :
- Set the number of simulation times in advance. Obtain the initial MLEs of parameters from the original sample .
- step 2 :
- Using the estimates to generate a new sample of size n. Denote it as .
- step 3 :
- Estimate MLEs based on .
- step 4 :
- Repeat step 2 and 3 times and gain .
- step 5 :
- Sort in ascending order and represent them as . Similarly, is the arranged sample in ascending order. Then the Boot-p interval of and can be written as and , where is the rounded number.
5. Simulation Results
- (1)
- Among all methods, there is a common tendency that all the estimation values are approaching true value and meanwhile mean square errors are decreasing as the sample size n and observed failure numbers m increase.
- (2)
- Generally, the estimates using Bayes method are more accurate compared with ML estimates from their estimated values and MSE. That is because MLEs are only calculated on the basis of data while Bayesian method also takes prior information of unknown parameters in into account. For inappropriate prior distribution and hyperparameters selection, Bayesian estimates may have larger bias. The simulation results show that our prior distribution selection is relatively suitable.
- (3)
- In Bayesian estimation, the estimates under SELF perform slightly better than under LLF and GELF, which illustrates that symmetric loss function is a suitable choice in these cases. Both LLF and GELF with have smaller estimate values than with . But between LLF and GELF, the results show little difference.
- (4)
- As for setting expected time, ML estimates under are lower than under . However, the effects of and are not obvious in Bayesian estimation. Meanwhile, different censoring schemes also show no significant tendency.
- (5)
- In the respect of computation techniques, importance sampling provides estimation values that are closer to true value and smaller MSEs than Lindley approximation.
- (1)
- The interval mean length is narrower and coverage rate is higher as sample size n and observed failure numbers m increase.
- (2)
- Although the results of Aymptotic confidence interval is not very satisfactory, Boot-p intervals and HPD credible intervals obtain highly good performances with lower ML and closer CL.
- (3)
- For different censoring scheme, the first type scheme performs less ML when obtaining asymptotic confidence intervals than other intervals when while in cases, the regulation is contrary.
6. Real Data Analysis
- (1)
- The results are relatively close between Bayesian estimates under importance sampling and estimates under Lindley approximation. Particularly, their estimates of parameter are very similar no matter what the expexted time T and censoring scheme R are. However, for the value of parameter , estimations using importance sampling are lower than that using Lindley method.
- (2)
- Compared with Bayesian estimator using importance sampling, the estimation values of applying MLE are relatively smaller except the situation of , while MLE results of are close when but become higher when .
- (3)
- According to the interval estimation results Table, the lower and higher bound of Boot-p interval are larger slightly. Generally, in the respect of interval length, the results of HPD credible interval are better than the other two.
7. Optimal Censoring Scheme
- (1)
- With the rising number of sample size, the determinant and trace of variance-covariance matrix decrease.
- (2)
- Setting a larger value for T can reduce the trace and determinant of slightly.
- (3)
- The first kind of censoring scheme is always the optimal censoring scheme no matter depending on Criteria I or Criteria II except when , , . However, according to the column named , the total expected experiment time of under different T or are always the largest among the three kinds of censoring schemes. Thurs costs more time on carrying out the test.
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A. Proof of MLE Existence
- Assume thatFor given
- (a)
- When , then , and . Thus, we havewhere , .
- , then when , . In addition, , . So .
- , then when , . Therefore .
- , then .
In conclusion, whatever the value of is,In Equation (A1), m is a positive constant. Hence . - (b)
- When , then , , so that , . Here we haveas a result, .
To sum up, when , is positive and when , is negative.Hence the solution of Equation (12) exists.
Appendix B. Inference of Density Function of X(i) under TN(μ, τ)
Appendix C. Inference of the Probability Mass Function of J
Appendix D. Lindley Approximation Expressions
Appendix E. Selected Computational Codes
library(numDeriv)
-
library(numDeriv)
-
###generate adaptive type II censoring data
-
adapttype2<−function(mu,tau,R,T){
-
m<−length(R)
-
n=sum(R)+m
-
W1<−runif(m)
-
V1<−rep(0,m)
-
U1<−rep(0,m)
-
x = rep(0,m)
-
for(i in 1:m)
-
{
-
V1[i]<−W1[i]^(1/(i + sum(R[(m − i + 1):m])))
-
}
-
for(i in 1:m)
-
{
-
U1[i]<−1-prod(V1[m:(m − i + 1)])
-
}
-
for(i in 1:m){
-
x[i] = sqrt(tau)∗qnorm(1-(1-U1[i])∗pnorm(mu/sqrt(tau)))+mu
-
}
-
i = 1
-
repeat{
-
if(x[i]>T){j = i − 1
-
break}
-
else{i = i + 1}
-
if(i==m){j=m
-
break}
-
}
-
begin = j + 2
-
if(j == 0){begin = 1}
-
if((j + 2) > m){end = m}
-
else{end = j + 1}
-
if(end == m)
-
{return(x)}
-
len = length(begin:m)
-
W2<−runif(len)
-
V2<−rep(0,len)
-
U2<−rep(0,len)
-
for(i in 1:len)
-
{
-
V2[i]<−W2[i]^(1/i)
-
}
-
for(i in 1:len)
-
{
-
U2[i]<−1-prod(V2[len:(len-i + 1)])
-
}
-
xi = function(x,mu,tau){(x-mu)/sqrt(tau)}
-
for(i in begin:m){
-
if(begin == 1)
-
{x[i] = qnorm(U2[i]∗(1-pnorm(xi(x[j+1],mu,tau)))
-
+pnorm(xi(x[j+1],mu,tau)))∗sqrt(tau)+mu
-
}
-
else{
-
x[i] = qnorm(U2[i-j-1]∗(1-pnorm(xi(x[j+1],mu,tau)))
-
+pnorm(xi(x[j+1],mu,tau)))∗sqrt(tau)+mu
-
}}
-
return(x)}
-
##MLE
-
Xii = function(xi,mu,tau){
-
(xi − mu)/sqrt(tau)}
-
Xi = function(mu,tau){
-
mu/sqrt(tau)}
-
mu = 2
-
tau = 1
-
num=1
-
T = 1
-
p = 0.2
-
covermu = 0
-
covertau = 0
-
z = qnorm(1 − p/2)
-
sim = 1000
-
leftmu = rep(0,sim)
-
rightmu = rep(0,sim)
-
lefttau = rep(0,sim)
-
righttau = rep(0,sim)
-
mumle = rep(0,sim)
-
taumle = rep(0,sim)
-
repeat{
-
R1 = rep(c(1,1,0),6)
-
R=R1
-
datax = adapttype2(mu,tau,R,T)
-
i = 1
-
m = length(R)
-
n = sum(R) + m
-
repeat{if(datax[i]>T){j = i − 1
-
break}
-
else{i = i + 1}
-
if(i == m){j = m
-
break}
-
}
-
mu1 = mu
-
tau1 = tau
-
test = try({
-
mle<−function(datax,R,x){
-
loglike = −sum(Xii(datax,x[1],x[2])^2/2)-0.5∗m∗log(x[2])-n∗log(pnorm(Xi(x[1],x[2])))
-
+sum(R[1:j]∗log(1-pnorm(Xii(datax[1:j],x[1],x[2]))))
-
+(n-m-sum(R[1:j]))*log(1-pnorm(Xii(datax[m],x[1],x[2])))
-
return(−loglike)
-
}
-
a=optim(c(mu1,tau1),mle,datax = datax,R=R1,method=’L-BFGS-B’,lower = c(0.01,0.01),
-
control=list(trace = FALSE,maxit = 1000))
-
mumle[num] = a$par[1]
-
taumle[num] = a$par[2]
-
})
-
if(“try-error” %in% class(test) | taumle[num]>1.3 | mumle[num]>2.3){next}
-
else{
-
h1 = hessian(func = mle,x = c(a$par[1],a$par[2]),datax = datax,R=R1)
-
var = solve(h1)
-
leftmu[num] = mumle[num] − z∗sqrt(var[1,1])
-
rightmu[num] = mumle[num] + z∗sqrt(var[1,1])
-
lefttau[num] = taumle[num] − z∗sqrt(var[2,2])
-
righttau[num] = taumle[num] + z∗sqrt(var[2,2])
-
if(mu>leftmu[num] & mu<rightmu[num]){covermu = covermu + 1}
-
if(tau>lefttau[num] & tau<righttau[num]){covertau = covertau + 1}
-
num = num + 1}
-
if(num>sim){break}
-
}
-
mean(mumle)
-
mean((mumle − mu)^2)
-
mean(taumle)
-
mean((taumle − tau)^2)
-
mean(leftmu)
-
mean(rightmu)
-
mean(lefttau)
-
mean(righttau)
-
covermu/sim
-
covertau/sim
-
mean(rightmu-leftmu)
-
###Bayesian~estimation
-
###SELF
-
a = 0.6
-
b = 0.3
-
c = 37
-
d = 0.6
-
p = 1
-
q = 1
-
SIM = 1000
-
sim = 5000
-
mum = 2
-
taum = 1
-
R = rep(c(1,1,0),10)
-
T = 3
-
i = 1
-
repeat{if(datax[i]>T){j = i − 1
-
break}
-
else{i = i + 1}
-
if(i == m){J = m
-
break}
-
}
-
muappro = rep(0,SIM)
-
tauappro = rep(0,SIM)
-
mullfappro = rep(0,SIM)
-
taullfappro = rep(0,SIM)
-
mugelfappro = rep(0,SIM)
-
taugelfappro = rep(0,SIM)
-
muleft = rep(0,SIM)
-
muright = rep(0,SIM)
-
mulen = rep(0,SIM)
-
tauleft = rep(0,SIM)
-
tauright = rep(0,SIM)
-
taulen = rep(0,SIM)
-
mucover = 0
-
taucover = 0
-
for(i in 1:SIM){
-
mu = rep(0,sim)
-
tau = rep(0,sim)
-
Q = rep(0,sim)
-
datax = adapttype2(mum,taum,R,T)
-
m = length(R)
-
n = sum(R)+m
-
IG1 = c + m/2
-
IG2 = 1/2∗(a^2∗b+d+sum(datax^2))
-
for(j in 1:sim){
-
tau[j] = rinvgamma(1,IG1,IG2)
-
TN1 = (sum(datax)+a∗b)/(b+m)
-
TN2 = tau[j]/(b+m)
-
mu[j] = rtruncnorm(1,0,Inf,TN1,sqrt(TN2))
-
re = 1
-
for(k in 1:J){
-
re = re∗(1-pnorm((datax[k] − mu[j])/sqrt(tau[j])))^R[k]}
-
Q[j] = pnorm((sum(datax)+a∗b)/(sqrt(tau[j]∗(b+m))))/pnorm(a∗sqrt(b)
-
/sqrt(tau[j]))∗pnorm(mu[j]/sqrt(tau[j]))^(−n)∗re∗(1 − pnorm((datax[m] − mu[j])
-
/sqrt(tau[j])))^(n-m-sum(R[1:J]))}
-
muappro[i] = sum(mu∗Q)/sum(Q)
-
tauappro[i] = sum(tau∗Q)/sum(Q)
-
mullfappro[i] = −1/p∗log(sum(exp(−p∗mu)∗Q)/sum(Q))
-
taullfappro[i] = −1/p∗log(sum(exp(−p∗tau)∗Q)/sum(Q))
-
mugelfappro[i] = (sum(mu^(−q)*Q)/sum(Q))^(−1/q)
-
taugelfappro[i] = (sum(tau^(−q)*Q)/sum(Q))^(−1/q)
-
summu = 0
-
prob = 0.05
-
muord = order(mu)
-
for(nu in 1:sim){
-
summu = summu + Q[muord[nu]]/sum(Q)
-
if(summu >= prob){
-
break}
-
}
-
muinterval = matrix(0,nu-1,2)
-
len = rep(0,nu − 1)
-
for(k in 1:(nu − 1)){
-
summ2 = 0
-
l = k
-
repeat{
-
summ2 = summ2 + Q[muord[l]]/sum(Q)
-
l = l + 1
-
if(summ2>=1-prob){
-
break}
-
}
-
l = l − 1
-
len[k] = summ2
-
muinterval[k,1] = mu[muord[k]]
-
muinterval[k,2] = mu[muord[l]]
-
}
-
muinterval = muinterval[is.na(muinterval[,2]) == 0,]
-
minvalue = min(muinterval[,2]-muinterval[,1])
-
judge1 = (muinterval[,2] − muinterval[,1]) == minvalue
-
muleft[i] = muinterval[judge1][1]
-
muright[i] = muinterval[judge1][2]
-
mulen[i] = muright[i] − muleft[i]
-
sumtau = 0
-
prob = 0.05
-
tauord = order(tau)
-
for(nu in 1:sim){
-
sumtau = sumtau+Q[tauord[nu]]/sum(Q)
-
if(sumtau >= prob){
-
break}
-
}
-
tauinterval = matrix(0,nu-1,2)
-
len = rep(0,nu − 1)
-
for(k in 1:(nu − 1)){
-
summ2 = 0
-
l = k
-
repeat{
-
summ2 = summ2 + Q[tauord[l]]/sum(Q)
-
l = l + 1
-
if(summ2 >= 1 − prob){
-
break}
-
}
-
l = l − 1
-
len[k] = summ2
-
tauinterval[k,1] = tau[tauord[k]]
-
tauinterval[k,2] = tau[tauord[l]]
-
}
-
tauinterval = tauinterval[is.na(tauinterval[,2])==0,]
-
minvalue = min(tauinterval[,2]-tauinterval[,1])
-
judge2 = (tauinterval[,2]-tauinterval[,1])==minvalue
-
tauleft[i] = tauinterval[judge2][1]
-
tauright[i] = tauinterval[judge2][2]
-
taulen[i] = tauright[i]-tauleft[i]
-
if(mum >= muleft[i] & mum <= muright[i]){mucover = mucover + 1}
-
if(taum >= tauleft[i] & taum <= tauright[i]){taucover = taucover + 1}
-
}
-
mean(muleft)
-
mean(muright)
-
mean(mulen)
-
mucover/SIM
-
mean(tauleft)
-
mean(tauright)
-
mean(taulen)
-
taucover/SIM
-
mean(muappro)
-
mean((muappro − mum)^2)
-
mean(tauappro)
-
mean((tauappro − taum)^2)
-
mean(mullfappro)
-
mean((mullfappro − mum)^2)
-
mean(taullfappro)
-
mean((taullfappro − taum)^2)
-
mean(mugelfappro)
-
mean((mugelfappro − mum)^2)
-
mean(taugelfappro)
-
mean((taugelfappro − taum)^2)
References
- Barr, D.R.; Sherrill, E.T. Mean and Variance of Truncated Normal Distributions. Am. Statian. 1999, 53, 357–361. [Google Scholar]
- Lodhi, C.; Tripathi, Y.M.; Rastogi, M.K. Estimating the parameters of a truncated normal distribution under progressive type II censoring. Commun. Stat. Simul. Comput. 2019, 1–25. [Google Scholar] [CrossRef]
- Pender, J. The truncated normal distribution: Applications to queues with impatient customers. Oper. Res. Lett. 2015, 43, 40–45. [Google Scholar] [CrossRef]
- Horrace, W.C. Moments of the truncated normal distribution. J. Product. Anal. 2015, 43, 133–138. [Google Scholar] [CrossRef]
- Balakrishnan, N.; Aggarwala, R. Progressive Censoring; Birkhäuser: Boston, MA, USA, 2000. [Google Scholar]
- Balakrishnan, N.; Cramer, E. The Art of Progressive Censoring; Birkhäuser: New York, NY, USA, 2014. [Google Scholar]
- Ng, H.K.T.; Kundu, D.; Ping, S.C. Statistical analysis of exponential lifetimes under an adaptive Type-II progressive censoring scheme. Nav. Res. Logist. 2009, 56, 687–698. [Google Scholar] [CrossRef] [Green Version]
- Cramer, E.; Iliopoulos, G. Adaptive progressive Type-II censoring. Test Off. J. Span. Soc. Stats Oper. Res. 2010, 19, 342–358. [Google Scholar] [CrossRef]
- Hemmati, F.; Khorram, E. On adaptive progressively Type-II censored competing risks data. Commun. Stat.-Simul. Comput. 2016, 46, 4671–4693. [Google Scholar] [CrossRef]
- Chen, S.; Gui, W. Statistical Analysis of a Lifetime Distribution with a Bathtub-Shaped Failure Rate Function under Adaptive Progressive Type-II Censoring. Mathematics 2020, 8, 670. [Google Scholar] [CrossRef]
- Varian, H.R. A Third Remark on the Number of Equilibria of an Economy. Econometrica 1975, 43, 985–986. [Google Scholar] [CrossRef]
- Sultan, K.S.; Alsadat, N.H.; Kundu, D. Bayesian and maximum likelihood estimations of the inverse Weibull parameters under progressive type-II censoring. J. Stat. Comput. Simul. 2014, 84, 2248–2265. [Google Scholar] [CrossRef]
- Xie, Y.; Gui, W. Statistical Inference of the Lifetime Performance Index with the Log-Logistic Distribution Based on Progressive First-Failure-Censored Data. Symmetry 2020, 12, 937. [Google Scholar] [CrossRef]
- Khatun, N.; Matin, M.A. A Study on LINEX Loss Function with Different Estimating Methods. Open J. Stat. 2020, 10, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Calabria, R.; Pulcini, G. An engineering approach to Bayes estimation for the Weibull distribution. Microelectron. Reliab. 1994, 34, 789–802. [Google Scholar] [CrossRef]
- Lindley, D.V. Approximate Bayesian methods. Trab. De Estad. Y De Investig. Oper. 1980, 31, 223–245. [Google Scholar] [CrossRef]
- Lawless, J.F. Statistical MODELS and Methods for Lifetime Data; John Wiley & Sons: New York, NY, USA, 1982. [Google Scholar]
- Gupta, R.D.; Kundu, D. Exponentiated Exponential Family: An Alternative to Gamma and Weibull Distributions. Biom. J. 2015, 43, 117–130. [Google Scholar] [CrossRef]
- Rao, G.S. Estimation of Reliability in Multicomponent Stress-strength Based on Generalized Exponential Distribution. Rev. Colomb. De Estadística 2012, 35, 67–76. [Google Scholar]
- Lee, S.; Noh, Y.; Chung, Y. Inverted exponentiated Weibull distribution with applications to lifetime data. Commun. Stat. Appl. Methods 2017, 24, 227–240. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.D.; Kundu, D. On the comparison of Fisher information of the Weibull and GE distributions. J. Statal Plan. Inference 2006, 136, 3130–3144. [Google Scholar] [CrossRef]
- Pradhan, B.; Kundu, D. Inference and optimal censoring schemes for progressively censored Birnbaum–Saunders distribution. J. Stat. Plan. Inference 2013, 143, 1098–1108. [Google Scholar] [CrossRef]
- Singh, S.; Tripathi, Y.M.; Wu, S.J. On estimating parameters of a progressively censored lognormal distribution. J. Stat. Comput. Simul. 2013, 85, 1071–1089. [Google Scholar] [CrossRef]

| T | R | |||||
|---|---|---|---|---|---|---|
| 1 | (30,12) | (6,6,0*16) | 2.1481 | 0.0389 | 1.1417 | 0.0445 |
| ((1,1,0)*6) | 1.8934 | 0.0392 | 0.9253 | 0.0499 | ||
| (40,16) | (8,8,0*22) | 2.1223 | 0.0371 | 1.1389 | 0.0436 | |
| ((1,1,0)*8) | 1.8939 | 0.0389 | 0.9230 | 0.0495 | ||
| (50,20) | (10,10,0*28) | 2.0802 | 0.0379 | 1.1367 | 0.0431 | |
| ((1,1,0)*10) | 1.9025 | 0.0375 | 0.9208 | 0.0495 | ||
| 3 | (30,12) | (6,6,0*16) | 1.8849 | 0.0292 | 0.9207 | 0.204 |
| ((1,1,0)*6) | 1.8657 | 0.0540 | 0.8395 | 0.0586 | ||
| (40,16) | (8,8,0*22) | 1.8853 | 0.0290 | 0.9293 | 0.0193 | |
| ((1,1,0)*8) | 1.9034 | 0.0471 | 0.8941 | 0.0380 | ||
| (50,20) | (10,10,0*28) | 1.8934 | 0.0279 | 0.9374 | 0.0188 | |
| ((1,1,0)*10) | 1.9225 | 0.0372 | 0.9723 | 0.0258 |
| T | R | SELF | LLF | GELF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EV | MSE | EV | MSE | EV | MSE | EV | MSE | EV | MSE | ||||
| 1 | (30,12) | (6,6,0*16) | 1.9420 | 0.0476 | 1.9325 | 0.0481 | 2.1018 | 0.0264 | 1.9322 | 0.0485 | 2.0760 | 0.0273 | |
| 0.8782 | 0.0404 | 0.8924 | 0.0578 | 0.8644 | 0.0337 | 0.8839 | 0.0596 | 0.8567 | 0.0359 | ||||
| (1,1,0)*6 | 1.9630 | 0.0513 | 1.9533 | 0.0595 | 1.1203 | 0.0486 | 1.9530 | 0.0560 | 1.1107 | 0.0464 | |||
| 1.1994 | 0.0564 | 1.1827 | 0.0489 | 1.2498 | 0.0456 | 1.1720 | 0.0452 | 1.2374 | 0.0697 | ||||
| (40,16) | (8,8,0*22) | 1.9711 | 0.0162 | 1.9600 | 0.0165 | 2.0483 | 0.0257 | 1.9597 | 0.0167 | 2.0494 | 0.0261 | ||
| 0.8865 | 0.0386 | 0.9105 | 0.0328 | 0.9821 | 0.0208 | 0.9011 | 0.0344 | 0.9773 | 0.0206 | ||||
| (1,1,0)*8 | 1.9708 | 0.0413 | 1.9572 | 0.0512 | 2.0587 | 0.0270 | 1.9568 | 0.0542 | 2.0706 | 0.0479 | |||
| 1.1242 | 0.0473 | 1.1097 | 0.0419 | 1.0602 | 0.0323 | 1.0993 | 0.0401 | 1.0507 | 0.0314 | ||||
| (50,20) | (10,10,0*28) | 2.0006 | 0.0159 | 1.9900 | 0.0158 | 2.0046 | 0.0039 | 1.9899 | 0.0160 | 1.9928 | 0.0037 | ||
| 0.8995 | 0.0166 | 1.0081 | 0.0117 | 0.9879 | 0.0204 | 0.9974 | 0.0140 | 0.9867 | 0.0214 | ||||
| (1,1,0)*10 | 1.9783 | 0.0195 | 1.9690 | 0.0198 | 2.0194 | 0.0276 | 1.9688 | 0.0200 | 2.0195 | 0.0279 | |||
| 1.0178 | 0.0274 | 1.0145 | 0.0268 | 1.0293 | 0.0220 | 1.0117 | 0.0266 | 1.0198 | 0.0217 | ||||
| 3 | (30,12) | (6,6,0*16) | 1.9420 | 0.0364 | 1.9309 | 0.0371 | 1.9586 | 0.0360 | 1.9305 | 0.0375 | 1.9584 | 0.0363 | |
| 0.8776 | 0.0233 | 0.8713 | 0.0246 | 1.1339 | 0.0537 | 0.8636 | 0.0266 | 1.1404 | 0.0371 | ||||
| (1,1,0)*6 | 1.9507 | 0.0299 | 1.9400 | 0.0307 | 2.0979 | 0.0429 | 1.9396 | 0.0310 | 2.0981 | 0.0432 | |||
| 0.9618 | 0.0245 | 0.9522 | 0.0242 | 1.1531 | 0.0441 | 0.9430 | 0.0251 | 1.1559 | 0.0461 | ||||
| (40,16) | (8,8,0*22) | 1.9505 | 0.0268 | 1.9412 | 0.0274 | 1.9815 | 0.0225 | 1.9408 | 0.0277 | 1.9812 | 0.0228 | ||
| 0.9209 | 0.0226 | 0.9137 | 0.0233 | 1.0608 | 0.0182 | 0.9056 | 0.0246 | 1.0509 | 0.0169 | ||||
| (1,1,0)*8 | 1.9708 | 0.0241 | 1.9572 | 0.0212 | 2.0341 | 0.0221 | 1.9568 | 0.0218 | 2.0339 | 0.0218 | |||
| 0.9714 | 0.0250 | 0.9627 | 0.0248 | 1.0867 | 0.0365 | 0.9538 | 0.0257 | 1.0717 | 0.0349 | ||||
| (50,20) | (10,10,0*28) | 2.0202 | 0.0275 | 2.0332 | 0.0207 | 2.0050 | 0.0055 | 2.0227 | 0.0197 | 2.0049 | 0.0056 | ||
| 1.0156 | 0.0220 | 1.0104 | 0.0114 | 1.0241 | 0.0170 | 1.0052 | 0.0119 | 1.0374 | 0.0135 | ||||
| (1,1,0)*10 | 1.9993 | 0.0260 | 1.9911 | 0.0157 | 2.0139 | 0.0171 | 1.9911 | 0.0159 | 2.0138 | 0.0173 | |||
| 0.9815 | 0.0242 | 0.9724 | 0.0239 | 1.0675 | 0.0216 | 0.9635 | 0.0245 | 1.0594 | 0.0270 | ||||
| T | R | SELF | LLF | GELF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EV | MSE | EV | MSE | EV | MSE | EV | MSE | EV | MSE | ||||
| 1 | (30,12) | (6,6,0*16) | 1.9472 | 0.0392 | 1.9350 | 0.0396 | 1.9672 | 0.0609 | 1.9346 | 0.0401 | 1.9639 | 0.0696 | |
| 0.8971 | 0.0667 | 0.9031 | 0.0686 | 1.1401 | 0.0740 | 0.9137 | 0.0720 | 1.1292 | 0.0738 | ||||
| (1,1,0)*6 | 1.9118 | 0.0582 | 1.8993 | 0.0520 | 2.0753 | 0.0550 | 1.8986 | 0.0507 | 2.0755 | 0.0552 | |||
| 1.1763 | 0.0843 | 1.1577 | 0.0740 | 1.1385 | 0.0434 | 1.1472 | 0.0688 | 1.1369 | 0.0475 | ||||
| (40,16) | (8,8,0*22) | 1.9726 | 0.0228 | 1.9631 | 0.0231 | 1.9810 | 0.0218 | 1.9629 | 0.0233 | 1.9808 | 0.0212 | ||
| 0.9210 | 0.0548 | 0.9392 | 0.0540 | 1.1021 | 0.0709 | 0.9392 | 0.0541 | 1.0906 | 0.0654 | ||||
| (1,1,0)*8 | 1.9243 | 0.0325 | 1.9131 | 0.0337 | 2.0347 | 0.0412 | 1.9125 | 0.0341 | 2.0220 | 0.0395 | |||
| 1.1148 | 0.0417 | 1.2100 | 0.0433 | 1.0867 | 0.0373 | 1.2034 | 0.0458 | 1.0863 | 0.0370 | ||||
| (50,20) | (10,10,0*28) | 2.0397 | 0.0218 | 2.0222 | 0.0225 | 2.0570 | 0.0200 | 2.0224 | 0.0225 | 2.0568 | 0.0196 | ||
| 0.9564 | 0.0115 | 0.9479 | 0.0120 | 1.0676 | 0.0253 | 0.9390 | 0.0131 | 1.0571 | 0.0262 | ||||
| (1,1,0)*8 | 1.9390 | 0.0281 | 1.9245 | 0.0348 | 2.0153 | 0.0214 | 1.9240 | 0.0489 | 1.9969 | 0.0207 | |||
| 1.0813 | 0.0340 | 1.0708 | 0.0319 | 1.9527 | 0.0373 | 1.0718 | 0.0317 | 1.0430 | 0.0368 | ||||
| 3 | (30,12) | (6,6,0*16) | 1.9118 | 0.0410 | 1.8933 | 0.424 | 1.2046 | 0.0896 | 1.8918 | 0.0425 | 1.2144 | 0.0898 | |
| 0.9695 | 0.0426 | 0.9711 | 0.0438 | 1.1675 | 0.0608 | 0.9705 | 0.0461 | 1.1783 | 0.0470 | ||||
| (1,1,0)*6 | 1.9602 | 0.0222 | 1.9469 | 0.0228 | 2.0932 | 0.0738 | 1.9464 | 0.0232 | 2.0941 | 0.0774 | |||
| 0.8807 | 0.0986 | 0.8990 | 0.0986 | 1.1623 | 0.0407 | 0.8988 | 0.0990 | 1.1516 | 0.0375 | ||||
| (40,16) | (8,8,0*22) | 1.9315 | 0.0327 | 1.9175 | 0.0343 | 0.9064 | 0.0292 | 1.9167 | 0.0348 | 0.9062 | 0.0296 | ||
| 1.0042 | 0.0321 | 0.9951 | 0.0311 | 1.1559 | 0.0461 | 0.9928 | 0.0326 | 1.1567 | 0.0575 | ||||
| (1,1,0)*8 | 1.9775 | 0.0169 | 1.9628 | 0.0176 | 2.0509 | 0.0544 | 1.9623 | 0.0232 | 2.0515 | 0.0535 | |||
| 0.9001 | 0.0276 | 0.9197 | 0.0265 | 1.1046 | 0.0369 | 0.9100 | 0.0268 | 1.1073 | 0.0394 | ||||
| (50,20) | (10,10,0*28) | 1.9521 | 0.0246 | 1.9356 | 0.0256 | 2.0491 | 0.0122 | 1.9349 | 0.0261 | 2.0491 | 0.0118 | ||
| 1.0041 | 0.0178 | 0.9934 | 0.0173 | 1.0736 | 0.0211 | 0.9833 | 0.0176 | 1.0808 | 0.0297 | ||||
| (1,1,0)*10 | 2.0022 | 0.0041 | 1.9832 | 0.0139 | 2.0221 | 0.0400 | 1.9833 | 0.0140 | 2.0123 | 0.0123 | |||
| 0.9302 | 0.0260 | 0.9417 | 0.0253 | 1.0711 | 0.0185 | 0.9436 | 0.0256 | 1.0593 | 0.0165 | ||||
| T | R | Asymptotic Interval | boot-p Interval | HPD Interval | |||||
|---|---|---|---|---|---|---|---|---|---|
| ML | CR | ML | CR | ML | CR | ||||
| 1 | (30,12) | (6,6,0*16) | 0.7051 | 0.8526 | 0.6564 | 0.8524 | 0.6240 | 0.8688 | |
| 1.2807 | 0.8976 | 0.9509 | 0.8887 | 0.9502 | 0.8548 | ||||
| (1,1,0)*6 | 0.5661 | 0.7897 | 0.7719 | 0.8976 | 0.7027 | 0.9679 | |||
| 1.2702 | 0.8526 | 1.7133 | 0.9161 | 1.7402 | 0.9211 | ||||
| (40,16) | (8,8,0*22) | 0.5869 | 0.8900 | 0.5681 | 0.9237 | 0.5693 | 0.8739 | ||
| 0.9909 | 0.9128 | 0.8337 | 0.9380 | 0.7506 | 0.8676 | ||||
| (1,1,0)*8 | 0.4996 | 0.8648 | 0.6698 | 0.9377 | 0.6888 | 0.9637 | |||
| 1.1521 | 0.8723 | 1.5411 | 0.9046 | 1.5419 | 0.9486 | ||||
| (50,20) | (10,10,0*28) | 0.5150 | 0.8932 | 0.5071 | 0.9368 | 0.4980 | 0.8273 | ||
| 0.8397 | 0.9170 | 0.7508 | 0.9444 | 0.5322 | 0.8787 | ||||
| (1,1,0)*10 | 0.4541 | 0.9085 | 0.5994 | 0.9624 | 0.6016 | 0.9251 | |||
| 1.0714 | 0.9231 | 1.4105 | 0.9387 | 1.4032 | 0.9316 | ||||
| 3 | (30,12) | (6,6,0*16) | 0.6784 | 0.8998 | 0.6560 | 0.9860 | 0.6516 | 0.8934 | |
| 0.9468 | 0.9198 | 0.9504 | 0.9000 | 0.9523 | 0.8892 | ||||
| (1,1,0)*6 | 0.5133 | 0.9080 | 0.5983 | 0.8340 | 0.6001 | 0.9225 | |||
| 0.9138 | 0.8496 | 1.0507 | 0.8953 | 1.0341 | 0.9165 | ||||
| (40,16) | (8,8,0*22) | 0.5811 | 0.9023 | 0.5667 | 0.9171 | 0.6098 | 0.8717 | ||
| 0.9468 | 0.9182 | 0.8327 | 0.9153 | 0.7143 | 0.8856 | ||||
| (1,1,0)*8 | 0.4522 | 0.9018 | 0.5141 | 0.9568 | 0.4824 | 0.9385 | |||
| 0.8231 | 0.8658 | 0.9323 | 0.9723 | 0.9348 | 0.9649 | ||||
| (50,20) | (10,10,0*28) | 0.5153 | 0.9188 | 0.5070 | 0.9256 | 0.5652 | 0.9095 | ||
| 0.8296 | 0.9268 | 0.7521 | 0.9356 | 0.6791 | 0.9101 | ||||
| (1,1,0)*10 | 0.4107 | 0.8876 | 0.4568 | 0.9799 | 0.4593 | 0.9474 | |||
| 0.7617 | 0.8836 | 0.8560 | 0.9216 | 0.8398 | 0.9178 | ||||
| Complete Data | 0.1788, 0.2892, 0.3300, 0.4152, 0.4212, 0.4560, 0.4840, 0.5184, 0.5196, 0.5412, 0.5556, 0.6780, 0.6864, 0.6888, 0.8412, 0.9312, 0.9864, 1.0512, 1.0584, 1.2792, 1.2804, 1.7340. |
| 0.1788, 0.4560, 0.4840, 0.5184, 0.5196, 0.5412, 0.5556, 0.6780, 0.6864, 0.6864, 0.6888, 0.8412, 0.9312, 0.9864, 1.0512, 1.0584, 1.2792, 1.2804, 1.7340. | |
| 0.1788, 0.4560, 0.5556, 0.6780, 0.6864, 0.6864, 0.6888, 0.8412, 0.9312, 0.9864, 1.0512, 1.0584, 1.2792, 1.2804, 1.7340. | |
| 0.1788, 0.3300, 0.4212, 0.4560, 0.4840, 0.5184, 0.5196, 0.5412, 0.5556, 0.6780, 0.6864, 0.6864, 0.6888, 0.8412, 0.9312, 0.9864, 1.0512, 1.0584, 1.2792, 1.2804, 1.7340. | |
| 0.1788, 0.3300, 0.4212, 0.4560, 0.5184, 0.5412, 0.5556, 0.6864, 0.6888, 0.8412, 0.9864, 1.0512, 1.0584, 1.2792, 1.2804, 1.7340. |
| T | R | MLE | Bayes-importance sampling | ||||||
| SELF | LLF | GELF | |||||||
| 0.4 | (23,15) | (4,4,0*13) | 0.8088 | 1.0179 | 1.0115 | 1.0247 | 1.0050 | 1.0184 | |
| 0.1728 | 0.1873 | 0.1869 | 0.1870 | 0.1828 | 0.1866 | ||||
| (23,14) | ((1,1,0*4),1,0) | 0.8890 | 0.8835 | 0.8777 | 0.8910 | 0.8699 | 0.8848 | ||
| 0.1339 | 0.1756 | 0.1752 | 0.1766 | 0.1713 | 0.1762 | ||||
| 0.9 | (23,15) | (4,4,0*13) | 0.8396 | 1.2052 | 1.1995 | 1.2118 | 1.1955 | 1.2058 | |
| 0.2930 | 0.1908 | 0.1903 | 0.1911 | 0.1861 | 0.1907 | ||||
| (23,14) | ((1,1,0*4),1,0) | 0.8869 | 0.9927 | 0.9877 | 0.9976 | 0.9824 | 0.9925 | ||
| 0.2535 | 0.1683 | 0.1679 | 0.1685 | 0.1640 | 0.1681 | ||||
| Bayes-Lindley Approximation | |||||||||
| SELF | LLF | GELF | |||||||
| 0.4 | (23,15) | (4,4,0*13) | 1.0184 | 1.0008 | 1.1067 | 0.9910 | 1.0075 | ||
| 0.2586 | 0.2575 | 0.2602 | 0.2501 | 0.2590 | |||||
| (23,14) | ((1,1,0*4),1,0) | 0.8656 | 0.8560 | 0.8765 | 0.8415 | 0.8671 | |||
| 0.2454 | 0.2443 | 0.2457 | 0.2370 | 0.2446 | |||||
| 0.9 | (23,15) | (4,4,0*13) | 1.2162 | 1.2080 | 1.2264 | 1.2025 | 1.2184 | ||
| 0.2615 | 0.2603 | 0.2632 | 0.2529 | 0.2619 | |||||
| (23,14) | ((1,1,0*4),1,0) | 0.9985 | 0.9908 | 1.0122 | 0.9825 | 1.0043 | |||
| 0.2337 | 0.2328 | 0.2354 | 0.2259 | 0.2344 | |||||
| T | R | Asymptotic Interval | Boot-p Interval | HPD Interval | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LB | UB | ML | LB | UB | ML | LB | UB | ML | |||
| 0.4 | (4,4,0*13) | 0.6163 | 1.0013 | 0.3849 | 0.7978 | 1.2182 | 0.4203 | 0.8003 | 1.2464 | 0.4460 | |
| 0.0459 | 0.2997 | 0.2538 | 0.1072 | 0.4152 | 0.3079 | 0.1331 | 0.2486 | 0.1155 | |||
| ((1,1,0*4),1,0) | 0.7041 | 1.0739 | 0.3698 | 0.8436 | 1.2601 | 0.4165 | 0.6666 | 1.0873 | 0.4208 | ||
| 0.0773 | 0.1904 | 0.1131 | 0.1290 | 0.4557 | 0.3267 | 0.1234 | 0.2307 | 0.1074 | |||
| 0.9 | (4,4,0*13) | 0.5975 | 1.0818 | 0.4843 | 0.7545 | 1.2495 | 0.4949 | 1.0015 | 1.4206 | 0.4191 | |
| 0.0000 | 0.6531 | 0.6531 | 0.1493 | 0.6821 | 0.5328 | 0.1341 | 0.2502 | 0.1161 | |||
| ((1,1,0*4),1,0) | 0.6348 | 1.1389 | 0.5040 | 0.6511 | 1.2273 | 0.5762 | 0.7885 | 1.1742 | 0.3857 | ||
| 0.0028 | 0.5041 | 0.5013 | 0.1902 | 0.8585 | 0.6683 | 0.1186 | 0.2248 | 0.1062 | |||
| T | (n,m) | Scheme | Criteria I | Optimal Plan(I) | Criteria II | Optimal Plan(II) | E() |
|---|---|---|---|---|---|---|---|
| 1 | (30,12) | (3*4,0*14) | 0.2722 | 0.0145 | 3.5264 | ||
| (1,1,0)*6 | 0.3762 | 0.0187 | 2.8765 | ||||
| (0*14,3*4) | 0.3747 | 0.0177 | 2.8634 | ||||
| (40,16) | (4*4,0*20) | 0.1841 | 0.0071 | 3.7767 | |||
| (1,1,0)*8 | 0.3056 | 0.0117 | 2.9185 | ||||
| (0*20,4*4) | 0.3038 | 0.0110 | 2.9083 | ||||
| (50,20) | (5*4,0*26) | 0.1362 | 0.0040 | 3.9394 | |||
| (1,1,0)*10 | 0.2578 | 0.0080 | 2.9486 | ||||
| (0*26,5*4) | 0.2565 | 0.0076 | 2.9421 | ||||
| 3 | (30,12) | (3*4,0*14) | 0.2040 | 0.0098 | 3.7690 | ||
| (1,1,0)*6 | 0.2181 | 0.0096 | 3.3055 | ||||
| (0*14,3*4) | 0.2344 | 0.0101 | 2.4204 | ||||
| (40,16) | (4*4,0*20) | 0.1542 | 0.0055 | 3.8994 | |||
| (1,1,0)*8 | 0.1781 | 0.0061 | 3.3466 | ||||
| (0*20,4*4) | 0.1906 | 0.0061 | 2.3948 | ||||
| (50,20) | (5*4,0*26) | 0.1280 | 0.0037 | 4.0243 | |||
| (1,1,0)*10 | 0.1431 | 0.0038 | 3.4533 | ||||
| (0*26,5*4) | 0.1618 | 0.0042 | 2.3903 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Gui, W. Estimation of Unknown Parameters of Truncated Normal Distribution under Adaptive Progressive Type II Censoring Scheme. Mathematics 2021, 9, 49. https://doi.org/10.3390/math9010049
Chen S, Gui W. Estimation of Unknown Parameters of Truncated Normal Distribution under Adaptive Progressive Type II Censoring Scheme. Mathematics. 2021; 9(1):49. https://doi.org/10.3390/math9010049
Chicago/Turabian StyleChen, Siqi, and Wenhao Gui. 2021. "Estimation of Unknown Parameters of Truncated Normal Distribution under Adaptive Progressive Type II Censoring Scheme" Mathematics 9, no. 1: 49. https://doi.org/10.3390/math9010049





