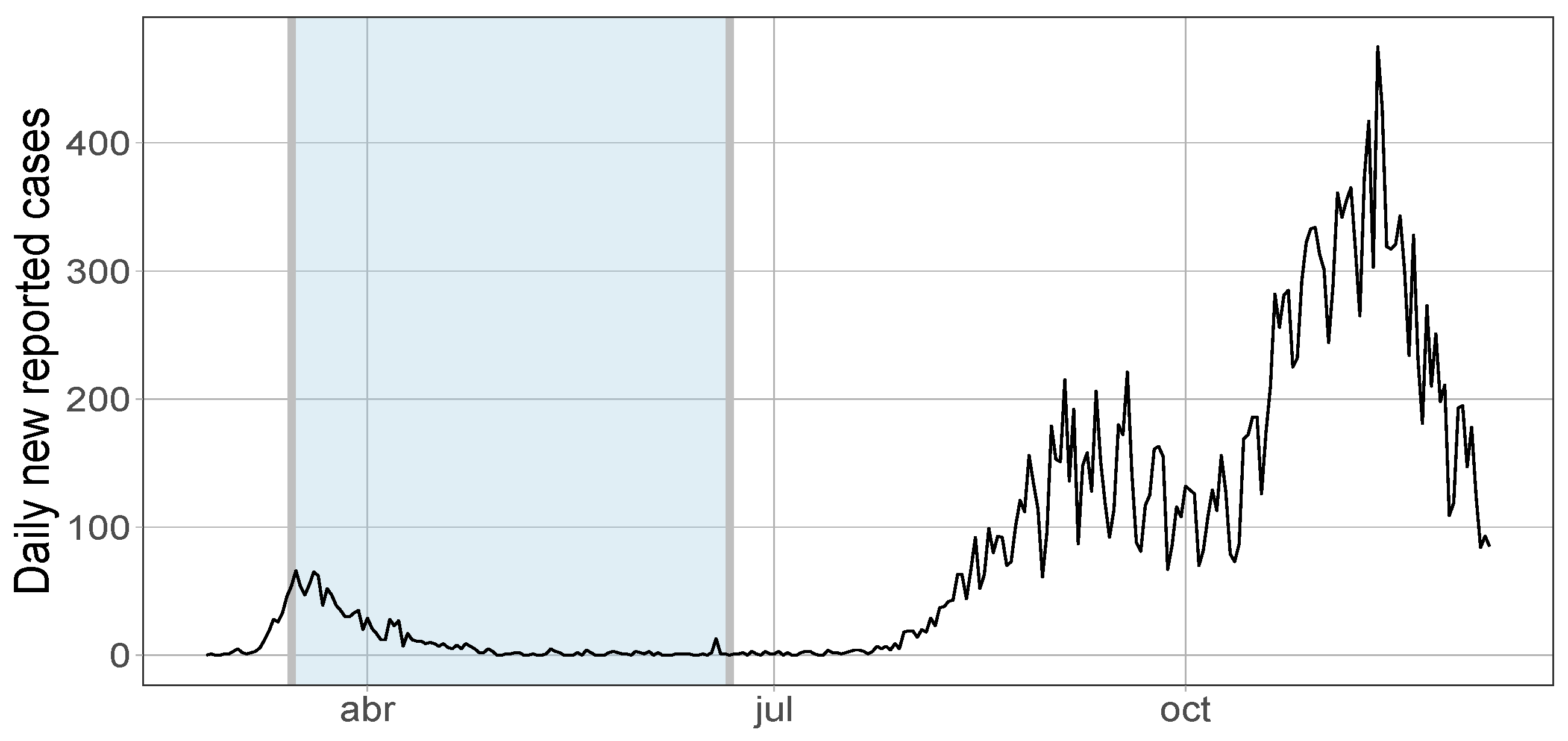
In this section we will illustrate our method analyzing real data associated to the COVID-19 in a specific region located at the south of Spain, the province of Cádiz. We collect data from the Spanish National Network for Epidemiological Surveillance (RENAVE, by its Spanish initials). At this moment, we would like to emphasize the difficulty of choosing the time zero as we mentioned in Remark 2. Here we consider the time zero on 25 February where the first case of the COVID-19 pandemic was confirmed in Andalusia which also coincides with the closest day to the epidemic growth. Data reflect the total number of confirmed cases with SARS-CoV-2, namely all those who have a positive test on Polymerase Chain Reaction (PCR) plus those positive in a rapid antibody test made in laboratory. We discard individuals having positive test using other methods, like antigen detection or Enzyme-Linked ImmunoSorbent Assay (ELISA).
At this moment, we evaluate the hyper-parameters of the different marginal prior distributions given in (
6)–(
8). The population of the province of Cádiz is estimated at
p = 1,240,155 inhabitants, so the value of
is
. Just considering the confirmed cases per 100,000 people shown in
Figure 1 for the Spanish ACs at the beginning of the pandemic and changing the scale by
we obtain the Maximum Likelihood Estimate (MLE) for the mean and the scale parameters in (
6), i.e.,
and
, respectively. The value
was determined because it was not found any case where the number of infections at the beginning were bigger. We would like to emphasize that in this first wave
M had little informative value to obtain the posterior distribution of
b as we have checked by taking different values for
M and just observing the posterior expected quantity for
and its posterior standard deviation equal to
shown in
Table 1. In other words, the result of the estimation is essentially independent of the choice of
M in this case. Finally, to bound the values of
and
, we take into account the highest and the lowest values found in Spain and other countries, as it is seen in [
34,
35,
37]. Those values are also reasonable with the observed range in Figure 1 described in [
33].
3.1. Forecasts for the Characteristics of Interest at Different Scenarios
In order to evaluate our model, we will estimate the functionals given in (
9)–(
11) at different stages of the pandemic. As a natural question, we first are interested in evaluating the benefits of the first lockdown imposed by the Spain’s central government. Second, we will locate our estimates during the lockdown and close to the end of the State of Alarm to verify not only that predictions are quite accurate, but also how daily new cases decrease. Finally, just observing the evolution of our estimates after the easing of Spain’s lockdown restrictions, we will be able to detect the beginning of the second and third waves by the increase of the daily number of new reported contagious.
3.1.1. First Scenario: The Benefits of the Lockdown
The lockdown in Spain was imposed on 14 March 2020. Therefore, in order to evaluate the benefits of that decision in the province of Cádiz, we will first consider T, the ending day, as 15 March 2020. The idea is to make daily predictions of the following week, from 16 March 2020 to 22 March 2020. Moreover, it is worth mentioning that week was close to the date of the maximum number of daily new reported cases of COVID-19 in the first wave. We are aware that the classical Gompertz curve is a poor model in the early stages of an epidemic. However one of the advantages of the Bayesian approach is the incorporation of prior information which leads to a better inference for small samples.
Figure 3a shows the observed time series of the daily cumulative cases up to
T—to feed the Bayesian model—and a week after
T (brown). Likewise, it shows a set of 500 Gompertz curves obtained by an i.i.d. random sample of size 500 from the posterior distribution
(grey). It is remarkable the band of the Gompertz curves leads us to predict the trend of daily cumulative positive cases of SARS-CoV-2 by incorporating variability.
Figure 3b shows the observed time series of the daily cases up to
T (black) and a week after
T (brown). It also shows the expectation (blue) and the
CI (blue dash line) of
as forecasts of the expected number of new daily cases of COVID-19 where
.
At first glance, a change in trend can be observed between the predictions of the expected values (which continues an upward trend) and the observed data after T, which begins a downward trend. For that reason, it seems that the lockdown imposed by the authorities was beneficial to control the initial evolution of the pandemic by reducing the daily number of expected new cases.
Regarding to the parameters of interest, it is remarkable that in case of no restrictions—no government interventions- after
T, we estimate that the
CI for the parameter
a—maximum number of infected—would lie on the interval
having a posterior mean of 27,766.41 inhabitants, see
Table 1. As the population size in Cádiz is 1,240,155 inhabitants, no restrictions could mean that approximately the
of population would be infected by the disease. Of course, this number could have meant the collapse of the health system and would have caused a much higher number of deaths. Additionally, the posterior mean of the time to reach the peak would have been
days, letting the effect of the pandemic considerably would have dragged on. Fortunately it was not the case.
3.1.2. Second Scenario: The Evolution of the Pandemic during the Lockdown
Now we will evaluate the goodness of fit of our model by making predictions during the lockdown period. For such a purpose, we will consider T, the ending day, as 3 May 2020. As in the first scenario, the idea is to make daily predictions for the following week, from 4 May 2020 to 10 May 2020. It is worth mentioning that the decrease of the number of new daily cases during the lockdown was the reason why Spanish authorities justified the end of the lockdown on 21 June 2020.
Analogously to
Figure 3 and
Figure 4a shows the time series of observed daily cumulative cases up to
T (black) and a week after
T (brown). Moreover, shows a band of Gompertz curves obtained from an i.i.d. random sample of the posterior distribution
(grey).
Moreover, analogously to
Figure 3 and
Figure 4b shows the observed time series of daily new cases (black) up to
T and a week after
T (brown). It also shows the forecasts of the expected number of new daily cases as the posterior mean of
(blue) and its
CI (blue dash line). In addition, we also compute the
CI of
(red band) and
(green band), where
. Those quantiles lead us to measure where the middle
of the daily new cases lies.
At first glance, the trend of both daily and cumulative expected values are quite similar to the observed data which implies that our model fits reasonably well the observations.
Table 2 shows a summary of the Bayesian estimates of the main parameters. As a first conclusion, it seems the lockdown had a direct effect on the estimates compared to the values given in
Table 1. Now the posterior mean of the maximum cumulative number of confirmed cases in the province of Cádiz, parameter
a, is about
people, close to the official cumulative number of confirmed cases at the end of the State of Alarm and having a
CI of
. Therefore, we estimate that about
of the population of the province of Cádiz was detected as a confirmed case of COVID-19 in the first wave and until the end of the lockdown. Taking into account that less than one out of ten cases was detected in the first wave, as it is described in [
44], our result seems consistent with seroprevalence studies made in Spain, where it was determined that
of inhabitant in the province of Cádiz presented IgG antibody against SARS-CoV2. Additionally, we also estimate that
days having a
CI
. All those estimates are close to the official data provided by RENAVE which predicts the peak in 20 days from 25 February 2020. To sum up, we would like to emphasize that a direct computation shows that the effect of the lockdown reduced the number of infected cases by about
.
In Spain, the end of the lockdown was on 21 June 2020 and our model fits reasonable well during that period and forecasts stop being as good after lockdown. It is apparent the easing of restrictions lead to a new change in the trend and the arrival of a new wave. We will see in the following scenario how we can predict it.
3.2. Detecting the Beginning of a New Wave
As we have mentioned, the model fits well the evolution of the number of new cases during the lockdown. By considering T, the end of the lockdown, as 21 June, we next propose a classical tool to detect the beginning of a future wave based on the percentile of the number of daily new cases. For such a purpose, we first estimate daily quantiles by the posterior mean of , where , i.e., for the first 42 days—6 weeks—after the lockdown. Second, for the ith week we count the cumulative number of confirmed cases where the observed daily number of contagious exceeding the estimate of the daily quantile, and we will denote it by , . For example, means that just one day the observed new daily cases exceed the estimate of the daily quantile in the first week after the lockdown. It is apparent that is a risk measure that takes values from 0 to 7, , and the larger the value, the greater the probability of a new wave.
Table 3 shows the values of
,
, in the province of Cádiz. Note the first week ranges from 22 June to 28 June and the sixth week from 27 July to 2 August. It is apparent that easing COVID-19 restrictions after the lockdown leads to more spreading of coronavirus in just a few weeks.
Again we face the problem to establish the time zero as mentioned in Remark 2. The value
in
Table 3 implies that in all days in the 5th week the observed new daily cases exceed the estimate of the 99% daily quantile. Therefore, in order to make predictions in the second wave we have considered the initial date as 27 July, five weeks after the lockdown was finished, and
T, the ending day, as 13 September 2020. We would like to emphasize that Spain had one of the most restrictive lockdown in the world in the first wave. After the lockdown people were afraid of going back to normal. We think this was the main reason of slow growth at the beginning of July. However, little by little people in summer were more confidence and jointly to an attempt to save the tourist season, infections started growth again at the end of July. It is apparent that initial conditions in the first and second waves are different. Therefore the value of the hyper-parameter
was determined taking into account that the initial cases of the second waves are, in some sense, determined by the cases at the end of the first wave. Finally, we consider the same prior information for the parameters
a and
c in order to have more prior variability.
The parameters of interest are shown in
Table 4. Note the second wave can be interpreted as an intermediate scenario between having no restrictions and the lockdown. Now the posterior mean of the maximum number of infected in the province of Cádiz,
a, is about 14,980.964 people and we also estimate that
. We recall that differences between the first and second wave can be attributed to both a higher rate of contagious and an increase of the number of people tested as described in Remark 4.
As a complementary study, Andalusia is divided into eight provinces, namely Almeria, Granada, Jaén, Málaga, Sevilla, Córdoba, Cádiz and Huelva. We compute the evolution of the risk measure given in
Table 3 for all of them. In order to make predictions, we only should take in account they have different population size, i.e., different
= P/100,000 in Expression (
6).
Table 5 shows the population sizes,
P, of the eight Andalusian provinces (population size according to the Instituto Nacional de Estadística
https://www.ine.es/up/9Gq4uzeUiT).
Figure 5 shows the evolution of the risk measure in Andalusia by using a color map. This figures allows us to make inter-provincial comparisons and detect how the effects of COVID-19 vary between provinces and territories.
To conclude our analysis, we have studied the evolution of the confirmed cases in the province of Cádiz during the autumn period. We first fix the beginning of the second wave as 27 July and T as 20 September. By using a similar argument as in the detection of the second wave, we compute the measure for the following four weeks, and 4, obtaining 3, 3, 4 and 6, respectively. It seems a third wave appears in the fourth week from the beginning of the second wave. In addition, that fourth week coincides with a vacation period in Spain. Therefore, we finally establish the beginning of the third wave as 11 October. In contrast to the second wave, the third wave appears before flattening the second curve.
By using data from 11 October to 8 December 2020—the submission date of this work—we present in
Table 6 the parameters of interest of the third wave. Again the hyper-parameter
has been modified because the third wave started having higher initial values at time zero.
Finally, we present in
Figure 6 the band of Gompertz curve for the second wave (green) and the band of Gompertz curve for the third wave (orange) obtained from an i.i.d. random sample of the posterior distributions. It is apparent that models fit well data. From the interpretation of
as the width (duration) of a wave and just observing the estimates of the parameter
c in
Table 2,
Table 4 and
Table 6, it is apparent that the duration of the second wave (if it were not interrupted by the third) would be more than twice longer that the duration of the first one and the third wave seems to be a bit shorter than the second one.