Deep Learning on Histopathology Images for Breast Cancer Classification: A Bibliometric Analysis
Abstract
:1. Introduction
2. Breast Cancer Image Classification
2.1. Bibliometric Analysis of Breast Cancer Studies
2.2. Breast Cancer Image Database
2.3. Breast Cancer Image Classification using Deep Learning Approaches
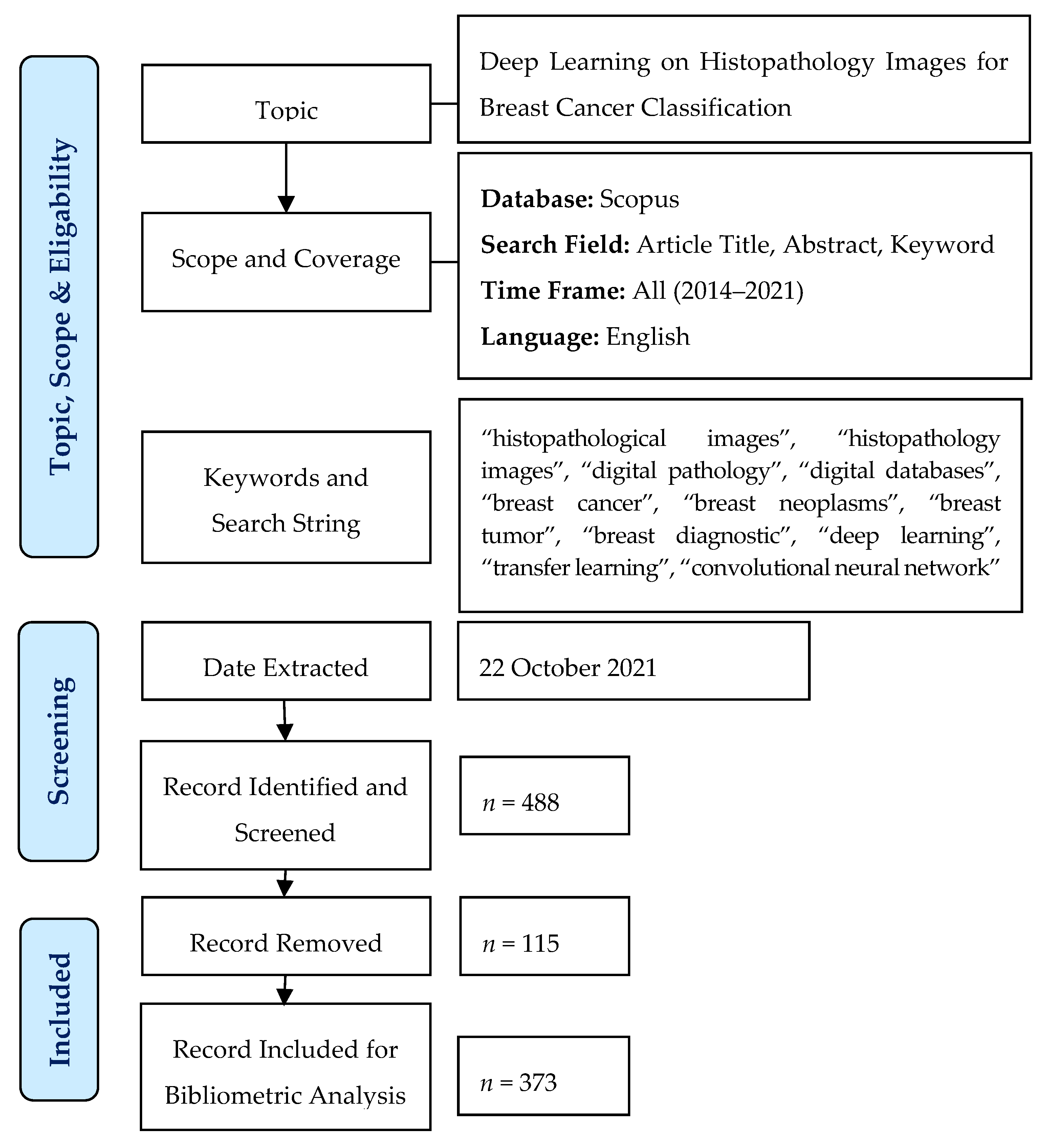
3. Materials and Methods
3.1. Bibliometric Analysis
3.2. Data Collection
4. Results and Discussion
4.1. Overview on Document and Source Type
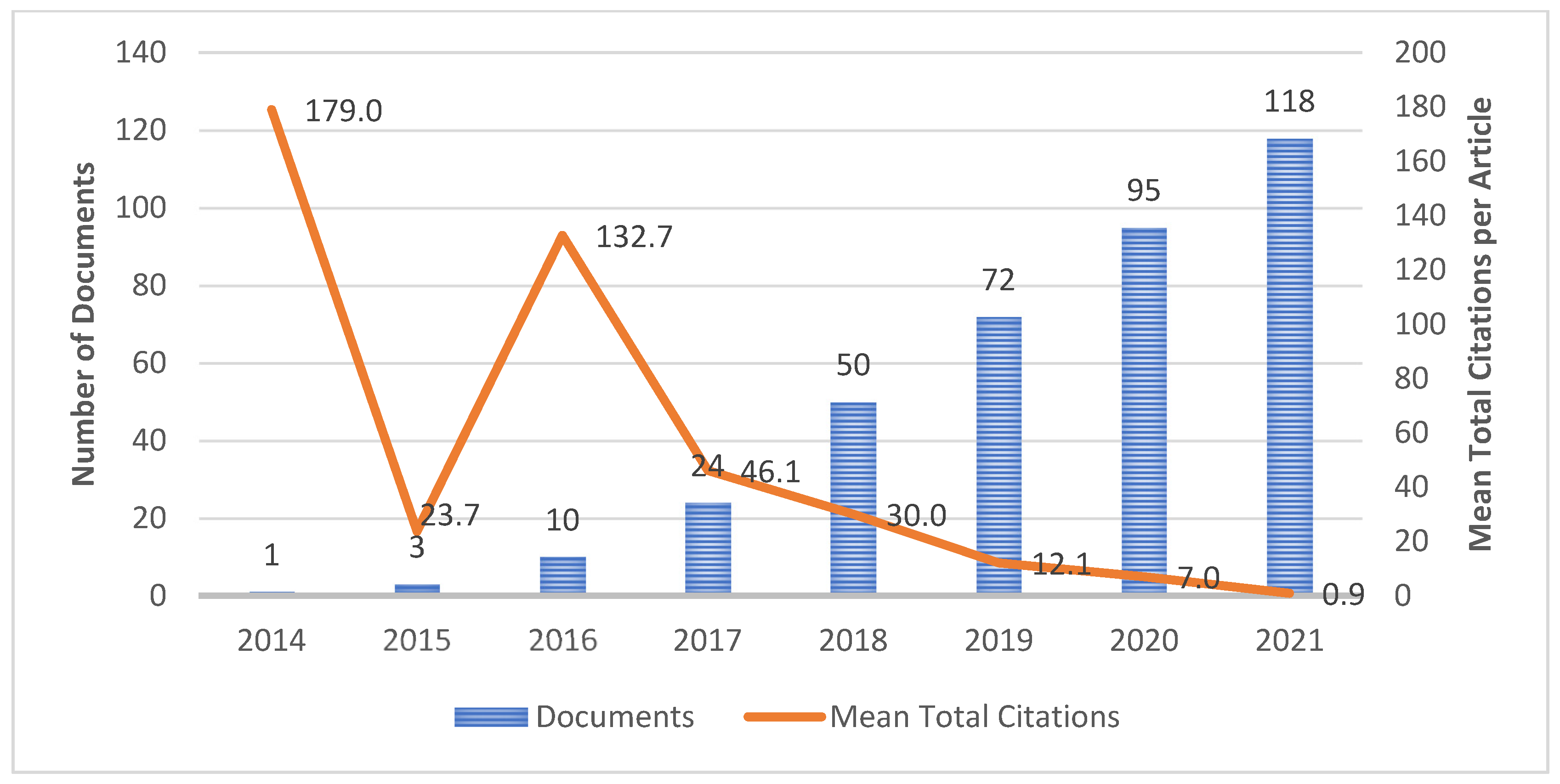
4.2. Publication Growth
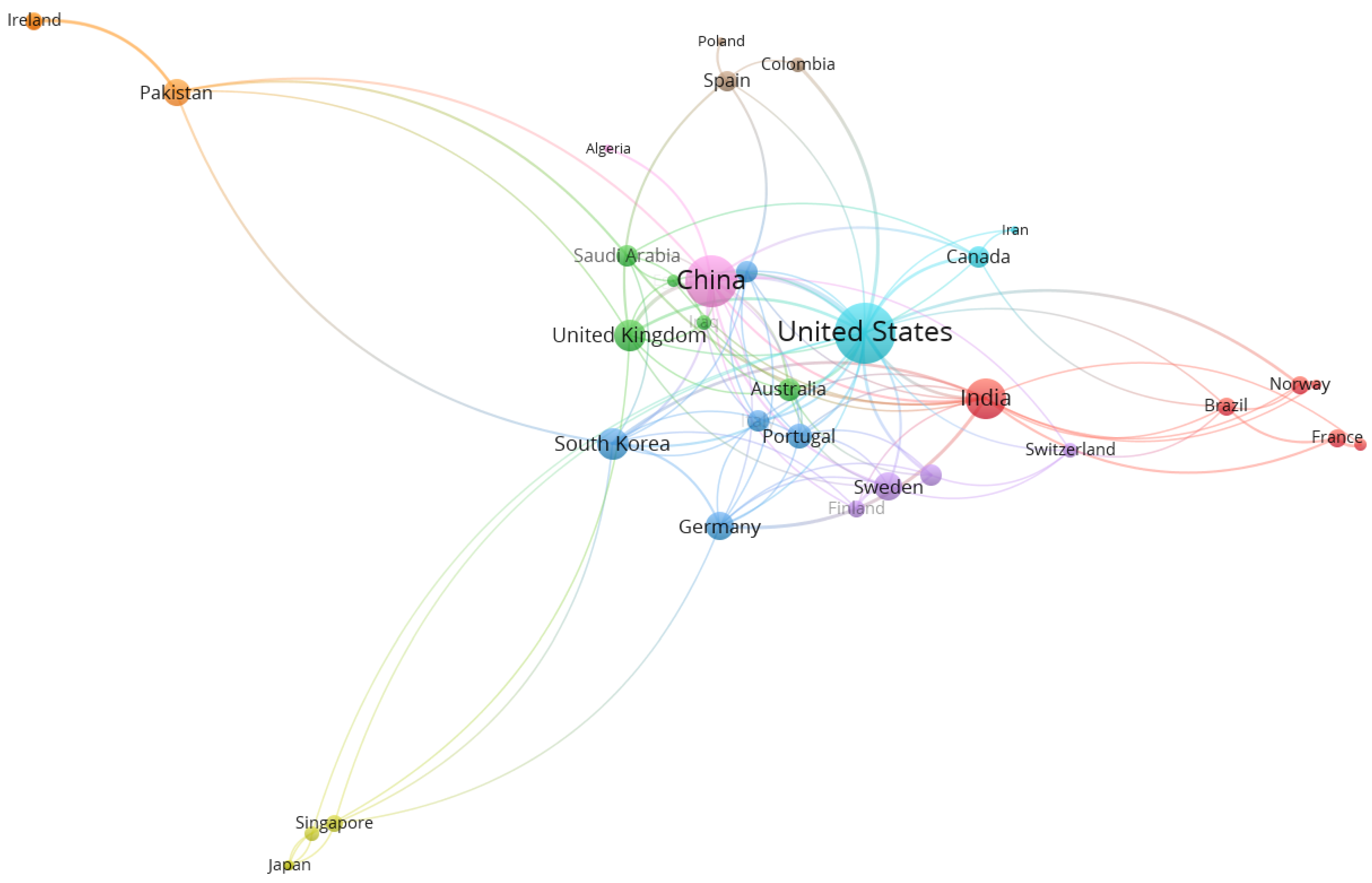
4.3. Country Network Analysis
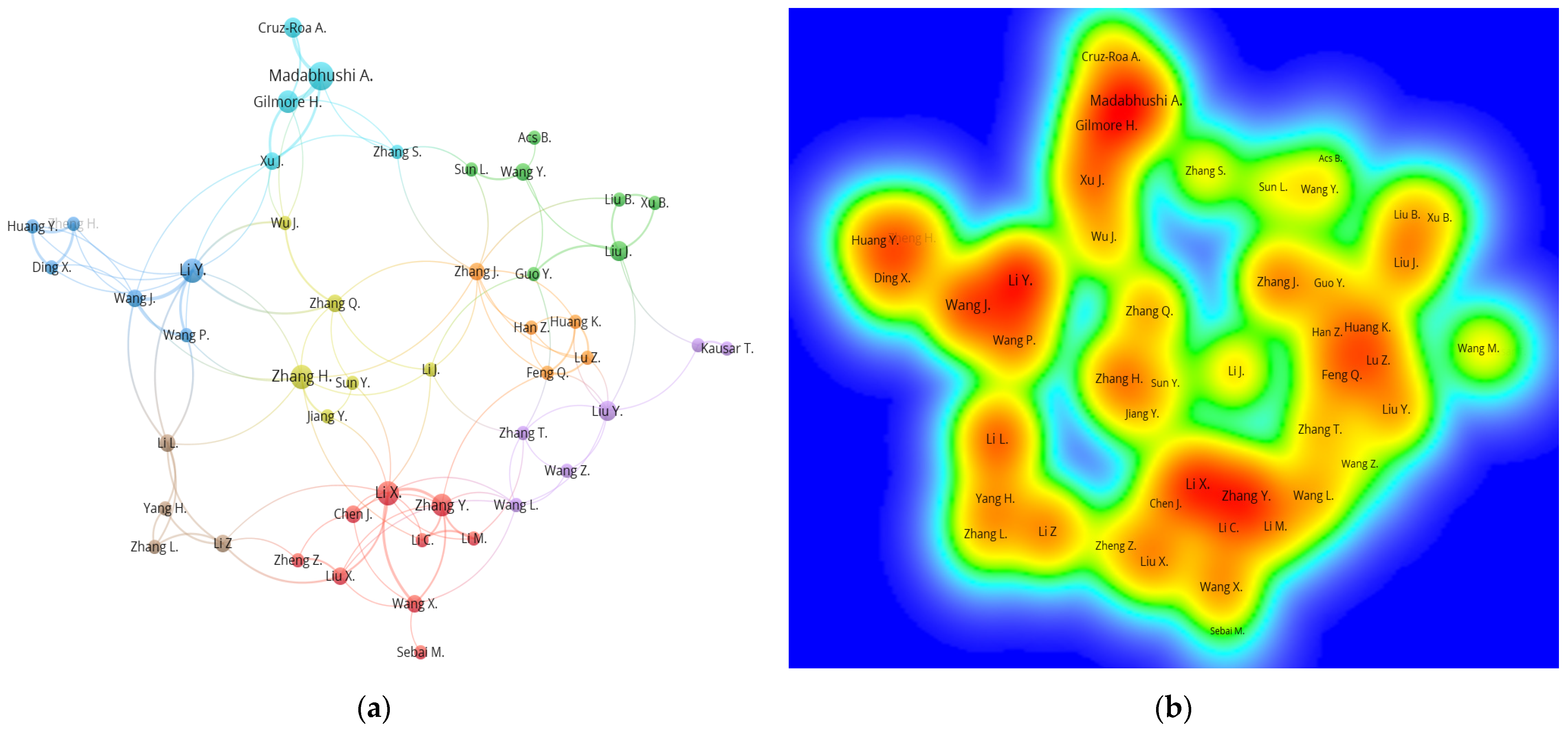
4.4. Author Network Analysis
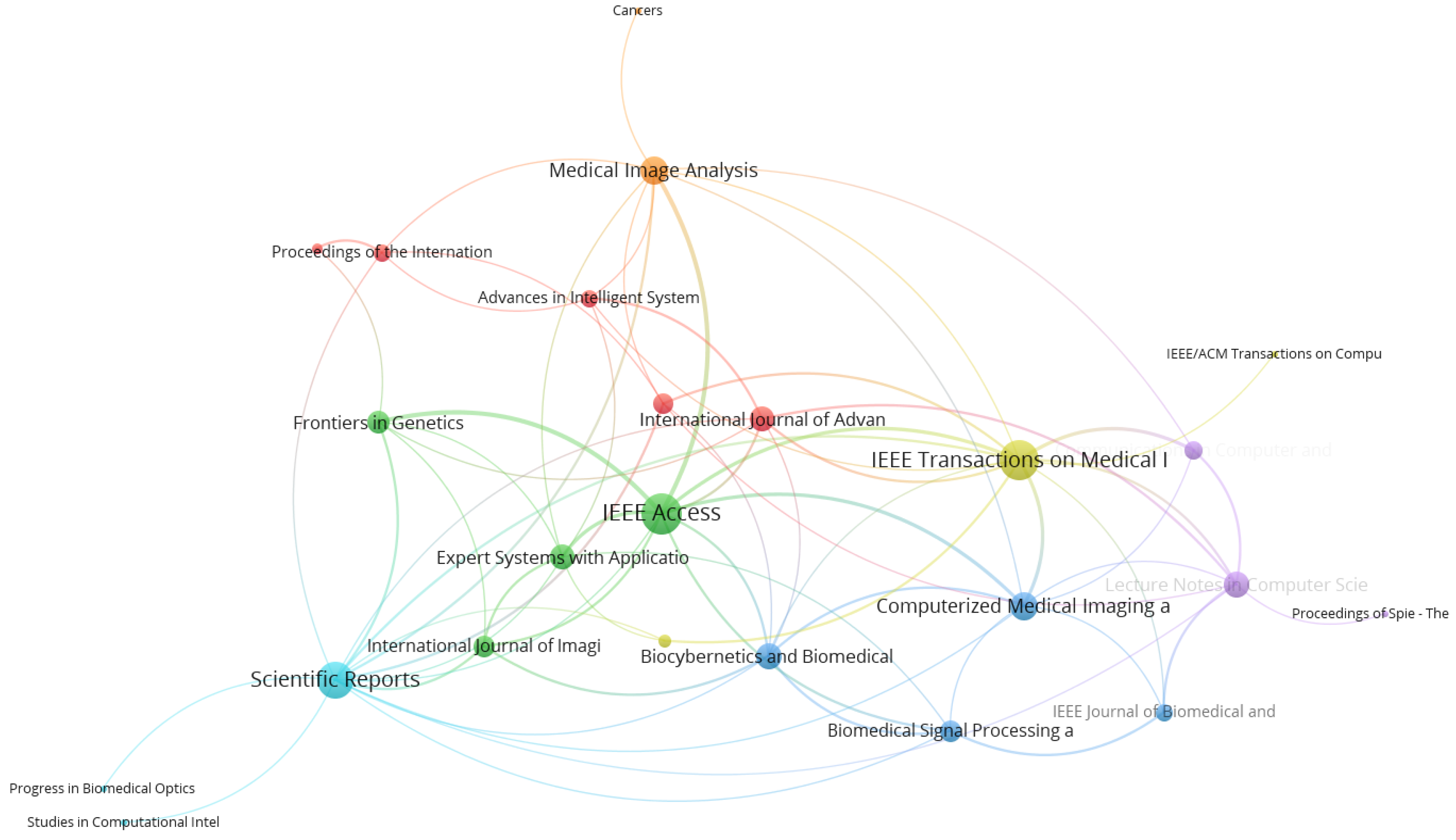
4.5. Journal Network Analysis
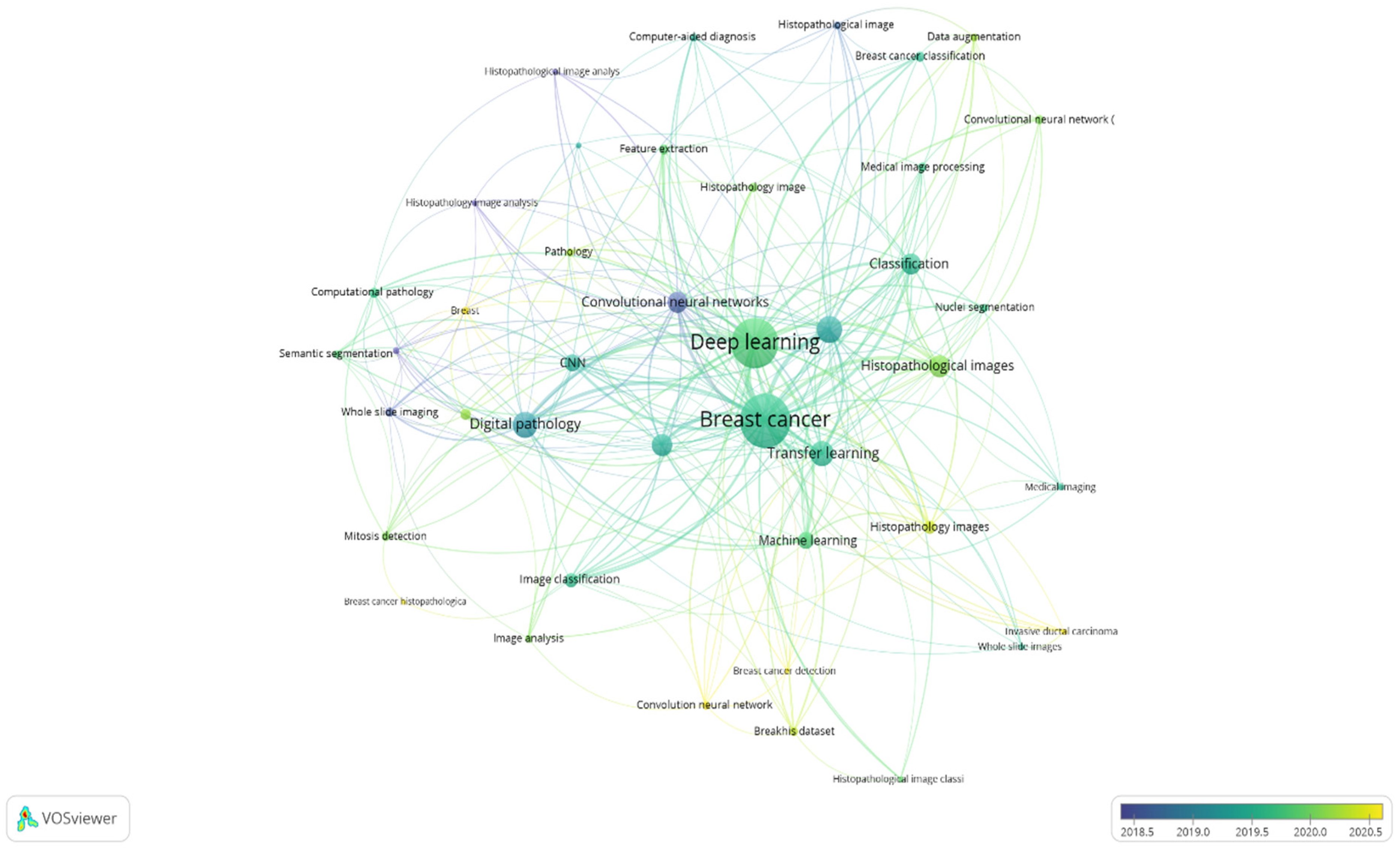
4.6. Co-Occurrence Analysis of Author Keywords
4.7. Computational Method for Histopathology Images
4.8. Challenges and Future Directions
4.8.1. Large Image Size
4.8.2. Color Variations
4.8.3. Insufficient Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nenclares, P.; Harrington, K.J. The Biology of Cancer. Medicine 2020, 48, 67–72. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Momenimovahed, Z.; Salehiniya, H. Epidemiological Characteristics of and Risk Factors for Breast Cancer in the World. Breast Cancer Targets Ther. 2019, 11, 151–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelwahab Yousef, A.J. Male Breast Cancer: Epidemiology and Risk Factors. Semin. Oncol. 2017, 44, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Nahid, A.-A.; Kong, Y. Involvement of Machine Learning for Breast Cancer Image Classification: A Survey. Comput. Math. Methods Med. 2017, 2017, 3781951. [Google Scholar] [CrossRef] [PubMed]
- Skandalakis, J.E. Embryology and Anatomy of the Breast. In Breast Augmentation; Shiffman, M., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 3–24. ISBN 978-3-540-78948-2. [Google Scholar]
- Bombonati, A.; Sgroi, D.C. The Molecular Pathology of Breast Cancer Progression. J. Pathol. 2011, 223, 308–318. [Google Scholar] [CrossRef] [Green Version]
- Gajdosova, V.; Lorencova, L.; Kasak, P.; Tkac, J. Electrochemical Nanobiosensors for Detection of Breast Cancer Biomarkers. Sensors 2020, 20, 4022. [Google Scholar] [CrossRef] [PubMed]
- Spanhol, F.A. Automatic Breast Cancer Classification from Histopathological Images: A Hybrid Approach. Ph.D. Thesis, Federal University of Parana, Curitiba, Brazil, 2018. [Google Scholar]
- Liu, Y.; Ren, L.; Cao, X.; Tong, Y. Breast Tumors Recognition Based on Edge Feature Extraction Using Support Vector Machine. Biomed. Signal Process. Control 2020, 58, 101825. [Google Scholar] [CrossRef]
- Danch-Wierzchowska, M.; Borys, D.; Bobek-Bilewicz, B.; Jarzab, M.; Swierniak, A. Simplification of Breast Deformation Modelling to Support Breast Cancer Treatment Planning. Biocybern. Biomed. Eng. 2016, 36, 531–536. [Google Scholar] [CrossRef]
- Mewada, H.K.; Patel, A.V.; Hassaballah, M.; Alkinani, M.H.; Mahant, K. Spectral–Spatial Features Integrated Convolution Neural Network for Breast Cancer Classification. Sensors 2020, 20, 4747. [Google Scholar] [CrossRef] [PubMed]
- Kiambe, K.; Kiambe, K. Breast Histopathological Image Feature Extraction with Convolutional Neural Networks for Classification. ICSES Trans. Image Process. Pattern Recognit. (ITIPPR) 2018, 4, 4–12. [Google Scholar] [CrossRef]
- Mathew, T.; Kini, J.R.; Rajan, J. Computational Methods for Automated Mitosis Detection in Histopathology Images: A Review. Biocybern. Biomed. Eng. 2021, 41, 64–82. [Google Scholar] [CrossRef]
- Zhu, C.; Song, F.; Wang, Y.; Dong, H.; Guo, Y.; Liu, J. Breast Cancer Histopathology Image Classification through Assembling Multiple Compact CNNs. BMC Med. Inform. Decis. Mak. 2019, 19, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valieris, R.; Amaro, L.; de Toledo Osório, C.A.B.; Bueno, A.P.; Mitrowsky, R.A.R.; Carraro, D.M.; Nunes, D.N.; Dias-Neto, E.; da Silva, I.T. Deep Learning Predicts Underlying Features on Pathology Images with Therapeutic Relevance for Breast and Gastric Cancer. Cancers 2020, 12, 3687. [Google Scholar] [CrossRef]
- Lagree, A.; Mohebpour, M.; Meti, N.; Saednia, K.; Lu, F.I.; Slodkowska, E.; Gandhi, S.; Rakovitch, E.; Shenfield, A.; Sadeghi-Naini, A.; et al. A Review and Comparison of Breast Tumor Cell Nuclei Segmentation Performances Using Deep Convolutional Neural Networks. Sci. Rep. 2021, 11, 8025. [Google Scholar] [CrossRef]
- Choudhary, T.; Mishra, V.; Goswami, A.; Sarangapani, J. A Transfer Learning with Structured Filter Pruning Approach for Improved Breast Cancer Classification on Point-of-Care Devices. Comput. Biol. Med. 2021, 134, 104432. [Google Scholar] [CrossRef]
- Kozegar, E.; Soryani, M.; Behnam, H.; Salamati, M.; Tan, T. Computer Aided Detection in Automated 3-D Breast Ultrasound Images: A Survey. Artif. Intell. Rev. 2020, 53, 1919–1941. [Google Scholar] [CrossRef] [Green Version]
- Murtaza, G.; Shuib, L.; Abdul Wahab, A.W.; Mujtaba, G.; Mujtaba, G.; Nweke, H.F.; Al-garadi, M.A.; Zulfiqar, F.; Raza, G.; Azmi, N.A. Deep Learning-Based Breast Cancer Classification through Medical Imaging Modalities: State of the Art and Research Challenges. Artif. Intell. Rev. 2020, 53, 1655–1720. [Google Scholar] [CrossRef]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-Classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef] [PubMed]
- Rezaeilouyeh, H.; Mollahosseini, A.; Mahoor, M.H. Microscopic Medical Image Classification Framework via Deep Learning and Shearlet Transform. J. Med. Imaging 2016, 3, 044501. [Google Scholar] [CrossRef]
- Pizarro, R.A.; Cheng, X.; Barnett, A.; Lemaitre, H.; Verchinski, B.A.; Goldman, A.L.; Xiao, E.; Luo, Q.; Berman, K.F.; Callicott, J.H.; et al. Automated Quality Assessment of Structural Magnetic Resonance Brain Images Based on a Supervised Machine Learning Algorithm. Front. Neuroinform. 2016, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Farjam, R.; Soltanian-Zadeh, H.; Zoroofi, R.A.; Jafari-Khouzani, K. Tree-Structured Grading of Pathological Images of Prostate. In Proceedings of the SPIE 5747, Medical Imaging 2005: Image Processing, San Diego, CA, USA, 29 April 2005; pp. 1–12. [Google Scholar] [CrossRef]
- Wang, P.; Hu, X.; Li, Y.; Liu, Q.; Zhu, X. Automatic Cell Nuclei Segmentation and Classification of Breast Cancer Histopathology Images. Signal Process. 2016, 122, 1–13. [Google Scholar] [CrossRef]
- Lopez Martinez, R.E.; Sierra, G. Research Trends in the International Literature on Natural Language Processing, 2000–2019—A Bibliometric Study. J. Scientometr. Res. 2020, 9, 310–318. [Google Scholar] [CrossRef]
- Ahmi, A.; Mohamad, R. Bibliometric Analysis of Global Scientific Literature on Web Accessibility. Int. J. Recent Technol. Eng. 2019, 7, 250–258. [Google Scholar]
- Marczewska, M.; Kostrzewski, M. Sustainable Business Models: A Bibliometric Performance Analysis. Energies 2020, 13, 6062. [Google Scholar] [CrossRef]
- de las Heras-Rosas, C.; Herrera, J.; Rodríguez-Fernández, M. Organisational Commitment in Healthcare Systems: A Bibliometric Analysis. Int. J. Environ. Res. Public Health 2021, 18, 2271. [Google Scholar] [CrossRef]
- Ozen Çınar, İ. Bibliometric Analysis of Breast Cancer Research in the Period 2009–2018. Int. J. Nurs. Pract. 2020, 26, e12845. [Google Scholar] [CrossRef] [PubMed]
- Salod, Z.; Singh, Y. A Five-Year (2015 to 2019) Analysis of Studies Focused on Breast Cancer Prediction Using Machine Learning: A Systematic Review and Bibliometric Analysis. J. Public Health Res. 2020, 9, 65–75. [Google Scholar] [CrossRef]
- Joshi, S.A.; Bongale, A.M.; Bongale, A. Breast Cancer Detection from Histopathology Images Using Machine Learning Techniques: A Bibliometric Analysis. Libr. Philos. Pract. 2021, 5376, 1–29. [Google Scholar]
- Erickson, B.J.; Korfiatis, P.; Akkus, Z.; Kline, T.L. Machine Learning for Medical Imaging. Radiographics 2017, 37, 505–515. [Google Scholar] [CrossRef]
- Bengio, Y. Learning Deep Architectures for AI. In Foundations and Trends® in Machine Learning; University of California: Berkeley, CA, USA, 2009; Volume 2, pp. 1–127. [Google Scholar] [CrossRef]
- Li, X.; Shen, X.; Zhou, Y.; Wang, X.; Li, T.-Q. Classification of Breast Cancer Histopathological Images Using Interleaved DenseNet with SENet (IDSNet). PLoS ONE 2020, 15, e0232127. [Google Scholar] [CrossRef] [PubMed]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. BACH: Grand Challenge on Breast Cancer Histology Images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- Asri, H.; Mousannif, H.; Moatassime, H.A.; Noel, T. Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis. Procedia Comput. Sci. 2016, 83, 1064–1069. [Google Scholar] [CrossRef] [Green Version]
- Bharat, A.; Pooja, N.; Reddy, R.A. Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis. In Proceedings of the 2018 3rd International Conference on Circuits, Control, Communication and Computing (I4C), Bangalore, India, 3–5 October 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, Q.; Liang, W.; Zou, X. An Efficient Feature Selection Strategy Based on Multiple Support Vector Machine Technology with Gene Expression Data. Biomed Res. Int. 2018, 2018, 7538204. [Google Scholar] [CrossRef] [PubMed]
- Kharya, S.; Agrawal, S.; Soni, S. Naive Bayes Classifiers: A Probabilistic Detection Model for Breast Cancer. Int. J. Comput. Appl. 2014, 92, 26–31. [Google Scholar] [CrossRef]
- Nahar, J.; Chen, Y.P.P.; Ali, S. Kernel-Based Naive Bayes Classifier for Breast Cancer Prediction. J. Biol. Syst. 2007, 15, 17–25. [Google Scholar] [CrossRef]
- Rashmi, G.D.; Lekha, A.; Bawane, N. Analysis of Efficiency of Classification and Prediction Algorithms (Naïve Bayes) for Breast Cancer Dataset. In Proceedings of the 2015 International Conference on Emerging Research in Electronics, Computer Science and Technology (ICERECT), Mandya, India, 17–19 December 2015; pp. 108–113. [Google Scholar]
- Octaviani, T.L.; Rustam, Z. Random Forest for Breast Cancer Prediction. In Proceedings of the AIP Conference Proceedings, Depok, Indonesia, 30–31 October 2018; pp. 1–7. [Google Scholar] [CrossRef]
- Elgedawy, M.N. Prediction of Breast Cancer Using Random Forest, Support Vector Machines and Naïve Bayes. Int. J. Eng. Comput. Sci. 2017, 6, 19884–19889. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Wang, H.; Cruz-Roa, A.; Basavanhally, A.; Gilmore, H.; Shih, N.; Feldman, M.; Tomaszewski, J.; Gonzalez, F.; Madabhushi, A. Mitosis Detection in Breast Cancer Pathology Images by Combining Handcrafted and Convolutional Neural Network Features. J. Med. Imaging 2014, 1, 034003. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Daud, S.M.; Abas, H.; Ahmad, N.A.; Maarop, N. Breast Cancer Classification Using Deep Learning Approaches and Histopathology Image: A Comparison Study. IEEE Access 2020, 8, 187531–187552. [Google Scholar] [CrossRef]
- Fujita, H. AI-Based Computer-Aided Diagnosis (AI-CAD): The Latest Review to Read First. Radiol. Phys. Technol. 2020, 13, 6–19. [Google Scholar] [CrossRef]
- Lin, C.J.; Jeng, S.Y. Optimization of Deep Learning Network Parameters Using Uniform Experimental Design for Breast Cancer Histopathological Image Classification. Diagnostics 2020, 10, 662. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Mishra, C.; Gupta, D.L. Deep Machine Learning and Neural Networks: An Overview. IAES Int. J. Artif. Intell. 2017, 6, 66–73. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Nguyen, T.T.; Nguyen, N.C.; Le, T.T. Multiclass Breast Cancer Classification Using Convolutional Neural Network. In Proceedings of the 2019 International Symposium on Electrical and Electronics Engineering (ISEE), Ho Chi Minh City, Vietnam, 10–12 October 2019; pp. 130–134. [Google Scholar] [CrossRef]
- Bengio, Y.; Lee, H. Editorial Introduction to the Neural Networks Special Issue on Deep Learning of Representations. Neural Netw. 2015, 64, 1–3. [Google Scholar] [CrossRef]
- Hirra, I.; Ahmad, M.; Hussain, A.; Ashraf, M.U.; Saeed, I.A.; Qadri, S.F.; Alghamdi, A.M.; Alfakeeh, A.S. Breast Cancer Classification from Histopathological Images Using Patch-Based Deep Learning Modeling. IEEE Access 2021, 9, 24273–24287. [Google Scholar] [CrossRef]
- Zuluaga-Gomez, J.; Al Masry, Z.; Benaggoune, K.; Meraghni, S.; Zerhouni, N. A CNN-Based Methodology for Breast Cancer Diagnosis Using Thermal Images. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2021, 9, 131–145. [Google Scholar] [CrossRef]
- Hameed, Z.; Zahia, S.; Garcia-Zapirain, B.; Aguirre, J.J.; Vanegas, A.M. Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models. Sensors 2020, 20, 4373. [Google Scholar] [CrossRef] [PubMed]
- Pavithra, P.; Ravichandran, K.S.; Sekar, K.R.; Manikandan, R. The Effect of Thermography on Breast Cancer Detection—A Survey. Syst. Rev. Pharm. 2018, 9, 10–16. [Google Scholar] [CrossRef]
- Alom, M.Z.; Yakopcic, C.; Nasrin, M.S.; Taha, T.M.; Asari, V.K. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. J. Digit. Imaging 2019, 32, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Železnik, D.; Blažun Vošner, H.; Kokol, P. A Bibliometric Analysis of the Journal of Advanced Nursing, 1976–2015. J. Adv. Nurs. 2017, 73, 2407–2419. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Tang, M.; Luo, L.; Li, C.; Chiclana, F.; Zeng, X.J. A Bibliometric Analysis and Visualization of Medical Big Data Research. Sustainability 2018, 10, 166. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Hao, Z.; Zhao, S.; Gong, J.; Yang, F. Artificial Intelligence in Health Care: Bibliometric Analysis. J. Med. Internet Res. 2020, 22, e18228. [Google Scholar] [CrossRef]
- Bhattacharya, S. Some Salient Aspects of Machine Learning Research: A Bibliometric Analysis. J. Scientometr. Res. 2019, 8, 85–92. [Google Scholar] [CrossRef]
- Aria, M.; Cuccurullo, C. Bibliometrix: An R-Tool for Comprehensive Science Mapping Analysis. J. Informetr. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- van Eck, N.J.; Waltman, L. Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef] [Green Version]
- Wahid, R.; Ahmi, A.; Alam, A.S.A.F. Growth and Collaboration in Massive Open Online Courses: A Bibliometric Analysis. Int. Rev. Res. Open Distance Learn. 2020, 21, 292–322. [Google Scholar] [CrossRef]
- Baas, J.; Schotten, M.; Plume, A.; Côté, G.; Karimi, R. Scopus as a Curated, High-Quality Bibliometric Data Source for Academic Research in Quantitative Science Studies. Quant. Sci. Stud. 2020, 1, 377–386. [Google Scholar] [CrossRef]
- Tober, M. PubMed, ScienceDirect, Scopus or Google Scholar—Which Is the Best Search Engine for an Effective Literature Research in Laser Medicine? Med. Laser Appl. 2011, 26, 139–144. [Google Scholar] [CrossRef]
- Al-antari, M.A.; Han, S.-M.; Kim, T.-S. Evaluation of Deep Learning Detection and Classification towards Computer-Aided Diagnosis of Breast Lesions in Digital X-Ray Mammograms. Comput. Methods Programs Biomed. 2020, 196, 105584. [Google Scholar] [CrossRef] [PubMed]
- Swiderski, B.; Kurek, J.; Osowski, S.; Kruk, M.; Barhoumi, W. Deep Learning and Non-Negative Matrix Factorization in Recognition of Mammograms. In Proceedings of the Eighth International Conference on Graphic and Image Processing (ICGIP 2016), Tokyo, Japan, 29–31 October 2016; pp. 1–7. [Google Scholar] [CrossRef]
- Grover, S.; Dalton, N. Abstract to Publication Rate: Do All the Papers Presented in Conferences See the Light of Being a Full Publication? Indian J. Psychiatry 2020, 62, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ilan, J. Web of Science with the Conference Proceedings Citation Indexes: The Case of Computer Science. Scientometrics 2010, 83, 809–824. [Google Scholar] [CrossRef]
- Purnell, P.J. Conference Proceedings Publications in Bibliographic Databases: A Case Study of Countries in Southeast Asia. Scientometrics 2021, 126, 355–387. [Google Scholar] [CrossRef]
- Janowczyk, A.; Madabhushi, A. Deep Learning for Digital Pathology Image Analysis: A Comprehensive Tutorial with Selected Use Cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H.; et al. Development and Validation of a Deep Learning Algorithm for Improving Gleason Scoring of Prostate Cancer. NPJ Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef] [Green Version]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA J. Am. Med. Assoc. 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Diagnosis and Precision Oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Zakaria, R.; Ahmi, A.; Ahmad, A.H.; Othman, Z.; Azman, K.F.; Ab Aziz, C.B.; Ismail, C.A.N.; Shafin, N. Visualising and Mapping a Decade of Literature on Honey Research: A Bibliometric Analysis from 2011 to 2020. J. Apic. Res. 2021, 60, 359–368. [Google Scholar] [CrossRef]
- Bongaarts, J. United Nations Department of Economic and Social Affairs, Population DivisionWorld Family Planning 2020: Highlights, United Nations Publications, 2020. 46 P. Popul. Dev. Rev. 2020, 46, 857–858. [Google Scholar] [CrossRef]
- Peters, H.P.F.; van Raan, A.F.J. Co-Word-Based Science Maps of Chemical Engineering. Part II: Representations by Combined Clustering and Multidimensional Scaling. Res. Policy 1993, 22, 47–71. [Google Scholar] [CrossRef]
- Waltman, L.; van Eck, N.J.; Noyons, E.C.M. A Unified Approach to Mapping and Clustering of Bibliometric Networks. J. Informetr. 2010, 4, 629–635. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Singh, S.K.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.K.; Chauhan, H.; Shrivastava, S.; Singh, R.K. Deep Feature Learning for Histopathological Image Classification of Canine Mammary Tumors and Human Breast Cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Satapathy, S.C.; Guttery, D.S.; Górriz, J.M.; Wang, S.H. Improved Breast Cancer Classification Through Combining Graph Convolutional Network and Convolutional Neural Network. Inf. Process. Manag. 2021, 58, 102439. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, N.; Das, I.; Maulik, U. Understanding Deep Learning Techniques for Image Segmentation. ACM Comput. Surv. 2019, 52, 1–35. [Google Scholar] [CrossRef] [Green Version]
- Sudharshan, P.J.; Petitjean, C.; Spanhol, F.; Oliveira, L.E.; Heutte, L.; Honeine, P. Multiple Instance Learning for Histopathological Breast Cancer Image Classification. Expert Syst. Appl. 2019, 117, 103–111. [Google Scholar] [CrossRef]
- Xu, J.; Xiang, L.; Liu, Q.; Gilmore, H.; Wu, J.; Tang, J.; Madabhushi, A. Stacked Sparse Autoencoder (SSAE) for Nuclei Detection on Breast Cancer Histopathology Images. IEEE Trans. Med. Imaging 2016, 35, 119–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Deng, Z. Bibliometric Analysis of the Application of Convolutional Neural Network in Computer Vision. IEEE Access 2020, 8, 155417–155428. [Google Scholar] [CrossRef]
- e Fonseca, B.D.P.F.; Sampaio, R.B.; de Fonseca, M.V.; Zicker, F. Co-Authorship Network Analysis in Health Research: Method and Potential Use. Health Res. Policy Syst. 2016, 14, 34. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Wang, J.; Li, Y.; Li, P.; Li, L.; Jiang, M. Automatic Classification of Breast Cancer Histopathological Images Based on Deep Feature Fusion and Enhanced Routing. Biomed. Signal Process. Control 2021, 65, 102341. [Google Scholar] [CrossRef]
- Elmannai, H.; Hamdi, M.; AlGarni, A. Deep Learning Models Combining for Breast Cancer Histopathology Image Classification. Int. J. Comput. Intell. Syst. 2021, 14, 102341. [Google Scholar] [CrossRef]
- Chen, K.; Yao, Q.; Sun, J.; He, Z.; Yao, L.; Liu, Z. International Publication Trends and Collaboration Performance of China in Healthcare Science and Services Research. Isr. J. Health Policy Res. 2016, 5, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, S.; Ur Rehman, S.; Iqbal, A.; Farooq, R.K.; Shahid, A.; Ullah, M.I. Breast Cancer Research in Pakistan: A Bibliometric Analysis. SAGE Open 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Rangarajan, B.; Shet, T.; Wadasadawala, T.; Nair, N.S.; Sairam, R.M.; Hingmire, S.S.; Bajpai, J. Breast Cancer: An Overview of Published Indian Data. South Asian J. Cancer 2016, 5, 86–92. [Google Scholar] [CrossRef]
- Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdelalim, A.; Abdoli, A.; Abdollahpour, I.; Abdulle, A.S.M.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A.; González, F. High-Throughput Adaptive Sampling for Whole-Slide Histopathology Image Analysis (HASHI) via Convolutional Neural Networks: Application to Invasive Breast Cancer Detection. PLoS ONE 2018, 13, e0196828. [Google Scholar] [CrossRef]
- Lu, C.; Xu, H.; Xu, J.; Gilmore, H.; Mandal, M.; Madabhushi, A. Multi-Pass Adaptive Voting for Nuclei Detection in Histopathological Images. Sci. Rep. 2016, 6, 33985. [Google Scholar] [CrossRef]
- Xu, J.; Xiang, L.; Wang, G.; Ganesan, S.; Feldman, M.; Shih, N.N.; Gilmore, H.; Madabhushi, A. Sparse Non-Negative Matrix Factorization (SNMF) Based Color Unmixing for Breast Histopathological Image Analysis. Comput. Med. Imaging Graph. 2015, 46, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samb, B.; Desai, N.; Nishtar, S.; Mendis, S.; Bekedam, H.; Wright, A.; Hsu, J.; Martiniuk, A.; Celletti, F.; Patel, K.; et al. Prevention and Management of Chronic Disease: A Litmus Test for Health-Systems Strengthening in Low-Income and Middle-Income Countries. Lancet 2010, 376, 1785–1797. [Google Scholar] [CrossRef]
- Ghosh, P.; Azam, S.; Hasib, K.M.; Karim, A.; Jonkman, M.; Anwar, A. A Performance Based Study on Deep Learning Algorithms in the Effective Prediction of Breast Cancer. In Proceedings of the International Joint Conference on Neural Networks, Shenzhen, China, 18–22 July 2021; pp. 1–8. [Google Scholar] [CrossRef]
- Mahmood, T.; Li, J.; Pei, Y.; Akhtar, F.; Jia, Y.; Khand, Z.H. Breast Mass Detection and Classification Using Deep Convolutional Neural Networks for Radiologist Diagnosis Assistance. In Proceedings of the 2021 IEEE 45th Annual Computers, Software, and Applications Conference, COMPSAC 2021, Madrid, Spain, 12–16 July 2021; pp. 1918–1923. [Google Scholar] [CrossRef]
- Salama, W.M.; Elbagoury, A.M.; Aly, M.H. Novel Breast Cancer Classification Framework Based on Deep Learning. IET Image Process. 2020, 14, 3254–3259. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.C.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and Reproducible Invasive Breast Cancer Detection in Whole-Slide Images: A Deep Learning Approach for Quantifying Tumor Extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Wei, Q.; Dietrich, C.F. Lymph Node Metastasis Prediction from Primary Breast. Radiology 2019, 294, 19–24. [Google Scholar] [CrossRef]
- Bianconi, F.; Kather, J.N.; Reyes-Aldasoro, C.C. Evaluation of Colour Pre-Processing on Patch-Based Classification of H&E-Stained Images. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Cham, Switzerland, 2019; pp. 56–64. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yan, N.; Yang, H.; Zhu, L.; Zheng, Z.; Yang, X.; Zhang, X. A Data Augmentation Method for Deep Learning Based on Multi-Degree of Freedom (Dof) Automatic Image Acquisition. Appl. Sci. 2020, 10, 7755. [Google Scholar] [CrossRef]
- Zhou, F.; Yang, S.; Fujita, H.; Chen, D.; Wen, C. Deep Learning Fault Diagnosis Method Based on Global Optimization GAN for Unbalanced Data. Knowl.-Based Syst. 2020, 187, 104837. [Google Scholar] [CrossRef]
- Stephan, P.; Veugelers, R.; Wang, J. Reviewers are Blinkered by Bibliometrics. Nature 2017, 544, 411–412. [Google Scholar] [CrossRef] [PubMed]

| Magnification | 40× | 100× | 200× | 400× |
|---|---|---|---|---|
| Benign | 652 | 644 | 623 | 588 |
| Malignant | 1370 | 1437 | 1390 | 1232 |
| Document Type | Frequency | Percentage (n = 373) |
|---|---|---|
| Article | 181 | 48.53 |
| Conference paper | 155 | 41.55 |
| Conference review | 15 | 4.02 |
| Book chapter | 7 | 1.87 |
| Erratum | 1 | 0.27 |
| Note | 1 | 0.27 |
| Review | 12 | 3.22 |
| Short Survey | 1 | 0.27 |
| Total | 373 | 100.00 |
| Country | TLS 1 | Links | Documents | Citations | Cluster |
|---|---|---|---|---|---|
| United States | 51 | 21 | 68 | 2235 | 6 |
| China | 39 | 17 | 71 | 1586 | 9 |
| India | 26 | 16 | 80 | 674 | 1 |
| South Korea | 17 | 11 | 12 | 444 | 3 |
| United Kingdom | 17 | 10 | 20 | 190 | 2 |
| Germany | 14 | 10 | 14 | 239 | 3 |
| Sweden | 14 | 9 | 10 | 192 | 5 |
| Pakistan | 13 | 6 | 13 | 172 | 7 |
| Portugal | 12 | 10 | 5 | 157 | 3 |
| Australia | 10 | 7 | 13 | 201 | 2 |
| Author | TLS 1 | Links | Documents | Citations | Affiliation | APY 2 |
|---|---|---|---|---|---|---|
| Li Y. | 18 | 10 | 7 | 93 | Chongqing University, China | 2018 |
| Madabhushi A. | 15 | 5 | 9 | 912 | Case Western Reserve University, United States | 2017 |
| Li X. | 15 | 10 | 7 | 56 | Chongqing University of Posts and Telecommunications, China | 2020 |
| Wang J. | 14 | 8 | 4 | 31 | Chongqing University, China | 2020 |
| Gilmore H. | 13 | 5 | 6 | 891 | Case Western Reserve University, United States | 2017 |
| Zhang Y. | 13 | 8 | 6 | 38 | Nanjing University, China | 2019 |
| Li. L | 13 | 7 | 4 | 36 | Chongqing University, China | 2020 |
| Xu J. | 11 | 7 | 4 | 485 | Nanjing University, China | 2018 |
| Zhang H. | 10 | 9 | 7 | 130 | East China Jiaotong University, China | 2019 |
| Li Z. | 10 | 6 | 4 | 22 | Chongqing University of Posts and Telecommunications, China | 2020 |
| Affiliation | Research Focus | Document |
|---|---|---|
| Case Western Reserve University | Convolutional neural network, digital pathology, image classification | 9 |
| Indian Institute of Technology Kharagpur | Features, convolutional neural network, whole slide images | 7 |
| Shenzhen University | Image classification, convolutional neural network | 6 |
| Radboud University Medical Center | Deep learning, whole slide images | 6 |
| University of Toronto | Convolutional neural network, review analysis | 6 |
| Karolinska Institute | Convolutional neural network, classification, deep learning | 5 |
| Xiamen University | Segmentation, detection, convolutional neural network | 5 |
| Sunnybrook Health University | Deep learning-based, convolutional neural network, feature extraction | 5 |
| Southern Medical University | Deep learning, cancer staging, classification | 4 |
| Chongqing University | Features, convolutional neural network, image classification | 3 |
| Journal | TLS 1 | Links | Documents | Cit 2 |
|---|---|---|---|---|
| IEEE Access | 26 | 10 | 10 | 48 |
| IEEE Transactions on Medical Imaging | 24 | 13 | 5 | 703 |
| Scientific Reports | 21 | 16 | 10 | 451 |
| Computerized Medical Imaging and Graphics | 14 | 9 | 4 | 114 |
| Medical Image Analysis | 14 | 10 | 5 | 177 |
| Biocybernetics and Biomedical Engineering | 12 | 8 | 5 | 41 |
| Lecture Notes in Computer Science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) | 12 | 8 | 36 | 433 |
| Expert Systems with Applications | 11 | 8 | 3 | 117 |
| International Journal of Advanced Computer Science and Applications | 11 | 7 | 4 | 66 |
| Frontiers in Genetics | 10 | 6 | 3 | 71 |
| Biomedical Signal Processing and Control | 9 | 6 | 5 | 25 |
| International Journal of Imaging Systems and Technology | 9 | 5 | 6 | 27 |
| Journal of Medical Imaging | 8 | 6 | 3 | 249 |
| Communications in Computer and Information Science | 7 | 4 | 10 | 10 |
| Advances in Intelligent Systems and Computing | 6 | 5 | 8 | 16 |
| IEEE Journal of Biomedical and Health Informatics | 6 | 4 | 6 | 101 |
| Proceedings of the International Joint Conference on Neural Networks | 6 | 5 | 4 | 417 |
| Proceedings—International Symposium on Biomedical Imaging | 4 | 3 | 13 | 262 |
| Lecture Notes in Electrical Engineering | 3 | 2 | 3 | 0 |
| Cancers | 1 | 1 | 6 | 9 |
| References | Journal | Model/Method | IF 1 | H-Index | Cit 2 | Year |
|---|---|---|---|---|---|---|
| Cruz-Roa et al. [106] | Scientific Reports | CNN/ConvNet | 4.380 | 213 | 292 | 2017 |
| Wang H. et al. [51] | Journal of Medical Imaging | CNN and handcrafted features | 3.610 | 29 | 272 | 2014 |
| Han Z. et al. [24] | Scientific Reports | Structured based deep CNN | 4.380 | 213 | 210 | 2017 |
| Ghosh S. et al. [88] | ACM Computing Surveys | Deep learning, CNN | 10.282 | 163 | 126 | 2019 |
| Alom M. Z. et al. [63] | Journal of Digital Imaging | Deep CNN, Inception-v4, ResNet, Recurrent CNN | 4.056 | 58 | 123 | 2019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khairi, S.S.M.; Bakar, M.A.A.; Alias, M.A.; Bakar, S.A.; Liong, C.-Y.; Rosli, N.; Farid, M. Deep Learning on Histopathology Images for Breast Cancer Classification: A Bibliometric Analysis. Healthcare 2022, 10, 10. https://doi.org/10.3390/healthcare10010010
Khairi SSM, Bakar MAA, Alias MA, Bakar SA, Liong C-Y, Rosli N, Farid M. Deep Learning on Histopathology Images for Breast Cancer Classification: A Bibliometric Analysis. Healthcare. 2022; 10(1):10. https://doi.org/10.3390/healthcare10010010
Chicago/Turabian StyleKhairi, Siti Shaliza Mohd, Mohd Aftar Abu Bakar, Mohd Almie Alias, Sakhinah Abu Bakar, Choong-Yeun Liong, Nurwahyuna Rosli, and Mohsen Farid. 2022. "Deep Learning on Histopathology Images for Breast Cancer Classification: A Bibliometric Analysis" Healthcare 10, no. 1: 10. https://doi.org/10.3390/healthcare10010010
APA StyleKhairi, S. S. M., Bakar, M. A. A., Alias, M. A., Bakar, S. A., Liong, C.-Y., Rosli, N., & Farid, M. (2022). Deep Learning on Histopathology Images for Breast Cancer Classification: A Bibliometric Analysis. Healthcare, 10(1), 10. https://doi.org/10.3390/healthcare10010010






