Domain Heuristic Fusion of Multi-Word Embeddings for Nutrient Value Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Related Work
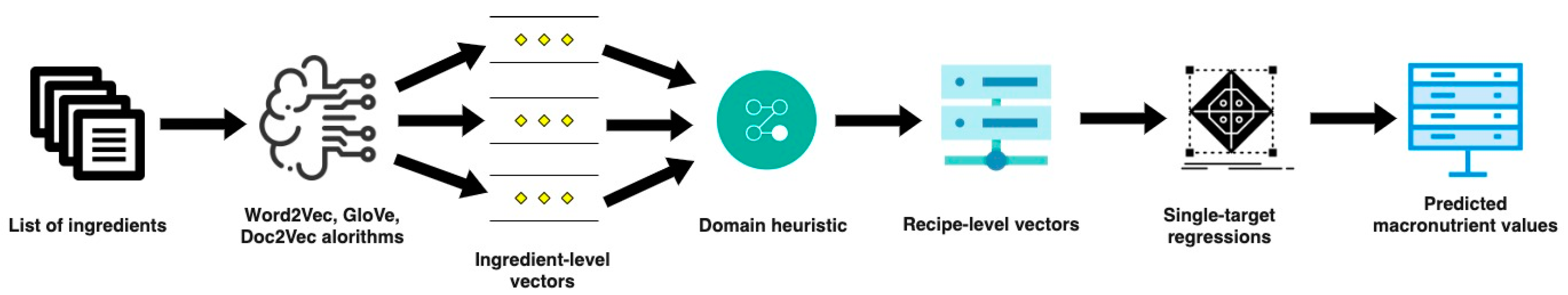
2.2. P-NUT
- Representation learning: Introduced by Mikolov et al. in 2013 [15] and Pennington et al. in 2014 [16], word embeddings have become indispensable for natural language processing (NLP) tasks in the past couple of years, and they have enabled various ML models that rely on vector representation as an input to benefit from these high-quality representations of text input. This kind of representation preserves more semantic and syntactic information of words, which leads to their status as being state-of-the-art in NLP.
- Unsupervised machine learning: Nutrient content exhibits notable variations between different types of foods. In a big dataset, including raw/simple and composite/recipe foods from various food groups, the content of macronutrients can have values from 0–100 per 100 . Needless to say, and as proven in [3,17], better models for macronutrient content prediction are built when grouping–clustering the instances according to domain-specific criteria [17].
- Supervised machine learning part: The final part of the P-NUT methodology is supervised ML, where separate predictive models are trained for the nutrients that we want to predict. The nutrient values are continuous data; therefore, the models are trained with single-target regression algorithms, in which, as input, we have the learned embeddings of the short text descriptions, clustered based on the chosen/available domain-specific criteria. Selecting the right ML algorithm for the purpose is challenging; the default accepted approach is selecting a few ML algorithms, setting the ranges for the hyper-parameters, hyper-parameter tuning, utilizing cross-fold validation to evaluate the estimators’ performances (with the same data in each iteration), and at the end, benchmarking the algorithms and selecting the best one(s). The most commonly used baselines for regression algorithms are the central tendency measures, i.e., mean and median of the train part of the dataset for all the predictions.
2.3. Domain-Specific Embeddings
2.4. Data
- recipe title: a short textual description of the recipe;
- recipe instruction: description of instructions for preparing the recipe;
- nutrient content: quantity of fat, protein, salt, saturates, and sugars per 100 g for the whole recipe, expressed in grams;
- ingredients: list of the ingredients needed;
- nutrient content of ingredients: for each ingredient, quantity in grams of fat, protein, saturates, sodium, and sugar per 100 g of the ingredient;
- quantity of each ingredient;
- units of measurement per each ingredient, by the household measurement system: cup, tablespoon, teaspoon, etc.;
- weight in grams per ingredient for the whole recipe.
3. Results
3.1. Data Preparation
3.2. Experimental Setup
- Generate embeddings on single ingredient level (multi-word non-contextualized embeddings) using the Word2Vec, GLoVe, and Doc2Vec algorithms [15,16,19,24]. For the Word2Vec and GloVe embeddings, we took into consideration different values for the dimension of the vectors and the sliding window size. For the vector dimensions, we chose [50, 100, 200]. For the Word2Vec embeddings, the two types of feature extraction available, CBOW and SG, were considered. For the chosen dimensions, we assigned different values, namely [2, 3, 5, 10], to the parameter called the ‘sliding’ window, which indicates the distance within a sentence between the current word and the word being predicted. With Word2Vec, combining the above-mentioned parameter values and the two heuristics, a total of 48 models were trained, while with GloVe, a total of 24 models were trained. When training the paragraph embeddings with Doc2Vec, we considered the same dimension sizes [50, 100, 200] and sliding window sizes [2, 3, 5, 10], as well as the two types of architectures, PV-DM and PV-DBOW, and we used the non-concatenative mode, meaning training separate models for the sum option and average option. Therefore, there were 48 Doc2Vec models trained in total.
- Generate embeddings on recipe level: use the above-defined domain heuristic (Equation (10)) to merge the embedding on single ingredient level.
- Apply single-target regression algorithms: building models for predicting the five given macronutrients: fat, protein, salt, saturates, and sugars. This part consisted of several steps:
- Selecting the regression algorithms: Linear regression, Ridge regression, Lasso regression, and ElasticNet regression (using the Scikit-learn library in Python [25]).
- Selecting ranges for the parameters for each algorithm and performing hyper-parameter tuning: A priori assignment of the ranges and values for all the parameters for all the regression algorithms. With GridSearchCV (from the Scikit-learn library in Python [25]), the best parameters for the model training were selected from all the combinations.
- Estimating the prediction error with k-fold cross-validation: We trained models with the previously selected best parameters and then evaluated them with cross-validation. To compare the regressors, the matched sample approach was chosen, using the same data in each iteration.
- Calculate domain-specific measure of accuracy: We defined the accuracy according to the appropriate tolerance levels for each nutrient, which were defined by international legalizations and regulations. In 2012, the European Commission Health and Consumers Directorate-General published a guidance document [26] in order to provide recommendations for calculating the acceptable differences between quantities of nutrients on the nutrient content labels of the food products and the ones established in Regulation EU 1169/2011 [27]. It is impossible for foods to contain the exact quantity of each nutrient on the printed labels; therefore, these tolerances for the food product labels are very important. These differences occur due to the natural variations of foods, and the variations occurring during production and the storage process. The accuracy is calculated according to the tolerance levels in Table 2.
3.3. Evaluation Outcomes
- Merge the single-ingredient embeddings with the domain-specific heuristic, perform the predictive modeling, obtain the results, and calculate the defined accuracy.
- Merge the single-ingredient embedding with conventional merging heuristics (sum and average) according to the merging heuristic used when obtaining the single-ingredient embeddings. In other words:
- The embedding on the recipe level is obtained by calculating an average of the embeddings on a single-ingredient level when they are obtained using Equation (4).
- The embedding on recipe level is obtained by summing the single-ingredient embeddings when they are obtained using Equation (5).
- Vector dimension: 100,
- Sliding window: 2,
- Architecture: PV-DBOW,
- Merging technique: sum.
4. Discussion and Future Work
- Normalizing the quantities: after the NER, the units (household measurements) need to be converted in grams, in order to have the same unit all across the dataset. This can be done using conversion tables [31], the problem that arises here is that these conversion tables are separate for liquid and dry ingredients, so the ingredients need to be separated into liquid and dry beforehand. This can be viewed as a classification problem.
- Map the extracted unit to the proper unit in the conversion table, which can be viewed as simple string matching, but since there are multiple ways of writing a single household measure unit, it can be viewed as a slightly more complex task than string matching, for example, mapping strings based on lexical similarity [32].
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ijaz, M.F.; Attique, M.; Son, Y. Data-Driven Cervical Cancer Prediction Model with Outlier Detection and Over-Sampling Methods. Sensors 2020, 20, 2809. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Diet, Nutrition, and the Prevention of Chronic Diseases: Report of a Joint WHO/FAO Expert Consultation; World Health Organization: Geneva, Switzerland, 2003; Volume 916. [Google Scholar]
- Ispirova, G.; Eftimov, T.; Koroušić Seljak, B. P-NUT: Predicting NUTrient Content from Short Text Descriptions. Mathematics 2020, 8, 1811. [Google Scholar] [CrossRef]
- Rand, W.M.; Pennington, J.A.; Murphy, S.P.; Klensin, J.C. Compiling Data for Food Composition Data Bases; United Nations University Press: Tokyo, Japan, 1991. [Google Scholar]
- Greenfield, H.; Southgate, D.A. Food Composition Data: Production, Management, and Use; Food and Agriculture Org.: Rome, Italy, 2003; ISBN 978-92-5-104949-5. [Google Scholar]
- Schakel, S.F.; Buzzard, I.M.; Gebhardt, S.E. Procedures for Estimating Nutrient Values for Food Composition Databases. J. Food Compos. Anal. 1997, 10, 102–114. [Google Scholar] [CrossRef] [Green Version]
- Machackova, M.; Giertlova, A.; Porubska, J.; Roe, M.; Ramos, C.; Finglas, P. EuroFIR Guideline on Calculation of Nutrient Content of Foods for Food Business Operators. Food Chem. 2018, 238, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Yunus, R.; Arif, O.; Afzal, H.; Amjad, M.F.; Abbas, H.; Bokhari, H.N.; Haider, S.T.; Zafar, N.; Nawaz, R. A Framework to Estimate the Nutritional Value of Food in Real Time Using Deep Learning Techniques. IEEE Access 2018, 7, 2643–2652. [Google Scholar] [CrossRef]
- Pouladzadeh, P.; Shirmohammadi, S.; Al-Maghrabi, R. Measuring Calorie and Nutrition from Food Image. IEEE Trans. Instrum. Meas. 2014, 63, 1947–1956. [Google Scholar] [CrossRef]
- Jiang, L.; Qiu, B.; Liu, X.; Huang, C.; Lin, K. DeepFood: Food Image Analysis and Dietary Assessment via Deep Model. IEEE Access 2020, 8, 47477–47489. [Google Scholar] [CrossRef]
- Samsung Health (S-Health). Available online: https://health.apps.samsung.com/terms (accessed on 12 April 2021).
- MyFitnessPal. Available online: https://www.myfitnesspal.com/ (accessed on 12 April 2021).
- Kaur, S.; Malik, K. Predicting and Estimating the Major Nutrients of Soil Using Machine Learning Techniques. In Soft Computing for Intelligent Systems; Springer: Cham, Germany, 2021; pp. 539–546. [Google Scholar]
- Wankhede, D.S. Analysis and Prediction of Soil Nutrients PH, N, P, K for Crop Using Machine Learning Classifier: A Review. In International Conference on Mobile Computing and Sustainable Informatics; Springer: Cham, Germany, 2020; pp. 111–121. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed Representations of Words and Phrases and Their Compositionality. In Proceedings of the 26th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013; pp. 3111–3119. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 25–29 October 2014; pp. 1532–1543. [Google Scholar]
- Ispirova, G.; Eftimov, T.; Seljak, B.K. Exploring Knowledge Domain Bias on a Prediction Task for Food and Nutrition Data. In Proceedings of the 2020 IEEE International Conference on Big Data (Big Data), Atlanta, GA, USA, 10–13 December 2020. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Le, Q.; Mikolov, T. Distributed Representations of Sentences and Documents. In Proceedings of the International Conference on Machine Learning, Beijing, China, 21 June 2014; pp. 1188–1196. [Google Scholar]
- Jiang, M.; Sanger, T.; Liu, X. Combining Contextualized Embeddings and Prior Knowledge for Clinical Named Entity Recognition: Evaluation Study. JMIR Med Inform. 2019, 7, e14850. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, X.; Hui, L.; Zou, L.; Li, H.; Xu, L.; Liu, W. Chinese Clinical Named Entity Recognition in Electronic Medical Records: Development of a Lattice Long Short-Term Memory Model With Contextualized Character Representations. JMIR Med. Inform. 2020, 8, e19848. [Google Scholar] [CrossRef] [PubMed]
- Rasmy, L.; Xiang, Y.; Xie, Z.; Tao, C.; Zhi, D. Med-BERT: Pretrained Contextualized Embeddings on Large-Scale Structured Electronic Health Records for Disease Prediction. NPJ Digit. Med. 2021, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Marin, J.; Biswas, A.; Ofli, F.; Hynes, N.; Salvador, A.; Aytar, Y.; Weber, I.; Torralba, A. Recipe1m+: A Dataset for Learning Cross-Modal Embeddings for Cooking Recipes and Food Images. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 187–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehurek, R.; Sojka, P. Gensim—Statistical Semantics in Python. NLP Cent. Fac. Inform. Masaryk Univ. 2011. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- European Commission Health and Consumers Directorate-General Guidance Document for Competent Authorities for the Control of Compliance with EU Legislation on: Regulation (EU) No 1169/2011 of the European Parliament and of the Council of 25 October 2011 on the Provision of Food Information to Consumers, Amending Regulations (EC) No 1924/2006 and (EC) No 1925/2006 of the European Parliament and of the Council, and Repealing Commission Directive 87/250/EEC, Council Directive 90/496/EEC, Commission Directive 1999/10/EC, Directive 2000/13/EC of the European Parliament and of the Council, Commission Directives 2002/67/EC and 2008/5/EC and Commission Regulation (EC) No 608/2004Devlin. Available online: https://ec.europa.eu/food/sites/food/files/safety/docs/labelling_nutrition-supplements-guidance_tolerances_1212_en.pdf (accessed on 13 April 2021).
- Commission, E. Regulation (EU) No 1169/2011 of the European Parliament and of the Council of 25 October 2011 on the Provision of Food Information to Consumers, Amending Regulations (EC) No 1924/2006 and (EC) No 1925/2006 of the European Parliament and of the Council, and Repealing Commission Directive 87/250/EEC, Council Directive 90/496/EEC, Commission Directive 1999/10/EC, Directive 2000/13/EC of the European Parliament and of the Council, Commission Directives 2002/67/EC and 2008/5/EC and Commission Regulation (EC) No 608/2004. Off. J. Eur. Union L 2011, 304, 18–63. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Popovski, G.; Kochev, S.; Korousic-Seljak, B.; Eftimov, T. FoodIE: A Rule-Based Named-Entity Recognition Method for Food Information Extraction. In Proceedings of the 8th International Conference on Pattern Recognition Applications and Methods, Prague, Czech Republic, 19–21 February 2019; pp. 915–922. [Google Scholar]
- Cenikj, G.; Popovski, G.; Stojanov, R.; Seljak, B.K.; Eftimov, T. BuTTER: BidirecTional LSTM for Food Named-Entity Recognition. In Proceedings of the 2020 IEEE International Conference on Big Data (Big Data), Atlanta, GA, USA, 10–13 December 2020; pp. 3550–3556. [Google Scholar]
- Lynn Wright Cooking Measurement Conversion Tables. Available online: https://www.saga.co.uk/magazine/food/cooking-tips/cooking-measurement-conversion-tables (accessed on 13 April 2021).
- Ispirova, G.; Eftimov, T.; Korousic-Seljak, B.; Korosec, P. Mapping Food Composition Data from Various Data Sources to a Domain-Specific Ontology. In Proceedings of the 9th International Conference on Knowledge Engineering and Ontology Development, Funchal, Portugal, 1–3 November 2017; pp. 203–210. [Google Scholar]


| Recipe Title | No-Bake Oatmeal Cookies | |||||
|---|---|---|---|---|---|---|
| Ingredients | “sugars, granulated”, “cocoa, dry powder, unsweetened”, “milk, fluid, 1% fat, without added vitamin a and vitamin d”, “butter, without salt”, “vanilla extract”, “peanut butter, smooth style, without salt”, “oats” | |||||
| Nutrients per 100 g | Energy | Fat | Protein | Salt | Saturates | Sugars |
| 378.64 | 35.40 | 3.81 | 0.06 | 21.01 | 8.59 | |
| Measure | Target | Algorithm | |||||
|---|---|---|---|---|---|---|---|
| With Domain Heuristic | Without Domain Heuristic | ||||||
| Word2Vec | GloVe | Doc2Vec | Word2Vec | GloVe | Doc2Vec | ||
| Maximal accuracy (in %) | Fat | 76.66 | 75.62 | 91.65 | 21.45 | 22.16 | 20.29 |
| Proteins | 90.57 | 88.79 | 97.98 | 55.47 | 54.54 | 52.67 | |
| Sugars | 73.38 | 76.35 | 88.14 | 25.73 | 25.81 | 22.97 | |
| Saturates | 72.78 | 73.66 | 95.95 | 24.00 | 24.71 | 20.58 | |
| Salt | 43.34 | 41.79 | 52.35 | 36.28 | 33.10 | 19.43 | |
| Minimal accuracy (in %) | Fat | 18.65 | 18.65 | 28.00 | 8.34 | 8.34 | 8.35 |
| Proteins | 56.60 | 56.60 | 56.57 | 27.20 | 27.20 | 27.22 | |
| Sugars | 17.03 | 17.03 | 28.54 | 7.90 | 7.90 | 7.90 | |
| Saturates | 30.52 | 30.52 | 45.27 | 8.34 | 8.34 | 8.35 | |
| Salt | 19.24 | 19.24 | 45.27 | 9.44 | 9.44 | 9.44 | |
| Mean accuracy (in %) | Fat | 37.96 | 48.03 | 68.99 | 12.54 | 13.22 | 14.70 |
| Proteins | 68.02 | 78.99 | 86.12 | 37.19 | 36.85 | 39.54 | |
| Sugars | 38.58 | 47.13 | 56.05 | 13.61 | 14.03 | 15.07 | |
| Saturates | 60.10 | 67.37 | 78.32 | 12.84 | 13.18 | 14.11 | |
| Salt | 26.25 | 27.49 | 31.01 | 16.07 | 15.65 | 18.37 | |
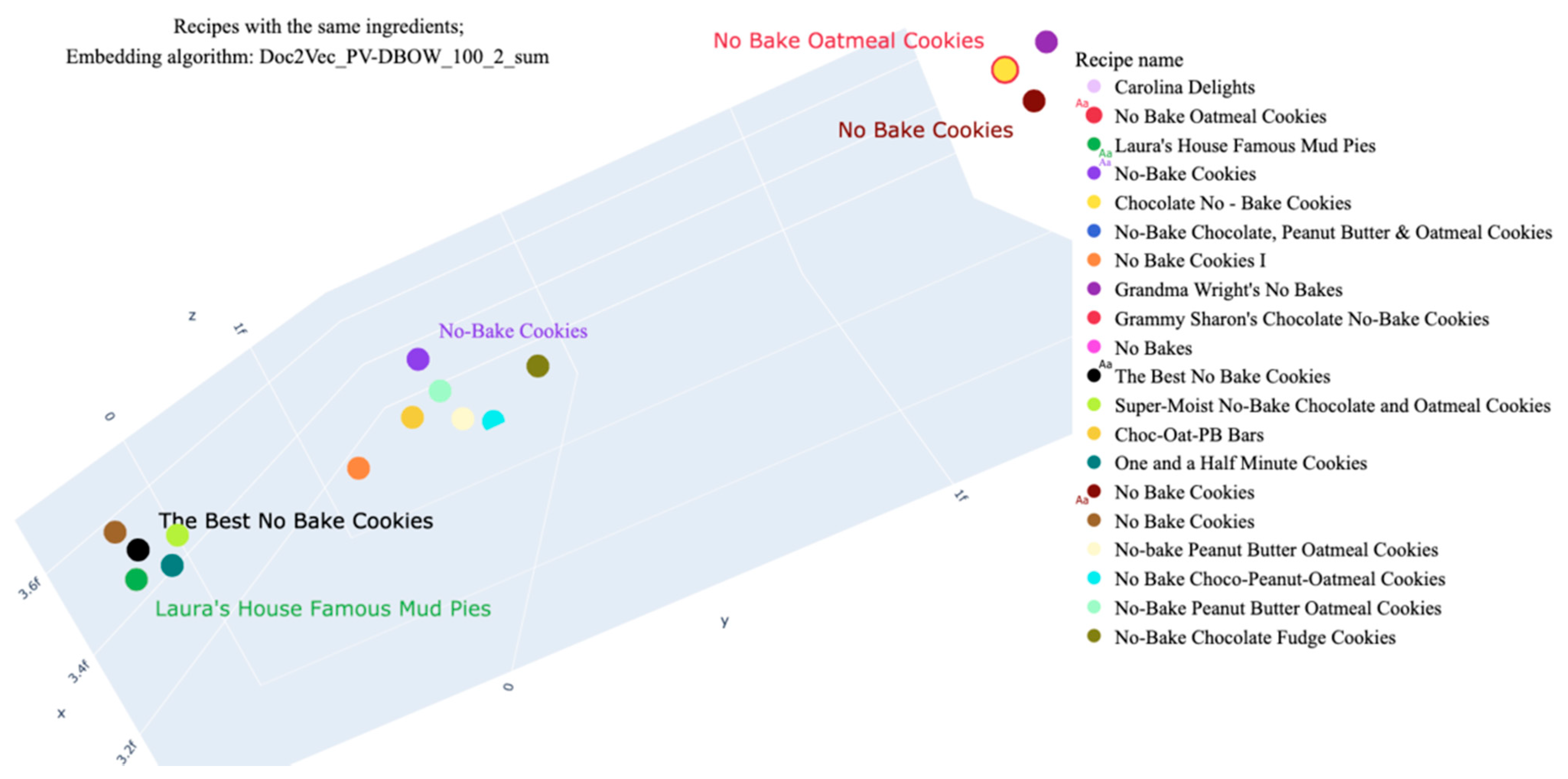
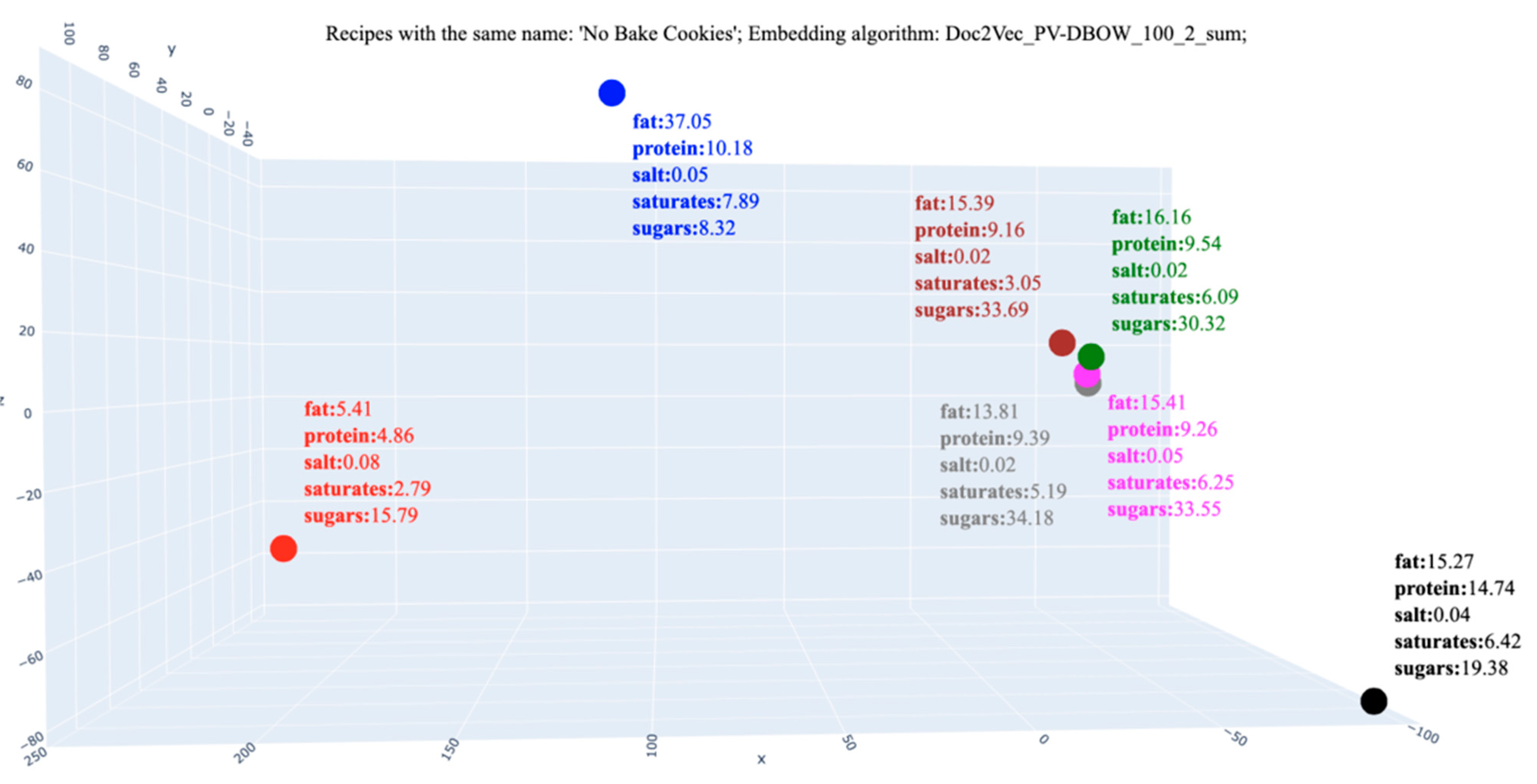
| Recipe Title | Ingredients | Nutrients Per 100 g | |||||
|---|---|---|---|---|---|---|---|
| Energy | Fat | Protein | Salt | Saturates | Sugars | ||
| The Best No Bake Cookies | “cocoa, dry powder, unsweetened”, “milk, fluid, 1% fat, without added vitamin a and vitamin d”, “oats”, “peanut butter, smooth style, without salt”, “sugars, granulated”, “butter, without salt”, “vanilla extract” | 378.64 | 35.40 | 3.81 | 0.06 | 21.01 | 8.59 |
| No Bake Oatmeal Cookies | 378.44 | 35.37 | 3.83 | 0.06 | 21.00 | 8.58 | |
| Laura’s House Famous Mud Pies | 385.10 | 15.58 | 4.45 | 0.02 | 8.05 | 50.80 | |
| No Bake Cookies | 323.39 | 15.26 | 14.74 | 0.03 | 6.41 | 19.38 | |
| No-Bake Cookies | 317.04 | 22.93 | 13.34 | 0.06 | 5.53 | 11.53 | |
| Chocolate No–Bake Cookies | 397.65 | 15.39 | 9.43 | 0.02 | 6.28 | 33.02 | |
| Recipe Title | The Best No Bake Cookies | No Bake Oatmeal Cookies | Laura’s House Famous Mud Pies | No Bake Cookies | No-Bake Cookies | Chocolate No–Bake Cookies | ||
|---|---|---|---|---|---|---|---|---|
| Nutrients per 100 g | Fat | A | 35.4 | 35.37 | 15.58 | 15.26 | 22.93 | 15.39 |
| DH | 32.28 | 32.92 | 14.38 | 15.32 | 25.54 | 14.47 | ||
| No DH | 23.11 | 12.44 | 29.33 | 16.78 | 16.78 | 8.79 | ||
| Protein | A | 3.81 | 3.83 | 4.45 | 14.74 | 13.34 | 9.43 | |
| DH | 3.59 | 4.38 | 4.40 | 12.29 | 9.12 | 7.26 | ||
| No DH | 8.96 | 19.67 | 2.34 | 12.11 | 7.34 | 15.67 | ||
| Salt | A | 0.06 | 0.06 | 0.02 | 0.03 | 0.06 | 0.02 | |
| DH | 0.03 | 0.03 | 0.15 | 0.06 | 0.30 | 0.39 | ||
| No DH | 1.78 | 0.30 | 0.20 | 0.28 | 0.20 | 0.38 | ||
| Saturates | A | 21.01 | 21.00 | 8.05 | 6.41 | 5.53 | 6.28 | |
| DH | 18.21 | 18.23 | 7.21 | 6.11 | 6.79 | 5.74 | ||
| No DH | 10.53 | 10.43 | 12.56 | 2.56 | 11.67 | 15.89 | ||
| Sugars | A | 8.59 | 8.58 | 50.80 | 19.38 | 11.53 | 33.02 | |
| DH | 10.88 | 10.88 | 50.62 | 22.36 | 11.30 | 34.46 | ||
| No DH | 27.45 | 18.99 | 24.75 | 2.33 | 21.74 | 27.85 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ispirova, G.; Eftimov, T.; Koroušić Seljak, B. Domain Heuristic Fusion of Multi-Word Embeddings for Nutrient Value Prediction. Mathematics 2021, 9, 1941. https://doi.org/10.3390/math9161941
Ispirova G, Eftimov T, Koroušić Seljak B. Domain Heuristic Fusion of Multi-Word Embeddings for Nutrient Value Prediction. Mathematics. 2021; 9(16):1941. https://doi.org/10.3390/math9161941
Chicago/Turabian StyleIspirova, Gordana, Tome Eftimov, and Barbara Koroušić Seljak. 2021. "Domain Heuristic Fusion of Multi-Word Embeddings for Nutrient Value Prediction" Mathematics 9, no. 16: 1941. https://doi.org/10.3390/math9161941
APA StyleIspirova, G., Eftimov, T., & Koroušić Seljak, B. (2021). Domain Heuristic Fusion of Multi-Word Embeddings for Nutrient Value Prediction. Mathematics, 9(16), 1941. https://doi.org/10.3390/math9161941







