Assessment of the Use of Patient Vital Sign Data for Preventing Misidentification and Medical Errors
Abstract
1. Introduction
2. Background
2.1. Patient Misidentification and Its Consequences
2.2. Artificial Intelligence, Expert Systems, and Neural Networks
2.3. Gradient Descent Trained Expert Systems
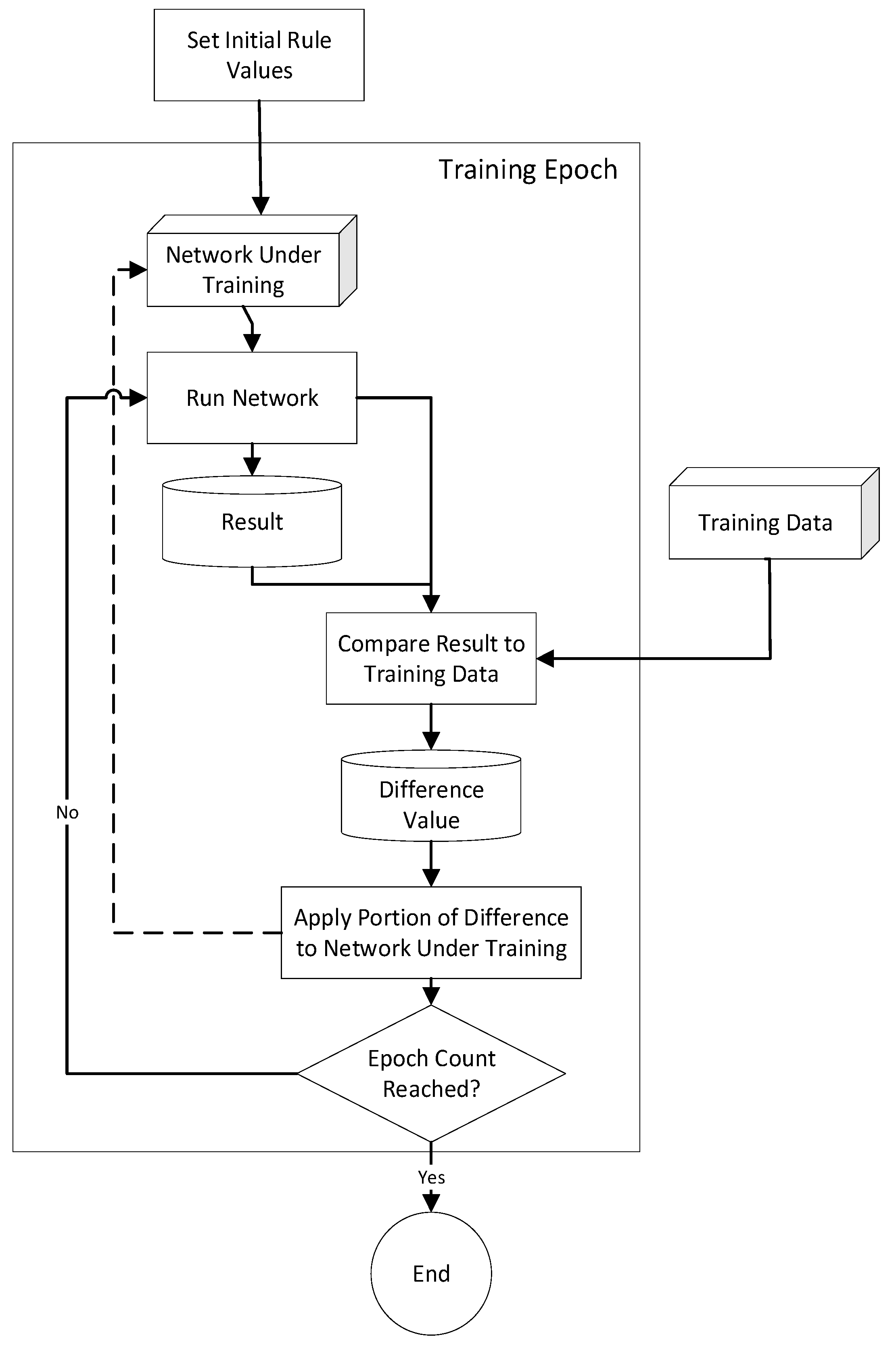
3. System Design
3.1. System Use Scenarios
3.2. System Operations
3.3. Limitations
4. Data and Analysis
4.1. First Network Design
4.2. Second Network Design
4.3. Third Network Design
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
SF:{24da3290-e934-4a9c-84e9-6a0d856e5073} = 000.393
would be presented as:
SF:{Heart Rate} = 000.393
SF:{Blood Pressure} = 000.459
SF:{Heart Rate} = 000.393
SF:{Respiratory Rate} = 000.222
SF:{End-tidal Carbon Dioxide} = 000.860
TR:{Blood Pressure} = 000.459>0001:0.05>{Output} = 000.500
PR:{Blood Pressure} = 000.500>>{Output}
PR:{Blood Pressure} = 000.443>>{Output}
SF:{Blood Pressure} = 000.500
SF:{Heart Rate} = 000.500
SF:{Respiratory Rate} = 000.500
SF:{End-tidal Carbon Dioxide} = 000.500
PR:{Blood Pressure} = 000.500>>{Output}
SF:{Blood Pressure} = 000.446
SF:{Heart Rate} = 000.399
SF:{Respiratory Rate} = 000.302
SF:{End-tidal Carbon Dioxide} = 000.812
PR:{Blood Pressure} = 000.446>>{Output}
Appendix B
Appendix B.1
Appendix B.1.1. Trials 1–10
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.512 | 0.529 | 0.037 | 0.053 | 0.023 |
| 0.492 | 0.523 | 0.044 | 0.064 | 0.032 |
| 0.654 | 0.538 | 0.129 | 0.144 | 0.110 |
| 0.476 | 0.538 | 0.064 | 0.083 | 0.053 |
| 0.514 | 0.527 | 0.034 | 0.052 | 0.024 |
| 0.533 | 0.535 | 0.045 | 0.061 | 0.035 |
| 0.464 | 0.523 | 0.059 | 0.077 | 0.044 |
| 0.532 | 0.538 | 0.056 | 0.073 | 0.042 |
| 0.517 | 0.538 | 0.050 | 0.067 | 0.042 |
| 0.684 | 0.538 | 0.153 | 0.168 | 0.134 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.501 | 0.548 | 0.054 | 0.076 | 0.038 |
| 0.492 | 0.523 | 0.044 | 0.064 | 0.032 |
| 0.723 | 0.584 | 0.153 | 0.190 | 0.125 |
| 0.706 | 0.584 | 0.142 | 0.178 | 0.118 |
| 0.502 | 0.553 | 0.055 | 0.076 | 0.039 |
| 0.500 | 0.572 | 0.082 | 0.114 | 0.067 |
| 0.464 | 0.523 | 0.059 | 0.077 | 0.044 |
| 0.511 | 0.584 | 0.099 | 0.137 | 0.079 |
| 0.529 | 0.568 | 0.090 | 0.123 | 0.072 |
| 0.889 | 0.584 | 0.305 | 0.341 | 0.268 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.529 | 0.039 | 0.055 | 0.025 |
| 0.500 | 0.523 | 0.043 | 0.064 | 0.030 |
| 0.500 | 0.538 | 0.052 | 0.068 | 0.045 |
| 0.500 | 0.538 | 0.052 | 0.068 | 0.045 |
| 0.500 | 0.527 | 0.039 | 0.055 | 0.024 |
| 0.500 | 0.535 | 0.047 | 0.062 | 0.036 |
| 0.500 | 0.523 | 0.043 | 0.064 | 0.030 |
| 0.500 | 0.538 | 0.052 | 0.068 | 0.045 |
| 0.500 | 0.538 | 0.052 | 0.068 | 0.045 |
| 0.500 | 0.538 | 0.052 | 0.068 | 0.045 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.498 | 0.529 | 0.040 | 0.056 | 0.027 |
| 0.492 | 0.523 | 0.044 | 0.064 | 0.032 |
| 0.708 | 0.538 | 0.172 | 0.187 | 0.153 |
| 0.701 | 0.538 | 0.167 | 0.182 | 0.148 |
| 0.494 | 0.527 | 0.041 | 0.057 | 0.027 |
| 0.502 | 0.535 | 0.046 | 0.061 | 0.036 |
| 0.464 | 0.523 | 0.059 | 0.077 | 0.044 |
| 0.526 | 0.538 | 0.053 | 0.071 | 0.041 |
| 0.546 | 0.538 | 0.061 | 0.079 | 0.045 |
| 0.815 | 0.538 | 0.277 | 0.292 | 0.259 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.500 | 0.012 | 0.007 | 0.020 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.717 | 0.064 | 0.060 | 0.067 |
| 0.486 | 0.010 | 0.004 | 0.026 |
| 0.482 | 0.033 | 0.011 | 0.049 |
| 0.536 | 0.004 | 0.011 | 0.019 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.508 | 0.024 | 0.015 | 0.041 |
| 0.515 | 0.002 | 0.004 | 0.011 |
| 0.584 | 0.100 | 0.056 | 0.126 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.493 | 0.007 | 0.002 | 0.016 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.792 | 0.069 | 0.059 | 0.080 |
| 0.716 | 0.010 | 0.004 | 0.026 |
| 0.487 | 0.015 | 0.016 | 0.051 |
| 0.524 | 0.024 | 0.004 | 0.044 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.485 | 0.026 | 0.002 | 0.049 |
| 0.516 | 0.013 | 0.009 | 0.024 |
| 0.709 | 0.180 | 0.130 | 0.302 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.500 | 0.000 | 0.005 | 0.007 |
| 0.527 | 0.027 | 0.019 | 0.056 |
| 0.717 | 0.217 | 0.213 | 0.221 |
| 0.486 | 0.014 | 0.002 | 0.028 |
| 0.482 | 0.018 | 0.003 | 0.034 |
| 0.536 | 0.036 | 0.022 | 0.052 |
| 0.474 | 0.026 | 0.017 | 0.043 |
| 0.508 | 0.008 | 0.010 | 0.017 |
| 0.515 | 0.015 | 0.005 | 0.021 |
| 0.584 | 0.084 | 0.058 | 0.128 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.500 | 0.002 | 0.005 | 0.007 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.717 | 0.010 | 0.005 | 0.013 |
| 0.486 | 0.215 | 0.199 | 0.229 |
| 0.482 | 0.012 | 0.009 | 0.029 |
| 0.536 | 0.035 | 0.020 | 0.050 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.508 | 0.018 | 0.010 | 0.036 |
| 0.515 | 0.032 | 0.026 | 0.041 |
| 0.584 | 0.231 | 0.187 | 0.257 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 33.8% | 37.0% | 31.2% |
| 78.1% | 200.6% | 41.2% |
| 49.5% | 60.8% | 41.4% |
| 15.5% | 48.8% | 4.8% |
| 94.5% | 93.6% | 45.5% |
| 8.2% | 55.3% | 17.8% |
| 17.6% | 43.6% | 9.1% |
| 43.0% | 56.4% | 35.5% |
| 4.0% | 16.5% | 9.6% |
| 65.2% | 75.2% | 41.4% |
| 40.9% | 68.78% | 27.75% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 13.7% | 21.4% | 5.5% |
| 78.1% | 200.6% | 41.2% |
| 45.2% | 63.7% | 31.1% |
| 7.1% | 22.0% | 2.3% |
| 27.8% | 67.1% | 41.7% |
| 29.8% | 65.2% | 3.7% |
| 17.6% | 43.6% | 9.1% |
| 26.6% | 35.9% | 3.2% |
| 14.8% | 19.1% | 11.7% |
| 59.1% | 88.4% | 48.6% |
| 32.0% | 62.7% | 19.8% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 0.11% | 20.70% | 12.98% |
| 61.26% | 187.18% | 28.79% |
| 420.59% | 495.51% | 313.93% |
| 27.12% | 41.27% | 4.49% |
| 47.57% | 62.57% | 13.13% |
| 77.64% | 144.56% | 35.23% |
| 58.96% | 66.93% | 57.34% |
| 14.67% | 37.08% | 14.00% |
| 28.09% | 46.07% | 8.11% |
| 162.56% | 287.64% | 84.75% |
| 89.86% | 138.95% | 57.28% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 5.63% | 28.17% | 8.89% |
| 78.14% | 200.63% | 41.25% |
| 5.61% | 8.48% | 2.94% |
| 129.04% | 134.32% | 126.10% |
| 30.33% | 49.66% | 33.94% |
| 75.23% | 140.07% | 32.97% |
| 17.65% | 43.63% | 9.12% |
| 34.52% | 50.35% | 23.09% |
| 51.31% | 56.48% | 51.59% |
| 83.27% | 88.03% | 72.15% |
| 51.07% | 79.98% | 40.20% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.037 | 0.012 | N | N | Y | Y | Y | 0 | 5 | 7 | 8 |
| 0.044 | 0.035 | N | N | N | Y | Y | 1 | 5 | 7 | 8 |
| 0.129 | 0.064 | N | N | N | N | Y | 0 | 0 | 0 | 2 |
| 0.064 | 0.010 | N | N | Y | Y | Y | 1 | 2 | 6 | 6 |
| 0.034 | 0.033 | N | N | N | Y | Y | 3 | 5 | 7 | 8 |
| 0.045 | 0.004 | Y | Y | Y | Y | Y | 0 | 2 | 6 | 8 |
| 0.059 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.056 | 0.024 | N | N | Y | Y | Y | 0 | 2 | 5 | 8 |
| 0.050 | 0.002 | Y | Y | Y | Y | Y | 2 | 3 | 5 | 8 |
| 0.153 | 0.100 | N | N | N | N | Y | 0 | 0 | 1 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.054 | 0.007 | Y | Y | Y | Y | Y | 1 | 3 | 5 | 7 |
| 0.044 | 0.035 | N | N | N | Y | Y | 1 | 5 | 7 | 8 |
| 0.153 | 0.069 | N | N | N | N | Y | 1 | 2 | 2 | 3 |
| 0.142 | 0.010 | N | Y | Y | Y | Y | 1 | 1 | 1 | 3 |
| 0.055 | 0.015 | N | N | Y | Y | Y | 1 | 3 | 5 | 6 |
| 0.082 | 0.024 | N | N | Y | Y | Y | 3 | 3 | 5 | 5 |
| 0.059 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.099 | 0.026 | N | N | N | Y | Y | 1 | 3 | 4 | 5 |
| 0.090 | 0.013 | Y | N | Y | Y | Y | 0 | 1 | 5 | 6 |
| 0.305 | 0.180 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.039 | 0.000 | Y | Y | Y | Y | Y | 1 | 4 | 7 | 7 |
| 0.043 | 0.027 | N | N | N | Y | Y | 1 | 2 | 7 | 8 |
| 0.052 | 0.217 | N | N | N | N | N | 2 | 6 | 7 | 8 |
| 0.052 | 0.014 | N | N | Y | Y | Y | 2 | 5 | 6 | 7 |
| 0.039 | 0.018 | N | N | Y | Y | Y | 1 | 6 | 7 | 7 |
| 0.047 | 0.036 | N | N | N | Y | Y | 1 | 5 | 6 | 8 |
| 0.043 | 0.026 | N | N | N | Y | Y | 1 | 2 | 7 | 8 |
| 0.052 | 0.008 | N | Y | Y | Y | Y | 1 | 5 | 6 | 7 |
| 0.052 | 0.015 | N | N | Y | Y | Y | 2 | 5 | 6 | 7 |
| 0.052 | 0.084 | N | N | N | N | Y | 2 | 6 | 7 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.040 | 0.002 | Y | Y | Y | Y | Y | 1 | 3 | 7 | 7 |
| 0.044 | 0.035 | N | N | N | Y | Y | 1 | 5 | 7 | 8 |
| 0.172 | 0.010 | Y | Y | Y | Y | Y | 0 | 0 | 0 | 0 |
| 0.167 | 0.215 | N | N | N | N | N | 0 | 1 | 1 | 2 |
| 0.041 | 0.012 | N | N | Y | Y | Y | 1 | 4 | 7 | 7 |
| 0.046 | 0.035 | N | N | N | Y | Y | 1 | 5 | 6 | 8 |
| 0.059 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.053 | 0.018 | N | N | Y | Y | Y | 1 | 3 | 5 | 8 |
| 0.061 | 0.032 | N | N | N | Y | Y | 0 | 0 | 4 | 8 |
| 0.277 | 0.231 | N | N | N | N | N | 0 | 0 | 0 | 1 |
Appendix B.1.2. Trials 11–20
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.750 | 0.704 | 0.046 | 0.074 | 0.014 |
| 0.674 | 0.714 | 0.041 | 0.064 | 0.037 |
| 0.679 | 0.706 | 0.043 | 0.068 | 0.031 |
| 0.679 | 0.696 | 0.024 | 0.058 | 0.035 |
| 0.734 | 0.704 | 0.032 | 0.056 | 0.016 |
| 0.728 | 0.694 | 0.035 | 0.067 | 0.019 |
| 0.728 | 0.704 | 0.028 | 0.052 | 0.019 |
| 0.750 | 0.704 | 0.046 | 0.070 | 0.017 |
| 0.706 | 0.708 | 0.024 | 0.050 | 0.022 |
| 0.684 | 0.699 | 0.033 | 0.061 | 0.027 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.982 | 0.913 | 0.069 | 0.185 | 0.012 |
| 0.861 | 0.901 | 0.063 | 0.153 | 0.087 |
| 0.911 | 0.901 | 0.053 | 0.163 | 0.053 |
| 0.911 | 0.901 | 0.053 | 0.163 | 0.053 |
| 0.984 | 0.909 | 0.075 | 0.212 | 0.018 |
| 0.959 | 0.902 | 0.067 | 0.179 | 0.020 |
| 0.959 | 0.908 | 0.059 | 0.180 | 0.023 |
| 1.000 | 0.885 | 0.115 | 0.241 | 0.034 |
| 0.933 | 0.894 | 0.063 | 0.184 | 0.036 |
| 0.889 | 0.901 | 0.058 | 0.158 | 0.066 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.699 | 0.199 | 0.231 | 0.168 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| 0.500 | 0.704 | 0.204 | 0.233 | 0.180 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.423 | 0.699 | 0.276 | 0.309 | 0.245 |
| 0.509 | 0.704 | 0.195 | 0.224 | 0.171 |
| 0.708 | 0.704 | 0.027 | 0.051 | 0.023 |
| 0.679 | 0.704 | 0.041 | 0.065 | 0.030 |
| 0.454 | 0.704 | 0.250 | 0.279 | 0.226 |
| 0.449 | 0.704 | 0.255 | 0.284 | 0.232 |
| 0.648 | 0.704 | 0.063 | 0.090 | 0.045 |
| 0.526 | 0.538 | 0.053 | 0.071 | 0.041 |
| 0.546 | 0.704 | 0.158 | 0.187 | 0.134 |
| 0.815 | 0.704 | 0.111 | 0.135 | 0.082 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.665 | 0.085 | 0.017 | 0.130 |
| 0.693 | 0.019 | 0.034 | 0.064 |
| 0.743 | 0.064 | 0.060 | 0.067 |
| 0.693 | 0.014 | 0.039 | 0.059 |
| 0.668 | 0.066 | 0.010 | 0.104 |
| 0.668 | 0.059 | 0.016 | 0.098 |
| 0.716 | 0.012 | 0.018 | 0.037 |
| 0.726 | 0.024 | 0.007 | 0.055 |
| 0.708 | 0.002 | 0.013 | 0.013 |
| 0.611 | 0.073 | 0.026 | 0.102 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.976 | 0.006 | 0.001 | 0.016 |
| 0.818 | 0.042 | 0.115 | 0.190 |
| 0.976 | 0.066 | 0.051 | 0.082 |
| 0.968 | 0.058 | 0.031 | 0.090 |
| 0.857 | 0.127 | 0.003 | 0.273 |
| 0.927 | 0.032 | 0.026 | 0.243 |
| 0.927 | 0.032 | 0.026 | 0.243 |
| 0.916 | 0.084 | 0.028 | 0.127 |
| 0.918 | 0.015 | 0.029 | 0.228 |
| 0.802 | 0.087 | 0.022 | 0.275 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.665 | 0.165 | 0.121 | 0.233 |
| 0.693 | 0.193 | 0.140 | 0.238 |
| 0.743 | 0.243 | 0.239 | 0.246 |
| 0.718 | 0.218 | 0.191 | 0.250 |
| 0.668 | 0.168 | 0.130 | 0.244 |
| 0.722 | 0.222 | 0.216 | 0.243 |
| 0.716 | 0.216 | 0.191 | 0.245 |
| 0.726 | 0.226 | 0.195 | 0.243 |
| 0.708 | 0.208 | 0.193 | 0.218 |
| 0.611 | 0.111 | 0.082 | 0.158 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.665 | 0.243 | 0.198 | 0.311 |
| 0.693 | 0.184 | 0.131 | 0.229 |
| 0.743 | 0.035 | 0.031 | 0.039 |
| 0.718 | 0.039 | 0.012 | 0.071 |
| 0.668 | 0.214 | 0.176 | 0.290 |
| 0.722 | 0.274 | 0.267 | 0.295 |
| 0.716 | 0.068 | 0.043 | 0.097 |
| 0.508 | 0.018 | 0.010 | 0.036 |
| 0.708 | 0.162 | 0.147 | 0.172 |
| 0.611 | 0.204 | 0.157 | 0.233 |
| % Err Avg | % MaxAX Avg | % Min Avg |
|---|---|---|
| 186.7% | 174.8% | 125.5% |
| 46.9% | 172.3% | 52.8% |
| 149.1% | 218.6% | 87.6% |
| 60.5% | 167.4% | 67.8% |
| 207.1% | 186.9% | 58.5% |
| 168.6% | 145.5% | 84.0% |
| 42.2% | 93.6% | 70.3% |
| 51.2% | 78.6% | 40.2% |
| 9.8% | 57.6% | 25.9% |
| 219.8% | 166.4% | 92.9% |
| 114.2% | 146.17% | 70.55% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 9.0% | 8.7% | 8.4% |
| 67.1% | 132.5% | 124.4% |
| 123.5% | 154.1% | 31.3% |
| 108.9% | 169.2% | 18.7% |
| 170.3% | 128.5% | 14.1% |
| 47.2% | 136.1% | 133.0% |
| 53.9% | 134.7% | 111.3% |
| 72.9% | 81.4% | 52.5% |
| 24.6% | 123.9% | 79.7% |
| 150.8% | 173.4% | 32.5% |
| 82.82% | 124.25% | 60.59% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 83.07% | 139.02% | 52.06% |
| 94.77% | 132.22% | 60.19% |
| 118.98% | 136.67% | 102.54% |
| 107.07% | 138.89% | 82.12% |
| 82.41% | 135.28% | 55.89% |
| 108.99% | 135.00% | 92.65% |
| 105.77% | 136.11% | 82.12% |
| 111.03% | 135.00% | 83.83% |
| 101.92% | 121.11% | 82.76% |
| 54.22% | 87.78% | 35.25% |
| 96.82% | 129.71% | 72.94% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 87.82% | 126.68% | 64.09% |
| 94.53% | 133.92% | 58.59% |
| 132.33% | 168.86% | 60.90% |
| 96.43% | 233.55% | 18.35% |
| 85.65% | 128.10% | 63.17% |
| 107.18% | 127.21% | 93.98% |
| 106.76% | 216.52% | 47.67% |
| 34.52% | 50.35% | 23.09% |
| 102.48% | 128.36% | 78.51% |
| 184.46% | 191.09% | 172.86% |
| 103.22% | 150.46% | 68.12% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.046 | 0.085 | N | N | N | N | Y | 0 | 4 | 7 | 8 |
| 0.041 | 0.019 | N | N | Y | Y | Y | 1 | 1 | 7 | 9 |
| 0.043 | 0.064 | N | N | N | N | Y | 0 | 2 | 7 | 9 |
| 0.024 | 0.014 | N | N | Y | Y | Y | 0 | 5 | 8 | 9 |
| 0.032 | 0.066 | N | N | N | N | Y | 3 | 6 | 8 | 8 |
| 0.035 | 0.059 | N | N | N | N | Y | 3 | 5 | 7 | 8 |
| 0.028 | 0.012 | N | N | Y | Y | Y | 4 | 6 | 7 | 8 |
| 0.046 | 0.024 | N | N | Y | Y | Y | 1 | 2 | 6 | 8 |
| 0.024 | 0.002 | Y | Y | Y | Y | Y | 0 | 6 | 8 | 9 |
| 0.033 | 0.073 | N | N | N | N | Y | 2 | 4 | 8 | 9 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.069 | 0.006 | N | Y | Y | Y | Y | 1 | 2 | 2 | 6 |
| 0.063 | 0.042 | N | N | N | Y | Y | 2 | 2 | 2 | 6 |
| 0.053 | 0.066 | N | N | N | N | Y | 2 | 3 | 3 | 8 |
| 0.053 | 0.058 | N | N | N | N | Y | 2 | 3 | 3 | 8 |
| 0.075 | 0.127 | N | N | N | N | N | 2 | 3 | 3 | 7 |
| 0.067 | 0.032 | N | N | N | Y | Y | 0 | 3 | 5 | 5 |
| 0.059 | 0.032 | N | N | N | Y | Y | 1 | 3 | 6 | 6 |
| 0.115 | 0.084 | N | N | N | N | Y | 0 | 1 | 2 | 4 |
| 0.063 | 0.015 | N | N | Y | Y | Y | 1 | 1 | 4 | 7 |
| 0.058 | 0.087 | N | N | N | N | Y | 0 | 0 | 5 | 9 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.276 | 0.243 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.195 | 0.184 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.027 | 0.035 | N | N | N | Y | Y | 2 | 6 | 8 | 9 |
| 0.041 | 0.039 | N | N | N | Y | Y | 0 | 2 | 6 | 9 |
| 0.250 | 0.214 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.255 | 0.274 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.063 | 0.068 | N | N | N | N | Y | 0 | 1 | 3 | 9 |
| 0.053 | 0.018 | N | N | Y | Y | Y | 1 | 3 | 5 | 8 |
| 0.158 | 0.162 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.111 | 0.204 | N | N | N | N | N | 0 | 0 | 0 | 6 |
Appendix B.1.3. Trials 21–30
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.552 | 0.532 | 0.053 | 0.074 | 0.044 |
| 0.492 | 0.532 | 0.053 | 0.073 | 0.033 |
| 0.557 | 0.532 | 0.055 | 0.078 | 0.045 |
| 0.456 | 0.532 | 0.076 | 0.095 | 0.050 |
| 0.512 | 0.532 | 0.049 | 0.069 | 0.032 |
| 0.640 | 0.557 | 0.085 | 0.105 | 0.073 |
| 0.464 | 0.532 | 0.068 | 0.087 | 0.044 |
| 0.633 | 0.560 | 0.079 | 0.098 | 0.067 |
| 0.467 | 0.532 | 0.065 | 0.084 | 0.042 |
| 0.734 | 0.588 | 0.153 | 0.168 | 0.134 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.552 | 0.532 | 0.053 | 0.074 | 0.044 |
| 0.492 | 0.532 | 0.053 | 0.073 | 0.033 |
| 0.557 | 0.532 | 0.055 | 0.078 | 0.045 |
| 0.456 | 0.532 | 0.076 | 0.095 | 0.050 |
| 0.512 | 0.532 | 0.049 | 0.069 | 0.032 |
| 0.599 | 0.559 | 0.061 | 0.085 | 0.047 |
| 0.464 | 0.532 | 0.068 | 0.087 | 0.044 |
| 0.598 | 0.561 | 0.062 | 0.085 | 0.045 |
| 0.467 | 0.532 | 0.065 | 0.084 | 0.042 |
| 0.889 | 0.591 | 0.297 | 0.335 | 0.258 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.600 | 0.557 | 0.059 | 0.078 | 0.050 |
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.600 | 0.560 | 0.058 | 0.076 | 0.050 |
| 0.600 | 0.532 | 0.085 | 0.112 | 0.068 |
| 0.889 | 0.591 | 0.297 | 0.335 | 0.258 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.552 | 0.532 | 0.053 | 0.074 | 0.044 |
| 0.492 | 0.532 | 0.053 | 0.073 | 0.033 |
| 0.557 | 0.532 | 0.055 | 0.078 | 0.045 |
| 0.456 | 0.532 | 0.076 | 0.095 | 0.050 |
| 0.512 | 0.532 | 0.049 | 0.069 | 0.032 |
| 0.600 | 0.557 | 0.059 | 0.078 | 0.050 |
| 0.464 | 0.532 | 0.068 | 0.087 | 0.044 |
| 0.605 | 0.560 | 0.060 | 0.079 | 0.053 |
| 0.467 | 0.532 | 0.065 | 0.084 | 0.042 |
| 0.815 | 0.588 | 0.227 | 0.242 | 0.209 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.018 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.555 | 0.002 | 0.017 | 0.021 |
| 0.472 | 0.016 | 0.010 | 0.019 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.623 | 0.017 | 0.027 | 0.051 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.549 | 0.083 | 0.071 | 0.098 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.634 | 0.100 | 0.056 | 0.126 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.018 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.555 | 0.002 | 0.017 | 0.021 |
| 0.472 | 0.016 | 0.010 | 0.019 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.597 | 0.002 | 0.040 | 0.051 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.513 | 0.085 | 0.069 | 0.102 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.716 | 0.173 | 0.130 | 0.252 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.073 | 0.066 | 0.085 |
| 0.527 | 0.073 | 0.045 | 0.082 |
| 0.555 | 0.045 | 0.027 | 0.064 |
| 0.472 | 0.128 | 0.126 | 0.135 |
| 0.481 | 0.119 | 0.106 | 0.133 |
| 0.623 | 0.023 | 0.011 | 0.067 |
| 0.474 | 0.126 | 0.117 | 0.143 |
| 0.549 | 0.051 | 0.039 | 0.065 |
| 0.475 | 0.126 | 0.126 | 0.126 |
| 0.634 | 0.034 | 0.008 | 0.078 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.018 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.555 | 0.002 | 0.017 | 0.021 |
| 0.472 | 0.016 | 0.010 | 0.019 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.623 | 0.023 | 0.011 | 0.067 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.549 | 0.056 | 0.044 | 0.070 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.634 | 0.181 | 0.137 | 0.207 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 45.6% | 48.6% | 39.6% |
| 65.5% | 194.8% | 36.2% |
| 3.4% | 36.7% | 26.4% |
| 21.1% | 36.9% | 10.0% |
| 61.6% | 63.4% | 54.3% |
| 19.8% | 48.9% | 37.2% |
| 15.4% | 43.6% | 8.1% |
| 105.8% | 106.3% | 99.6% |
| 12.3% | 19.2% | 9.5% |
| 65.2% | 75.2% | 41.4% |
| 41.6% | 67.36% | 36.23% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 45.6% | 48.6% | 39.6% |
| 65.5% | 194.8% | 36.2% |
| 3.4% | 36.7% | 26.4% |
| 21.1% | 36.9% | 10.0% |
| 61.6% | 63.4% | 54.3% |
| 3.0% | 107.8% | 47.2% |
| 15.4% | 43.6% | 8.1% |
| 137.1% | 152.8% | 120.1% |
| 12.3% | 19.2% | 9.5% |
| 58.1% | 75.1% | 50.5% |
| 42.31% | 77.89% | 40.19% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 85.57% | 97.42% | 75.21% |
| 86.58% | 72.54% | 65.68% |
| 53.48% | 56.96% | 39.85% |
| 151.00% | 185.24% | 119.72% |
| 139.89% | 156.46% | 117.94% |
| 39.02% | 132.99% | 14.52% |
| 148.05% | 172.69% | 127.28% |
| 87.84% | 85.21% | 77.43% |
| 147.97% | 185.24% | 111.70% |
| 53.87% | 167.20% | 9.36% |
| 99.33% | 131.20% | 75.87% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 45.63% | 48.55% | 39.64% |
| 65.46% | 194.79% | 36.15% |
| 3.37% | 36.71% | 26.43% |
| 21.05% | 36.89% | 10.03% |
| 61.62% | 63.40% | 54.35% |
| 39.97% | 134.72% | 13.96% |
| 15.41% | 43.63% | 8.07% |
| 93.53% | 88.44% | 82.73% |
| 12.28% | 19.16% | 9.50% |
| 79.59% | 85.55% | 65.47% |
| 43.79% | 75.18% | 34.63% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.053 | 0.024 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.053 | 0.035 | N | N | N | Y | Y | 0 | 4 | 6 | 8 |
| 0.055 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 4 | 8 |
| 0.076 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.049 | 0.030 | N | N | N | Y | Y | 0 | 2 | 7 | 8 |
| 0.085 | 0.017 | N | N | Y | Y | Y | 0 | 1 | 2 | 5 |
| 0.068 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.079 | 0.083 | N | N | N | N | Y | 0 | 2 | 3 | 5 |
| 0.065 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 7 |
| 0.153 | 0.100 | N | N | N | N | Y | 0 | 0 | 1 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.085 | 0.073 | N | N | N | N | Y | 0 | 1 | 2 | 5 |
| 0.085 | 0.073 | N | N | N | N | Y | 0 | 1 | 2 | 5 |
| 0.085 | 0.045 | N | N | N | Y | Y | 0 | 1 | 1 | 5 |
| 0.085 | 0.128 | N | N | N | N | N | 0 | 1 | 2 | 6 |
| 0.085 | 0.119 | N | N | N | N | N | 0 | 1 | 2 | 6 |
| 0.059 | 0.023 | N | N | Y | Y | Y | 1 | 1 | 2 | 9 |
| 0.085 | 0.126 | N | N | N | N | N | 0 | 1 | 2 | 6 |
| 0.058 | 0.051 | N | N | N | N | Y | 1 | 2 | 3 | 9 |
| 0.085 | 0.126 | N | N | N | N | N | 0 | 1 | 2 | 6 |
| 0.063 | 0.034 | N | N | N | Y | Y | 0 | 0 | 4 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.053 | 0.024 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.053 | 0.035 | N | N | N | Y | Y | 0 | 4 | 6 | 8 |
| 0.055 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 4 | 8 |
| 0.076 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.049 | 0.030 | N | N | N | Y | Y | 0 | 2 | 7 | 8 |
| 0.059 | 0.023 | N | N | Y | Y | Y | 1 | 1 | 2 | 9 |
| 0.068 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.060 | 0.056 | N | N | N | N | Y | 1 | 2 | 3 | 8 |
| 0.065 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 7 |
| 0.227 | 0.181 | N | N | N | N | N | 0 | 0 | 1 | 1 |
Appendix B.1.4. Trials 31–40
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.552 | 0.540 | 0.061 | 0.081 | 0.047 |
| 0.492 | 0.540 | 0.061 | 0.083 | 0.033 |
| 0.557 | 0.540 | 0.064 | 0.084 | 0.049 |
| 0.456 | 0.540 | 0.084 | 0.105 | 0.050 |
| 0.512 | 0.540 | 0.057 | 0.079 | 0.032 |
| 0.712 | 0.550 | 0.227 | 0.193 | 0.148 |
| 0.464 | 0.540 | 0.076 | 0.097 | 0.044 |
| 0.684 | 0.540 | 0.144 | 0.180 | 0.140 |
| 0.467 | 0.540 | 0.074 | 0.094 | 0.042 |
| 0.784 | 0.638 | 0.153 | 0.168 | 0.134 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.552 | 0.540 | 0.061 | 0.081 | 0.047 |
| 0.492 | 0.540 | 0.061 | 0.083 | 0.033 |
| 0.557 | 0.540 | 0.064 | 0.084 | 0.049 |
| 0.456 | 0.540 | 0.084 | 0.105 | 0.050 |
| 0.512 | 0.540 | 0.057 | 0.079 | 0.032 |
| 0.697 | 0.546 | 0.151 | 0.184 | 0.141 |
| 0.464 | 0.540 | 0.076 | 0.097 | 0.044 |
| 0.684 | 0.540 | 0.144 | 0.180 | 0.140 |
| 0.467 | 0.540 | 0.074 | 0.094 | 0.042 |
| 0.889 | 0.598 | 0.290 | 0.331 | 0.248 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.550 | 0.150 | 0.182 | 0.140 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.540 | 0.160 | 0.194 | 0.152 |
| 0.700 | 0.638 | 0.086 | 0.102 | 0.077 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.552 | 0.540 | 0.061 | 0.081 | 0.047 |
| 0.492 | 0.540 | 0.061 | 0.083 | 0.033 |
| 0.557 | 0.540 | 0.064 | 0.084 | 0.049 |
| 0.456 | 0.540 | 0.084 | 0.105 | 0.050 |
| 0.512 | 0.540 | 0.057 | 0.079 | 0.032 |
| 0.698 | 0.550 | 0.147 | 0.179 | 0.138 |
| 0.464 | 0.540 | 0.076 | 0.097 | 0.044 |
| 0.684 | 0.540 | 0.144 | 0.180 | 0.140 |
| 0.467 | 0.540 | 0.074 | 0.094 | 0.042 |
| 0.815 | 0.638 | 0.177 | 0.192 | 0.159 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.018 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.555 | 0.002 | 0.017 | 0.021 |
| 0.472 | 0.016 | 0.010 | 0.019 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.674 | 0.103 | 0.035 | 0.091 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.541 | 0.143 | 0.128 | 0.154 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.684 | 0.100 | 0.056 | 0.126 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.018 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.555 | 0.002 | 0.017 | 0.021 |
| 0.472 | 0.016 | 0.010 | 0.019 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.664 | 0.033 | 0.038 | 0.089 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.541 | 0.143 | 0.128 | 0.154 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.723 | 0.166 | 0.130 | 0.212 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.173 | 0.166 | 0.185 |
| 0.527 | 0.173 | 0.145 | 0.182 |
| 0.555 | 0.145 | 0.127 | 0.164 |
| 0.472 | 0.228 | 0.226 | 0.235 |
| 0.481 | 0.219 | 0.206 | 0.233 |
| 0.674 | 0.026 | 0.046 | 0.080 |
| 0.474 | 0.226 | 0.217 | 0.243 |
| 0.541 | 0.159 | 0.144 | 0.170 |
| 0.475 | 0.226 | 0.226 | 0.226 |
| 0.684 | 0.016 | 0.028 | 0.043 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.018 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.555 | 0.002 | 0.017 | 0.021 |
| 0.472 | 0.016 | 0.010 | 0.019 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.674 | 0.023 | 0.049 | 0.077 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.541 | 0.143 | 0.128 | 0.154 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.684 | 0.131 | 0.087 | 0.157 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 39.2% | 44.3% | 37.2% |
| 56.3% | 194.8% | 31.8% |
| 2.9% | 33.8% | 24.5% |
| 18.9% | 36.9% | 9.1% |
| 52.4% | 55.4% | 54.3% |
| 45.4% | 47.3% | 23.4% |
| 13.7% | 43.6% | 7.2% |
| 99.3% | 90.8% | 85.8% |
| 10.9% | 19.2% | 8.5% |
| 65.2% | 75.2% | 41.4% |
| 40.4% | 64.13% | 32.32% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 39.2% | 44.3% | 37.2% |
| 56.3% | 194.8% | 31.8% |
| 2.9% | 33.8% | 24.5% |
| 18.9% | 36.9% | 9.1% |
| 52.4% | 55.4% | 54.3% |
| 22.0% | 48.3% | 26.8% |
| 13.7% | 43.6% | 7.2% |
| 99.3% | 90.8% | 85.8% |
| 10.9% | 19.2% | 8.5% |
| 57.0% | 63.9% | 52.5% |
| 37.26% | 63.10% | 33.77% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 108.01% | 109.39% | 95.18% |
| 108.55% | 95.22% | 93.63% |
| 90.98% | 84.60% | 83.69% |
| 142.75% | 148.60% | 120.97% |
| 136.85% | 135.75% | 119.94% |
| 17.25% | 44.01% | 33.02% |
| 141.18% | 143.00% | 125.35% |
| 99.34% | 94.56% | 87.70% |
| 141.14% | 148.60% | 116.33% |
| 18.64% | 41.85% | 36.36% |
| 100.47% | 104.56% | 91.22% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 39.25% | 44.28% | 37.23% |
| 56.32% | 194.79% | 31.81% |
| 2.92% | 33.81% | 24.49% |
| 18.91% | 36.89% | 9.07% |
| 52.44% | 55.42% | 54.35% |
| 15.73% | 43.16% | 35.37% |
| 13.68% | 43.63% | 7.24% |
| 99.26% | 90.78% | 85.77% |
| 10.85% | 19.16% | 8.49% |
| 73.60% | 81.60% | 54.32% |
| 38.30% | 64.35% | 34.81% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.061 | 0.024 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.061 | 0.035 | N | N | N | Y | Y | 0 | 4 | 6 | 7 |
| 0.064 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 3 | 7 |
| 0.084 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.057 | 0.030 | N | N | N | Y | Y | 0 | 2 | 7 | 7 |
| 0.227 | 0.103 | Y | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.076 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.144 | 0.143 | N | N | N | N | N | 1 | 2 | 2 | 2 |
| 0.074 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 7 |
| 0.153 | 0.100 | N | N | N | N | Y | 0 | 0 | 1 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.061 | 0.024 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.061 | 0.035 | N | N | N | Y | Y | 0 | 4 | 6 | 7 |
| 0.064 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 3 | 7 |
| 0.084 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.057 | 0.030 | N | N | N | Y | Y | 0 | 2 | 7 | 7 |
| 0.151 | 0.033 | N | N | N | Y | Y | 0 | 1 | 1 | 1 |
| 0.076 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.144 | 0.143 | N | N | N | N | N | 1 | 2 | 2 | 2 |
| 0.074 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 7 |
| 0.290 | 0.166 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.160 | 0.173 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.160 | 0.173 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.160 | 0.145 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.160 | 0.228 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.160 | 0.219 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.150 | 0.026 | Y | N | N | Y | Y | 0 | 0 | 1 | 1 |
| 0.160 | 0.226 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.160 | 0.159 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.160 | 0.226 | N | N | N | N | N | 0 | 1 | 2 | 2 |
| 0.086 | 0.016 | N | N | Y | Y | Y | 1 | 1 | 1 | 5 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.061 | 0.024 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.061 | 0.035 | N | N | N | Y | Y | 0 | 4 | 6 | 7 |
| 0.064 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 3 | 7 |
| 0.084 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.057 | 0.030 | N | N | N | Y | Y | 0 | 2 | 7 | 7 |
| 0.147 | 0.023 | Y | N | Y | Y | Y | 0 | 0 | 1 | 1 |
| 0.076 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.144 | 0.143 | N | N | N | N | N | 1 | 2 | 2 | 2 |
| 0.074 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 7 |
| 0.177 | 0.131 | N | N | N | N | N | 1 | 1 | 1 | 1 |
Appendix B.1.5. Trials 41–50
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.551 | 0.528 | 0.050 | 0.073 | 0.038 |
| 0.492 | 0.521 | 0.045 | 0.065 | 0.030 |
| 0.679 | 0.563 | 0.129 | 0.144 | 0.110 |
| 0.501 | 0.563 | 0.064 | 0.083 | 0.053 |
| 0.512 | 0.527 | 0.045 | 0.064 | 0.032 |
| 0.591 | 0.550 | 0.058 | 0.074 | 0.050 |
| 0.464 | 0.527 | 0.063 | 0.082 | 0.044 |
| 0.585 | 0.555 | 0.057 | 0.073 | 0.046 |
| 0.467 | 0.527 | 0.061 | 0.079 | 0.042 |
| 0.709 | 0.563 | 0.153 | 0.168 | 0.134 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.550 | 0.525 | 0.051 | 0.073 | 0.037 |
| 0.492 | 0.527 | 0.049 | 0.068 | 0.033 |
| 0.723 | 0.587 | 0.149 | 0.186 | 0.120 |
| 0.706 | 0.587 | 0.139 | 0.174 | 0.113 |
| 0.512 | 0.527 | 0.045 | 0.064 | 0.032 |
| 0.549 | 0.565 | 0.060 | 0.086 | 0.046 |
| 0.464 | 0.527 | 0.063 | 0.082 | 0.044 |
| 0.549 | 0.574 | 0.073 | 0.104 | 0.057 |
| 0.467 | 0.527 | 0.061 | 0.079 | 0.042 |
| 0.889 | 0.587 | 0.301 | 0.337 | 0.263 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.550 | 0.528 | 0.050 | 0.073 | 0.038 |
| 0.550 | 0.527 | 0.050 | 0.073 | 0.039 |
| 0.550 | 0.563 | 0.053 | 0.070 | 0.041 |
| 0.550 | 0.563 | 0.053 | 0.070 | 0.041 |
| 0.550 | 0.527 | 0.050 | 0.073 | 0.039 |
| 0.550 | 0.550 | 0.043 | 0.059 | 0.030 |
| 0.550 | 0.527 | 0.050 | 0.073 | 0.039 |
| 0.550 | 0.555 | 0.045 | 0.061 | 0.033 |
| 0.550 | 0.527 | 0.050 | 0.073 | 0.039 |
| 0.550 | 0.563 | 0.053 | 0.070 | 0.041 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.550 | 0.528 | 0.050 | 0.073 | 0.038 |
| 0.492 | 0.527 | 0.049 | 0.068 | 0.033 |
| 0.708 | 0.563 | 0.152 | 0.167 | 0.133 |
| 0.701 | 0.563 | 0.147 | 0.162 | 0.128 |
| 0.512 | 0.527 | 0.045 | 0.064 | 0.032 |
| 0.551 | 0.550 | 0.043 | 0.059 | 0.030 |
| 0.464 | 0.527 | 0.063 | 0.082 | 0.044 |
| 0.561 | 0.555 | 0.047 | 0.063 | 0.036 |
| 0.467 | 0.527 | 0.061 | 0.079 | 0.042 |
| 0.815 | 0.563 | 0.252 | 0.267 | 0.234 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.024 | 0.017 | 0.036 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.742 | 0.064 | 0.060 | 0.067 |
| 0.511 | 0.010 | 0.004 | 0.026 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.584 | 0.007 | 0.023 | 0.031 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.534 | 0.050 | 0.040 | 0.066 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.609 | 0.100 | 0.056 | 0.126 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.526 | 0.024 | 0.017 | 0.035 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.792 | 0.069 | 0.059 | 0.080 |
| 0.716 | 0.010 | 0.004 | 0.026 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.561 | 0.012 | 0.017 | 0.052 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.497 | 0.052 | 0.032 | 0.072 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.712 | 0.176 | 0.130 | 0.277 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.023 | 0.017 | 0.035 |
| 0.527 | 0.023 | 0.005 | 0.032 |
| 0.742 | 0.192 | 0.188 | 0.196 |
| 0.511 | 0.039 | 0.023 | 0.053 |
| 0.481 | 0.069 | 0.056 | 0.083 |
| 0.584 | 0.034 | 0.010 | 0.064 |
| 0.474 | 0.076 | 0.067 | 0.093 |
| 0.534 | 0.016 | 0.005 | 0.031 |
| 0.475 | 0.076 | 0.076 | 0.076 |
| 0.609 | 0.059 | 0.033 | 0.103 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.527 | 0.023 | 0.017 | 0.035 |
| 0.527 | 0.035 | 0.027 | 0.064 |
| 0.742 | 0.035 | 0.031 | 0.038 |
| 0.511 | 0.190 | 0.174 | 0.204 |
| 0.481 | 0.030 | 0.018 | 0.044 |
| 0.584 | 0.033 | 0.009 | 0.063 |
| 0.474 | 0.010 | 0.007 | 0.019 |
| 0.534 | 0.026 | 0.016 | 0.042 |
| 0.475 | 0.008 | 0.008 | 0.008 |
| 0.609 | 0.206 | 0.162 | 0.232 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 47.4% | 48.5% | 44.8% |
| 76.2% | 213.8% | 40.6% |
| 49.5% | 60.8% | 41.4% |
| 15.5% | 48.8% | 4.8% |
| 67.5% | 68.3% | 54.3% |
| 11.4% | 46.6% | 42.2% |
| 16.5% | 43.6% | 8.6% |
| 88.6% | 90.9% | 87.0% |
| 13.1% | 19.2% | 10.1% |
| 65.2% | 75.2% | 41.4% |
| 45.1% | 71.57% | 37.52% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 46.2% | 48.4% | 45.6% |
| 71.2% | 194.8% | 38.8% |
| 46.3% | 66.4% | 31.8% |
| 7.2% | 23.0% | 2.3% |
| 67.5% | 68.3% | 54.3% |
| 19.8% | 112.4% | 20.1% |
| 16.5% | 43.6% | 8.6% |
| 70.7% | 69.6% | 55.9% |
| 13.1% | 19.2% | 10.1% |
| 58.6% | 82.0% | 49.5% |
| 41.71% | 72.77% | 31.70% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 46.37% | 47.87% | 43.10% |
| 46.83% | 43.00% | 14.16% |
| 362.63% | 477.41% | 268.19% |
| 73.60% | 75.61% | 56.17% |
| 137.21% | 144.14% | 112.63% |
| 80.26% | 215.29% | 16.59% |
| 151.06% | 172.46% | 126.96% |
| 34.53% | 51.28% | 15.77% |
| 150.91% | 194.34% | 103.07% |
| 111.22% | 251.53% | 46.36% |
| 119.46% | 167.29% | 80.30% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 46.38% | 47.87% | 43.11% |
| 71.24% | 194.79% | 38.80% |
| 22.82% | 28.50% | 18.29% |
| 129.59% | 135.78% | 126.24% |
| 67.54% | 68.32% | 54.35% |
| 78.36% | 211.92% | 15.35% |
| 16.46% | 43.63% | 8.56% |
| 55.92% | 66.63% | 45.06% |
| 13.14% | 19.16% | 10.09% |
| 81.61% | 86.91% | 69.16% |
| 58.31% | 90.35% | 42.90% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.050 | 0.024 | N | N | Y | Y | Y | 2 | 4 | 4 | 8 |
| 0.045 | 0.035 | N | N | N | Y | Y | 0 | 5 | 7 | 8 |
| 0.129 | 0.064 | N | N | N | N | Y | 0 | 0 | 0 | 2 |
| 0.064 | 0.010 | N | N | Y | Y | Y | 1 | 2 | 6 | 6 |
| 0.045 | 0.030 | N | N | N | Y | Y | 0 | 2 | 8 | 8 |
| 0.058 | 0.007 | Y | Y | Y | Y | Y | 1 | 2 | 2 | 9 |
| 0.063 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 8 |
| 0.057 | 0.050 | N | N | N | N | Y | 2 | 3 | 4 | 8 |
| 0.061 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 8 |
| 0.153 | 0.100 | N | N | N | N | Y | 0 | 0 | 1 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.051 | 0.024 | N | N | Y | Y | Y | 2 | 4 | 4 | 8 |
| 0.049 | 0.035 | N | N | N | Y | Y | 0 | 4 | 7 | 8 |
| 0.149 | 0.069 | N | N | N | N | Y | 1 | 2 | 2 | 3 |
| 0.139 | 0.010 | N | Y | Y | Y | Y | 1 | 1 | 1 | 3 |
| 0.045 | 0.030 | N | N | N | Y | Y | 0 | 2 | 8 | 8 |
| 0.060 | 0.012 | N | N | Y | Y | Y | 1 | 1 | 4 | 8 |
| 0.063 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 8 |
| 0.073 | 0.052 | N | N | N | N | Y | 1 | 2 | 3 | 6 |
| 0.061 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 8 |
| 0.301 | 0.176 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.050 | 0.023 | N | N | Y | Y | Y | 3 | 4 | 4 | 8 |
| 0.050 | 0.023 | N | N | Y | Y | Y | 2 | 4 | 4 | 8 |
| 0.053 | 0.192 | N | N | N | N | N | 1 | 4 | 6 | 9 |
| 0.053 | 0.039 | N | N | N | Y | Y | 1 | 4 | 5 | 8 |
| 0.050 | 0.069 | N | N | N | N | Y | 2 | 5 | 5 | 8 |
| 0.043 | 0.034 | N | N | N | Y | Y | 0 | 3 | 6 | 8 |
| 0.050 | 0.076 | N | N | N | N | Y | 2 | 5 | 5 | 8 |
| 0.045 | 0.016 | N | N | Y | Y | Y | 1 | 3 | 7 | 8 |
| 0.050 | 0.076 | N | N | N | N | Y | 2 | 5 | 5 | 8 |
| 0.053 | 0.059 | N | N | N | N | Y | 1 | 4 | 6 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.050 | 0.023 | N | N | Y | Y | Y | 3 | 4 | 4 | 8 |
| 0.049 | 0.035 | N | N | N | Y | Y | 0 | 4 | 7 | 8 |
| 0.152 | 0.035 | Y | N | N | Y | Y | 0 | 0 | 0 | 2 |
| 0.147 | 0.190 | N | N | N | N | N | 0 | 0 | 1 | 3 |
| 0.045 | 0.030 | N | N | N | Y | Y | 0 | 2 | 8 | 8 |
| 0.043 | 0.033 | N | N | N | Y | Y | 0 | 3 | 6 | 8 |
| 0.063 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 8 |
| 0.047 | 0.026 | N | N | N | Y | Y | 1 | 2 | 5 | 8 |
| 0.061 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 8 |
| 0.252 | 0.206 | N | N | N | N | N | 0 | 0 | 0 | 1 |
Appendix B.2
Appendix B.2.1. Trials 1–10
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.515 | 0.527 | 0.038 | 0.055 | 0.025 |
| 0.498 | 0.526 | 0.039 | 0.056 | 0.027 |
| 0.644 | 0.482 | 0.167 | 0.194 | 0.140 |
| 0.434 | 0.506 | 0.072 | 0.093 | 0.055 |
| 0.540 | 0.565 | 0.049 | 0.065 | 0.037 |
| 0.515 | 0.482 | 0.069 | 0.095 | 0.052 |
| 0.497 | 0.484 | 0.057 | 0.083 | 0.051 |
| 0.782 | 0.482 | 0.299 | 0.326 | 0.272 |
| 0.493 | 0.527 | 0.042 | 0.058 | 0.031 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.552 | 0.054 | 0.080 | 0.045 |
| 0.500 | 0.550 | 0.054 | 0.080 | 0.044 |
| 0.644 | 0.482 | 0.167 | 0.194 | 0.140 |
| 0.500 | 0.519 | 0.043 | 0.068 | 0.037 |
| 0.499 | 0.611 | 0.113 | 0.145 | 0.098 |
| 0.515 | 0.482 | 0.069 | 0.095 | 0.052 |
| 0.500 | 0.485 | 0.058 | 0.084 | 0.051 |
| 0.782 | 0.482 | 0.299 | 0.326 | 0.272 |
| 0.500 | 0.552 | 0.552 | 0.577 | 0.528 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.527 | 0.039 | 0.056 | 0.026 |
| 0.500 | 0.526 | 0.039 | 0.056 | 0.026 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| 0.500 | 0.506 | 0.043 | 0.063 | 0.031 |
| 0.500 | 0.565 | 0.074 | 0.089 | 0.060 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| 0.500 | 0.484 | 0.058 | 0.084 | 0.050 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| 0.500 | 0.527 | 0.039 | 0.056 | 0.026 |
| 0.500 | 0.527 | 0.039 | 0.056 | 0.026 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.497 | 0.527 | 0.040 | 0.057 | 0.028 |
| 0.503 | 0.526 | 0.038 | 0.056 | 0.025 |
| 0.637 | 0.482 | 0.161 | 0.188 | 0.134 |
| 0.511 | 0.603 | 0.096 | 0.119 | 0.086 |
| 0.500 | 0.565 | 0.074 | 0.089 | 0.061 |
| 0.515 | 0.482 | 0.069 | 0.095 | 0.052 |
| 0.476 | 0.484 | 0.053 | 0.077 | 0.053 |
| 0.803 | 0.482 | 0.321 | 0.347 | 0.293 |
| 0.508 | 0.527 | 0.038 | 0.055 | 0.024 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.505 | 0.011 | 0.006 | 0.016 |
| 0.526 | 0.028 | 0.009 | 0.055 |
| 0.673 | 0.029 | 0.004 | 0.049 |
| 0.454 | 0.020 | 0.010 | 0.032 |
| 0.509 | 0.030 | 0.018 | 0.044 |
| 0.479 | 0.036 | 0.052 | 0.101 |
| 0.537 | 0.040 | 0.022 | 0.062 |
| 0.498 | 0.283 | 0.275 | 0.301 |
| 0.498 | 0.005 | 0.001 | 0.008 |
| 0.493 | 0.007 | 0.000 | 0.049 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.496 | 0.004 | 0.001 | 0.012 |
| 0.540 | 0.040 | 0.013 | 0.087 |
| 0.673 | 0.029 | 0.004 | 0.049 |
| 0.520 | 0.020 | 0.010 | 0.032 |
| 0.526 | 0.027 | 0.052 | 0.073 |
| 0.479 | 0.036 | 0.052 | 0.101 |
| 0.538 | 0.038 | 0.020 | 0.059 |
| 0.498 | 0.283 | 0.275 | 0.301 |
| 0.499 | 0.499 | 0.494 | 0.501 |
| 0.493 | 0.007 | 0.000 | 0.049 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.505 | 0.005 | 0.001 | 0.009 |
| 0.526 | 0.026 | 0.007 | 0.053 |
| 0.673 | 0.173 | 0.148 | 0.193 |
| 0.454 | 0.046 | 0.034 | 0.055 |
| 0.509 | 0.009 | 0.005 | 0.022 |
| 0.479 | 0.021 | 0.067 | 0.086 |
| 0.537 | 0.037 | 0.019 | 0.059 |
| 0.498 | 0.002 | 0.007 | 0.019 |
| 0.498 | 0.002 | 0.001 | 0.006 |
| 0.473 | 0.027 | 0.000 | 0.068 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.505 | 0.008 | 0.002 | 0.012 |
| 0.526 | 0.022 | 0.003 | 0.049 |
| 0.673 | 0.037 | 0.012 | 0.056 |
| 0.523 | 0.012 | 0.001 | 0.031 |
| 0.509 | 0.010 | 0.004 | 0.023 |
| 0.479 | 0.036 | 0.052 | 0.101 |
| 0.537 | 0.061 | 0.043 | 0.083 |
| 0.498 | 0.305 | 0.296 | 0.322 |
| 0.498 | 0.010 | 0.007 | 0.014 |
| 0.493 | 0.007 | 0.000 | 0.049 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 27.70% | 29.53% | 23.87% |
| 70.90% | 202.50% | 15.41% |
| 17.50% | 34.72% | 2.07% |
| 27.42% | 57.86% | 11.10% |
| 61.63% | 67.74% | 46.98% |
| 52.08% | 105.62% | 99.33% |
| 70.41% | 121.02% | 26.55% |
| 94.78% | 100.94% | 92.22% |
| 11.62% | 24.74% | 1.18% |
| 11.63% | 56.30% | 0.00% |
| 44.57% | 80.10% | 31.87% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 7.69% | 14.97% | 1.82% |
| 74.54% | 197.27% | 16.44% |
| 17.50% | 34.72% | 2.07% |
| 45.34% | 86.74% | 15.32% |
| 24.17% | 73.72% | 35.86% |
| 52.08% | 105.62% | 99.33% |
| 65.84% | 117.44% | 23.66% |
| 94.78% | 100.94% | 92.22% |
| 90.34% | 94.90% | 85.62% |
| 11.63% | 56.30% | 0.00% |
| 48.39% | 88.26% | 37.23% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 12.00% | 36.23% | 1.83% |
| 66.56% | 203.43% | 12.00% |
| 290.89% | 379.31% | 171.79% |
| 106.19% | 107.20% | 87.30% |
| 12.65% | 36.61% | 5.08% |
| 34.77% | 132.02% | 99.25% |
| 63.55% | 116.53% | 22.44% |
| 3.24% | 22.05% | 13.79% |
| 5.03% | 10.89% | 3.02% |
| 69.02% | 121.74% | 0.00% |
| 66.39% | 116.60% | 41.65% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 19.00% | 43.77% | 3.26% |
| 58.30% | 199.51% | 6.00% |
| 22.79% | 41.88% | 6.13% |
| 12.48% | 35.57% | 0.84% |
| 13.26% | 37.19% | 4.49% |
| 52.08% | 105.62% | 99.33% |
| 115.35% | 156.04% | 55.70% |
| 95.13% | 100.87% | 92.70% |
| 25.98% | 30.27% | 25.49% |
| 11.63% | 56.30% | 0.00% |
| 42.60% | 80.70% | 29.39% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.038 | 0.011 | Y | N | Y | Y | Y | 0 | 4 | 7 | 8 |
| 0.039 | 0.028 | N | N | N | Y | Y | 2 | 5 | 7 | 8 |
| 0.167 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 0 |
| 0.072 | 0.020 | N | N | Y | Y | Y | 1 | 1 | 4 | 6 |
| 0.049 | 0.030 | N | N | N | Y | Y | 3 | 4 | 5 | 8 |
| 0.069 | 0.036 | N | N | N | Y | Y | 0 | 3 | 3 | 7 |
| 0.057 | 0.040 | N | N | N | Y | Y | 2 | 3 | 6 | 7 |
| 0.299 | 0.283 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.042 | 0.005 | Y | Y | Y | Y | Y | 2 | 4 | 7 | 7 |
| 0.060 | 0.007 | N | Y | Y | Y | Y | 1 | 2 | 5 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.054 | 0.004 | N | Y | Y | Y | Y | 2 | 3 | 5 | 7 |
| 0.054 | 0.040 | N | N | N | Y | Y | 3 | 4 | 5 | 7 |
| 0.167 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 0 |
| 0.043 | 0.020 | N | N | Y | Y | Y | 2 | 4 | 6 | 8 |
| 0.113 | 0.027 | Y | N | N | Y | Y | 0 | 0 | 2 | 4 |
| 0.069 | 0.036 | N | N | N | Y | Y | 0 | 3 | 3 | 7 |
| 0.058 | 0.038 | N | N | N | Y | Y | 2 | 3 | 6 | 7 |
| 0.299 | 0.283 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.552 | 0.499 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.060 | 0.007 | N | Y | Y | Y | Y | 1 | 2 | 5 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.039 | 0.005 | N | Y | Y | Y | Y | 1 | 3 | 7 | 7 |
| 0.039 | 0.026 | N | N | N | Y | Y | 2 | 4 | 7 | 8 |
| 0.060 | 0.173 | N | N | N | N | N | 2 | 3 | 6 | 8 |
| 0.043 | 0.046 | N | N | N | Y | Y | 1 | 4 | 7 | 8 |
| 0.074 | 0.009 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 7 |
| 0.060 | 0.021 | N | N | Y | Y | Y | 2 | 2 | 5 | 7 |
| 0.058 | 0.037 | N | N | N | Y | Y | 2 | 3 | 6 | 7 |
| 0.060 | 0.002 | Y | Y | Y | Y | Y | 1 | 2 | 5 | 7 |
| 0.039 | 0.002 | Y | Y | Y | Y | Y | 1 | 3 | 7 | 7 |
| 0.039 | 0.027 | N | N | N | Y | Y | 2 | 4 | 7 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.040 | 0.008 | N | Y | Y | Y | Y | 1 | 4 | 7 | 7 |
| 0.038 | 0.022 | N | N | Y | Y | Y | 2 | 5 | 7 | 8 |
| 0.161 | 0.037 | Y | N | N | Y | Y | 0 | 0 | 0 | 1 |
| 0.096 | 0.012 | Y | N | Y | Y | Y | 0 | 1 | 4 | 6 |
| 0.074 | 0.010 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 7 |
| 0.069 | 0.036 | N | N | N | Y | Y | 0 | 3 | 3 | 7 |
| 0.053 | 0.061 | N | N | N | N | Y | 2 | 5 | 6 | 7 |
| 0.321 | 0.305 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.038 | 0.010 | N | Y | Y | Y | Y | 1 | 5 | 7 | 8 |
| 0.060 | 0.007 | N | Y | Y | Y | Y | 1 | 2 | 5 | 7 |
Appendix B.2.2. Trials 11–20
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.900 | 0.923 | 0.048 | 0.091 | 0.039 |
| 0.809 | 0.926 | 0.117 | 0.168 | 0.077 |
| 0.894 | 0.923 | 0.050 | 0.095 | 0.042 |
| 0.809 | 0.927 | 0.119 | 0.168 | 0.080 |
| 0.934 | 0.912 | 0.050 | 0.101 | 0.027 |
| 0.894 | 0.934 | 0.054 | 0.101 | 0.045 |
| 1.000 | 0.922 | 0.078 | 0.124 | 0.025 |
| 0.932 | 0.923 | 0.042 | 0.084 | 0.024 |
| 0.956 | 0.933 | 0.032 | 0.073 | 0.021 |
| 0.934 | 0.923 | 0.042 | 0.084 | 0.024 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.900 | 0.923 | 0.048 | 0.091 | 0.039 |
| 0.809 | 0.922 | 0.114 | 0.166 | 0.084 |
| 0.894 | 0.923 | 0.050 | 0.095 | 0.042 |
| 0.894 | 0.923 | 0.050 | 0.095 | 0.042 |
| 0.934 | 0.923 | 0.042 | 0.084 | 0.024 |
| 0.894 | 0.923 | 0.050 | 0.094 | 0.042 |
| 0.894 | 0.923 | 0.050 | 0.094 | 0.042 |
| 1.000 | 0.923 | 0.077 | 0.121 | 0.025 |
| 0.956 | 0.923 | 0.042 | 0.084 | 0.025 |
| 0.934 | 0.923 | 0.042 | 0.084 | 0.024 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.902 | 0.402 | 0.468 | 0.354 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| 0.500 | 0.923 | 0.423 | 0.475 | 0.380 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.600 | 0.923 | 0.324 | 0.375 | 0.280 |
| 0.642 | 0.923 | 0.282 | 0.333 | 0.238 |
| 0.861 | 0.923 | 0.068 | 0.116 | 0.060 |
| 0.606 | 0.923 | 0.317 | 0.369 | 0.274 |
| 0.694 | 0.923 | 0.229 | 0.281 | 0.186 |
| 0.654 | 0.923 | 0.270 | 0.321 | 0.226 |
| 0.826 | 0.923 | 0.098 | 0.149 | 0.077 |
| 0.803 | 0.923 | 0.120 | 0.172 | 0.091 |
| 0.781 | 0.923 | 0.142 | 0.194 | 0.109 |
| 0.875 | 0.923 | 0.060 | 0.106 | 0.051 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.913 | 0.013 | 0.028 | 0.036 |
| 0.884 | 0.075 | 0.002 | 0.180 |
| 0.957 | 0.063 | 0.012 | 0.103 |
| 0.968 | 0.160 | 0.133 | 0.192 |
| 0.918 | 0.016 | 0.054 | 0.060 |
| 0.834 | 0.061 | 0.078 | 0.164 |
| 0.966 | 0.034 | 0.005 | 0.059 |
| 0.976 | 0.044 | 0.013 | 0.061 |
| 0.958 | 0.002 | 0.013 | 0.013 |
| 0.861 | 0.073 | 0.026 | 0.102 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.913 | 0.013 | 0.028 | 0.036 |
| 0.884 | 0.075 | 0.002 | 0.180 |
| 0.957 | 0.063 | 0.012 | 0.103 |
| 0.968 | 0.075 | 0.048 | 0.107 |
| 0.918 | 0.016 | 0.054 | 0.060 |
| 0.834 | 0.061 | 0.078 | 0.164 |
| 0.966 | 0.072 | 0.047 | 0.101 |
| 0.976 | 0.024 | 0.007 | 0.055 |
| 0.958 | 0.002 | 0.013 | 0.013 |
| 0.861 | 0.073 | 0.026 | 0.102 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.907 | 0.407 | 0.371 | 0.483 |
| 0.884 | 0.384 | 0.307 | 0.488 |
| 0.957 | 0.457 | 0.405 | 0.496 |
| 0.968 | 0.468 | 0.441 | 0.500 |
| 0.918 | 0.418 | 0.380 | 0.494 |
| 0.834 | 0.334 | 0.230 | 0.472 |
| 0.966 | 0.466 | 0.441 | 0.495 |
| 0.976 | 0.476 | 0.445 | 0.493 |
| 0.958 | 0.458 | 0.443 | 0.468 |
| 0.861 | 0.361 | 0.332 | 0.408 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.913 | 0.313 | 0.273 | 0.337 |
| 0.884 | 0.242 | 0.165 | 0.347 |
| 0.957 | 0.096 | 0.045 | 0.136 |
| 0.968 | 0.362 | 0.335 | 0.394 |
| 0.918 | 0.224 | 0.186 | 0.300 |
| 0.834 | 0.180 | 0.077 | 0.319 |
| 0.966 | 0.140 | 0.116 | 0.170 |
| 0.976 | 0.173 | 0.142 | 0.190 |
| 0.958 | 0.177 | 0.162 | 0.187 |
| 0.861 | 0.014 | 0.033 | 0.043 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 26.87% | 92.19% | 30.67% |
| 64.04% | 234.03% | 1.19% |
| 125.74% | 242.89% | 12.15% |
| 134.79% | 238.48% | 78.66% |
| 31.61% | 220.37% | 53.73% |
| 112.48% | 174.30% | 162.14% |
| 44.10% | 47.54% | 19.65% |
| 106.65% | 249.49% | 15.46% |
| 7.28% | 59.10% | 17.77% |
| 175.07% | 121.12% | 106.92% |
| 82.86% | 167.95% | 49.83% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 26.87% | 92.19% | 30.67% |
| 66.12% | 214.97% | 1.20% |
| 125.74% | 242.89% | 12.15% |
| 148.40% | 252.37% | 50.18% |
| 38.24% | 251.59% | 64.52% |
| 120.42% | 185.71% | 173.82% |
| 142.74% | 240.48% | 49.81% |
| 30.77% | 45.64% | 27.94% |
| 5.66% | 50.71% | 15.44% |
| 175.07% | 121.12% | 106.92% |
| 88.00% | 169.77% | 53.27% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 101.24% | 136.34% | 79.20% |
| 90.63% | 128.59% | 64.53% |
| 107.92% | 130.70% | 85.27% |
| 110.62% | 131.75% | 92.85% |
| 98.74% | 130.04% | 80.01% |
| 78.77% | 124.37% | 48.43% |
| 109.99% | 130.43% | 92.85% |
| 112.52% | 129.91% | 93.69% |
| 108.14% | 123.32% | 93.17% |
| 85.16% | 107.51% | 69.90% |
| 100.37% | 127.30% | 79.99% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 96.75% | 120.18% | 72.58% |
| 85.92% | 145.59% | 49.48% |
| 141.18% | 226.97% | 38.25% |
| 114.16% | 144.06% | 90.80% |
| 97.68% | 161.46% | 66.20% |
| 66.69% | 140.93% | 23.80% |
| 143.22% | 220.42% | 77.28% |
| 144.04% | 208.56% | 82.58% |
| 124.20% | 172.03% | 83.27% |
| 24.21% | 65.22% | 40.66% |
| 103.81% | 160.54% | 62.49% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.048 | 0.013 | Y | N | Y | Y | Y | 0 | 2 | 3 | 9 |
| 0.117 | 0.075 | N | N | N | N | Y | 0 | 0 | 0 | 1 |
| 0.050 | 0.063 | N | N | N | N | Y | 1 | 3 | 4 | 9 |
| 0.119 | 0.160 | N | N | N | N | N | 0 | 0 | 0 | 2 |
| 0.050 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 5 | 7 |
| 0.054 | 0.061 | N | N | N | N | Y | 0 | 3 | 3 | 9 |
| 0.078 | 0.034 | N | N | N | Y | Y | 0 | 1 | 4 | 6 |
| 0.042 | 0.044 | N | N | N | Y | Y | 0 | 3 | 7 | 9 |
| 0.032 | 0.002 | N | Y | Y | Y | Y | 2 | 5 | 7 | 8 |
| 0.042 | 0.073 | N | N | N | N | Y | 0 | 4 | 8 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.048 | 0.013 | Y | N | Y | Y | Y | 0 | 2 | 3 | 9 |
| 0.114 | 0.075 | N | N | N | N | Y | 0 | 0 | 1 | 2 |
| 0.050 | 0.063 | N | N | N | N | Y | 1 | 3 | 4 | 9 |
| 0.050 | 0.075 | N | N | N | N | Y | 1 | 3 | 4 | 9 |
| 0.042 | 0.016 | Y | N | Y | Y | Y | 0 | 3 | 6 | 8 |
| 0.050 | 0.061 | N | N | N | N | Y | 0 | 3 | 4 | 9 |
| 0.050 | 0.072 | N | N | N | N | Y | 0 | 3 | 4 | 9 |
| 0.077 | 0.024 | Y | N | Y | Y | Y | 0 | 0 | 4 | 6 |
| 0.042 | 0.002 | N | Y | Y | Y | Y | 1 | 4 | 6 | 8 |
| 0.042 | 0.073 | N | N | N | N | Y | 0 | 4 | 8 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.324 | 0.313 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.282 | 0.242 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.068 | 0.096 | N | N | N | N | Y | 1 | 2 | 3 | 6 |
| 0.317 | 0.362 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.229 | 0.224 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.270 | 0.180 | Y | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.098 | 0.140 | N | N | N | N | N | 1 | 1 | 2 | 5 |
| 0.120 | 0.173 | N | N | N | N | N | 0 | 0 | 1 | 3 |
| 0.142 | 0.177 | N | N | N | N | N | 0 | 0 | 0 | 2 |
| 0.060 | 0.014 | N | N | Y | Y | Y | 1 | 1 | 4 | 8 |
Appendix B.2.3. Trials 21–30
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.671 | 0.610 | 0.073 | 0.082 | 0.066 |
| 0.607 | 0.616 | 0.049 | 0.066 | 0.033 |
| 0.644 | 0.491 | 0.159 | 0.194 | 0.130 |
| 0.484 | 0.541 | 0.058 | 0.075 | 0.041 |
| 0.590 | 0.616 | 0.047 | 0.065 | 0.034 |
| 0.651 | 0.565 | 0.088 | 0.104 | 0.074 |
| 0.545 | 0.527 | 0.044 | 0.061 | 0.035 |
| 0.782 | 0.482 | 0.299 | 0.326 | 0.272 |
| 0.563 | 0.616 | 0.063 | 0.079 | 0.054 |
| 0.600 | 0.491 | 0.124 | 0.158 | 0.095 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.600 | 0.608 | 0.050 | 0.089 | 0.050 |
| 0.561 | 0.620 | 0.072 | 0.114 | 0.059 |
| 0.644 | 0.491 | 0.159 | 0.194 | 0.130 |
| 0.600 | 0.562 | 0.055 | 0.083 | 0.047 |
| 0.499 | 0.620 | 0.121 | 0.160 | 0.097 |
| 0.598 | 0.567 | 0.053 | 0.082 | 0.044 |
| 0.600 | 0.544 | 0.071 | 0.094 | 0.057 |
| 0.414 | 0.620 | 0.206 | 0.245 | 0.172 |
| 0.956 | 0.923 | 0.042 | 0.084 | 0.025 |
| 0.600 | 0.491 | 0.124 | 0.158 | 0.095 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.600 | 0.605 | 0.045 | 0.061 | 0.031 |
| 0.600 | 0.616 | 0.047 | 0.065 | 0.033 |
| 0.600 | 0.491 | 0.124 | 0.158 | 0.095 |
| 0.600 | 0.560 | 0.058 | 0.075 | 0.040 |
| 0.600 | 0.616 | 0.047 | 0.065 | 0.033 |
| 0.600 | 0.565 | 0.055 | 0.072 | 0.038 |
| 0.600 | 0.542 | 0.070 | 0.089 | 0.057 |
| 0.600 | 0.616 | 0.047 | 0.065 | 0.033 |
| 0.600 | 0.616 | 0.047 | 0.065 | 0.033 |
| 0.600 | 0.491 | 0.124 | 0.158 | 0.095 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.601 | 0.605 | 0.045 | 0.061 | 0.031 |
| 0.559 | 0.616 | 0.065 | 0.081 | 0.058 |
| 0.637 | 0.482 | 0.161 | 0.188 | 0.134 |
| 0.599 | 0.541 | 0.060 | 0.077 | 0.054 |
| 0.500 | 0.616 | 0.117 | 0.131 | 0.112 |
| 0.599 | 0.565 | 0.055 | 0.071 | 0.038 |
| 0.541 | 0.542 | 0.040 | 0.059 | 0.032 |
| 0.803 | 0.482 | 0.321 | 0.347 | 0.293 |
| 0.528 | 0.616 | 0.090 | 0.106 | 0.086 |
| 0.600 | 0.491 | 0.124 | 0.158 | 0.095 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.654 | 0.017 | 0.015 | 0.024 |
| 0.631 | 0.024 | 0.009 | 0.027 |
| 0.673 | 0.029 | 0.004 | 0.049 |
| 0.498 | 0.015 | 0.010 | 0.021 |
| 0.559 | 0.030 | 0.018 | 0.044 |
| 0.641 | 0.011 | 0.022 | 0.037 |
| 0.497 | 0.048 | 0.018 | 0.066 |
| 0.498 | 0.283 | 0.275 | 0.301 |
| 0.590 | 0.027 | 0.027 | 0.027 |
| 0.579 | 0.021 | 0.000 | 0.149 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.594 | 0.006 | 0.001 | 0.010 |
| 0.616 | 0.055 | 0.035 | 0.112 |
| 0.673 | 0.029 | 0.004 | 0.049 |
| 0.611 | 0.011 | 0.005 | 0.020 |
| 0.526 | 0.027 | 0.052 | 0.073 |
| 0.607 | 0.009 | 0.022 | 0.063 |
| 0.560 | 0.040 | 0.019 | 0.060 |
| 0.528 | 0.115 | 0.098 | 0.136 |
| 0.958 | 0.002 | 0.013 | 0.013 |
| 0.579 | 0.021 | 0.000 | 0.149 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.654 | 0.054 | 0.048 | 0.056 |
| 0.631 | 0.031 | 0.016 | 0.034 |
| 0.673 | 0.073 | 0.048 | 0.093 |
| 0.513 | 0.088 | 0.079 | 0.094 |
| 0.559 | 0.041 | 0.028 | 0.055 |
| 0.641 | 0.040 | 0.014 | 0.073 |
| 0.558 | 0.042 | 0.029 | 0.053 |
| 0.601 | 0.001 | 0.011 | 0.016 |
| 0.590 | 0.011 | 0.011 | 0.011 |
| 0.579 | 0.021 | 0.000 | 0.149 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.654 | 0.053 | 0.047 | 0.055 |
| 0.631 | 0.072 | 0.057 | 0.074 |
| 0.673 | 0.037 | 0.012 | 0.056 |
| 0.498 | 0.100 | 0.094 | 0.105 |
| 0.559 | 0.060 | 0.046 | 0.073 |
| 0.641 | 0.041 | 0.015 | 0.073 |
| 0.558 | 0.018 | 0.006 | 0.031 |
| 0.498 | 0.305 | 0.296 | 0.322 |
| 0.590 | 0.062 | 0.062 | 0.062 |
| 0.579 | 0.021 | 0.000 | 0.149 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 23.53% | 28.84% | 22.86% |
| 49.26% | 79.82% | 13.68% |
| 18.44% | 37.39% | 2.07% |
| 25.34% | 51.30% | 13.33% |
| 63.49% | 67.74% | 50.95% |
| 11.96% | 35.38% | 29.03% |
| 108.88% | 108.15% | 51.49% |
| 94.78% | 100.94% | 92.22% |
| 42.16% | 49.21% | 33.52% |
| 17.16% | 93.75% | 0.00% |
| 45.50% | 65.25% | 30.92% |
| % Err Avg | % Max Avg | %Min Avg |
|---|---|---|
| 12.55% | 11.27% | 2.08% |
| 75.76% | 187.87% | 30.78% |
| 18.44% | 37.39% | 2.07% |
| 20.85% | 42.62% | 5.99% |
| 22.47% | 74.47% | 32.58% |
| 16.27% | 143.76% | 27.33% |
| 56.22% | 63.03% | 32.43% |
| 55.46% | 79.09% | 39.86% |
| 5.66% | 50.71% | 15.44% |
| 17.16% | 93.75% | 0.00% |
| 30.08% | 78.40% | 18.86% |
| % Err Avg | % Max Avg | %Min Avg |
|---|---|---|
| 120.07% | 182.60% | 77.63% |
| 65.29% | 102.60% | 24.63% |
| 59.29% | 97.88% | 30.30% |
| 151.10% | 198.30% | 125.96% |
| 85.60% | 85.76% | 83.91% |
| 72.73% | 191.54% | 19.58% |
| 59.16% | 59.99% | 50.07% |
| 1.65% | 49.00% | 16.17% |
| 22.12% | 32.16% | 16.17% |
| 17.16% | 93.75% | 0.00% |
| 65.42% | 109.36% | 44.44% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 118.18% | 180.36% | 76.33% |
| 110.22% | 128.78% | 69.98% |
| 22.79% | 41.88% | 6.13% |
| 165.71% | 174.77% | 136.56% |
| 51.16% | 64.56% | 35.01% |
| 74.93% | 195.50% | 20.89% |
| 44.33% | 94.64% | 10.15% |
| 95.13% | 100.87% | 92.70% |
| 68.24% | 71.35% | 57.91% |
| 17.16% | 93.75% | 0.00% |
| 76.79% | 114.65% | 50.57% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.073 | 0.017 | Y | N | Y | Y | Y | 0 | 1 | 1 | 8 |
| 0.049 | 0.024 | N | N | Y | Y | Y | 1 | 4 | 6 | 8 |
| 0.159 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 1 |
| 0.058 | 0.015 | N | N | Y | Y | Y | 0 | 1 | 4 | 7 |
| 0.047 | 0.030 | N | N | N | Y | Y | 3 | 4 | 5 | 8 |
| 0.088 | 0.011 | N | N | Y | Y | Y | 1 | 1 | 1 | 4 |
| 0.044 | 0.048 | N | N | N | Y | Y | 2 | 3 | 6 | 8 |
| 0.299 | 0.283 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.063 | 0.027 | N | N | N | Y | Y | 1 | 2 | 5 | 7 |
| 0.124 | 0.021 | Y | N | Y | Y | Y | 0 | 0 | 0 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.045 | 0.054 | N | N | N | N | Y | 1 | 4 | 5 | 8 |
| 0.047 | 0.031 | N | N | N | Y | Y | 1 | 4 | 6 | 8 |
| 0.124 | 0.073 | N | N | N | N | Y | 0 | 1 | 1 | 2 |
| 0.058 | 0.088 | N | N | N | N | Y | 0 | 0 | 4 | 9 |
| 0.047 | 0.041 | N | N | N | Y | Y | 1 | 4 | 6 | 8 |
| 0.055 | 0.040 | N | N | N | Y | Y | 0 | 0 | 3 | 9 |
| 0.070 | 0.042 | N | N | N | Y | Y | 1 | 1 | 2 | 6 |
| 0.047 | 0.001 | Y | Y | Y | Y | Y | 0 | 3 | 6 | 8 |
| 0.047 | 0.011 | N | N | Y | Y | Y | 1 | 3 | 6 | 8 |
| 0.124 | 0.021 | Y | N | Y | Y | Y | 0 | 0 | 0 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.045 | 0.053 | N | N | N | N | Y | 1 | 4 | 5 | 8 |
| 0.065 | 0.072 | N | N | N | N | Y | 1 | 1 | 6 | 7 |
| 0.161 | 0.037 | Y | N | N | Y | Y | 0 | 0 | 0 | 1 |
| 0.060 | 0.100 | N | N | N | N | N | 1 | 2 | 3 | 8 |
| 0.117 | 0.060 | N | N | N | N | Y | 0 | 1 | 1 | 4 |
| 0.055 | 0.041 | N | N | N | Y | Y | 0 | 0 | 3 | 9 |
| 0.040 | 0.018 | N | N | Y | Y | Y | 2 | 3 | 5 | 8 |
| 0.321 | 0.305 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.090 | 0.062 | N | N | N | N | Y | 1 | 1 | 2 | 5 |
| 0.124 | 0.021 | Y | N | Y | Y | Y | 0 | 0 | 0 | 2 |
Appendix B.2.4. Trials 31–40
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.754 | 0.668 | 0.096 | 0.118 | 0.078 |
| 0.657 | 0.668 | 0.047 | 0.066 | 0.037 |
| 0.666 | 0.668 | 0.049 | 0.068 | 0.035 |
| 0.517 | 0.562 | 0.050 | 0.067 | 0.026 |
| 0.640 | 0.668 | 0.046 | 0.065 | 0.039 |
| 0.798 | 0.668 | 0.131 | 0.153 | 0.115 |
| 0.632 | 0.635 | 0.038 | 0.055 | 0.035 |
| 0.534 | 0.668 | 0.134 | 0.152 | 0.125 |
| 0.613 | 0.668 | 0.061 | 0.079 | 0.059 |
| 0.508 | 0.668 | 0.160 | 0.178 | 0.146 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.625 | 0.629 | 0.052 | 0.104 | 0.050 |
| 0.561 | 0.627 | 0.080 | 0.130 | 0.056 |
| 0.636 | 0.629 | 0.054 | 0.104 | 0.050 |
| 0.700 | 0.605 | 0.096 | 0.129 | 0.079 |
| 0.499 | 0.629 | 0.130 | 0.176 | 0.100 |
| 0.657 | 0.629 | 0.063 | 0.109 | 0.052 |
| 0.700 | 0.603 | 0.097 | 0.130 | 0.080 |
| 0.414 | 0.629 | 0.215 | 0.261 | 0.175 |
| 0.526 | 0.629 | 0.103 | 0.152 | 0.078 |
| 0.613 | 0.629 | 0.053 | 0.105 | 0.051 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.637 | 0.072 | 0.089 | 0.054 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.635 | 0.074 | 0.090 | 0.056 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.627 | 0.668 | 0.053 | 0.071 | 0.047 |
| 0.559 | 0.668 | 0.109 | 0.127 | 0.105 |
| 0.628 | 0.668 | 0.053 | 0.070 | 0.047 |
| 0.646 | 0.562 | 0.091 | 0.109 | 0.083 |
| 0.500 | 0.668 | 0.168 | 0.186 | 0.152 |
| 0.659 | 0.668 | 0.048 | 0.066 | 0.036 |
| 0.606 | 0.635 | 0.038 | 0.055 | 0.038 |
| 0.407 | 0.668 | 0.261 | 0.279 | 0.240 |
| 0.528 | 0.668 | 0.140 | 0.158 | 0.130 |
| 0.700 | 0.668 | 0.060 | 0.081 | 0.042 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.735 | 0.019 | 0.018 | 0.027 |
| 0.681 | 0.024 | 0.009 | 0.027 |
| 0.699 | 0.033 | 0.018 | 0.035 |
| 0.535 | 0.019 | 0.019 | 0.019 |
| 0.609 | 0.030 | 0.018 | 0.044 |
| 0.805 | 0.007 | 0.009 | 0.026 |
| 0.619 | 0.013 | 0.008 | 0.029 |
| 0.651 | 0.117 | 0.106 | 0.132 |
| 0.640 | 0.027 | 0.027 | 0.027 |
| 0.584 | 0.076 | 0.035 | 0.192 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.618 | 0.007 | 0.001 | 0.011 |
| 0.623 | 0.062 | 0.035 | 0.123 |
| 0.673 | 0.038 | 0.014 | 0.052 |
| 0.703 | 0.003 | 0.003 | 0.008 |
| 0.526 | 0.027 | 0.052 | 0.073 |
| 0.704 | 0.047 | 0.020 | 0.150 |
| 0.588 | 0.112 | 0.078 | 0.149 |
| 0.528 | 0.115 | 0.098 | 0.136 |
| 0.612 | 0.087 | 0.018 | 0.114 |
| 0.623 | 0.010 | 0.080 | 0.155 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.735 | 0.035 | 0.027 | 0.036 |
| 0.681 | 0.019 | 0.017 | 0.034 |
| 0.699 | 0.001 | 0.001 | 0.017 |
| 0.597 | 0.103 | 0.098 | 0.109 |
| 0.609 | 0.091 | 0.078 | 0.105 |
| 0.805 | 0.105 | 0.089 | 0.124 |
| 0.619 | 0.081 | 0.076 | 0.096 |
| 0.651 | 0.049 | 0.034 | 0.061 |
| 0.640 | 0.061 | 0.061 | 0.061 |
| 0.584 | 0.116 | 0.000 | 0.227 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.735 | 0.108 | 0.101 | 0.110 |
| 0.681 | 0.122 | 0.107 | 0.125 |
| 0.699 | 0.071 | 0.056 | 0.073 |
| 0.535 | 0.111 | 0.111 | 0.111 |
| 0.609 | 0.110 | 0.096 | 0.123 |
| 0.805 | 0.146 | 0.130 | 0.165 |
| 0.619 | 0.013 | 0.002 | 0.019 |
| 0.651 | 0.244 | 0.233 | 0.259 |
| 0.640 | 0.112 | 0.112 | 0.112 |
| 0.584 | 0.116 | 0.000 | 0.227 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 19.60% | 22.55% | 22.41% |
| 50.75% | 72.01% | 13.68% |
| 66.20% | 100.00% | 25.89% |
| 37.12% | 71.43% | 27.82% |
| 65.46% | 67.74% | 44.47% |
| 5.61% | 22.59% | 5.89% |
| 34.42% | 52.17% | 22.37% |
| 87.20% | 105.85% | 69.45% |
| 43.14% | 45.03% | 33.52% |
| 47.61% | 131.96% | 19.67% |
| 45.71% | 69.13% | 28.52% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 13.06% | 10.08% | 3.02% |
| 77.25% | 217.78% | 26.96% |
| 69.69% | 103.73% | 12.96% |
| 3.29% | 10.41% | 2.55% |
| 20.99% | 72.18% | 29.60% |
| 75.38% | 285.85% | 17.93% |
| 115.40% | 114.41% | 97.74% |
| 53.25% | 77.69% | 37.40% |
| 84.04% | 145.41% | 11.84% |
| 19.27% | 304.82% | 75.90% |
| 53.16% | 134.24% | 31.59% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 57.86% | 86.12% | 33.33% |
| 31.67% | 41.98% | 39.47% |
| 2.50% | 20.37% | 2.39% |
| 142.71% | 179.84% | 123.03% |
| 151.09% | 186.60% | 129.01% |
| 174.78% | 295.45% | 109.26% |
| 110.03% | 135.34% | 106.92% |
| 82.03% | 81.34% | 74.69% |
| 100.85% | 144.74% | 74.69% |
| 193.12% | 280.25% | 0.00% |
| 104.66% | 145.20% | 69.28% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 202.95% | 232.24% | 141.65% |
| 112.01% | 118.91% | 84.32% |
| 134.67% | 157.89% | 79.60% |
| 122.65% | 133.49% | 101.69% |
| 65.23% | 80.43% | 51.50% |
| 305.83% | 451.92% | 195.62% |
| 34.84% | 48.80% | 3.73% |
| 93.43% | 108.14% | 83.36% |
| 79.69% | 86.10% | 70.61% |
| 193.12% | 280.25% | 0.00% |
| 134.44% | 169.82% | 81.21% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.096 | 0.019 | Y | N | Y | Y | Y | 0 | 0 | 0 | 3 |
| 0.047 | 0.024 | N | N | Y | Y | Y | 1 | 4 | 6 | 8 |
| 0.049 | 0.033 | N | N | N | Y | Y | 0 | 2 | 5 | 8 |
| 0.050 | 0.019 | Y | N | Y | Y | Y | 0 | 3 | 5 | 8 |
| 0.046 | 0.030 | N | N | N | Y | Y | 3 | 4 | 5 | 8 |
| 0.131 | 0.007 | Y | Y | Y | Y | Y | 0 | 0 | 0 | 2 |
| 0.038 | 0.013 | N | N | Y | Y | Y | 1 | 2 | 6 | 8 |
| 0.134 | 0.117 | N | N | N | N | N | 0 | 0 | 0 | 2 |
| 0.061 | 0.027 | N | N | N | Y | Y | 1 | 2 | 5 | 7 |
| 0.160 | 0.076 | Y | N | N | N | Y | 0 | 0 | 0 | 0 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.052 | 0.007 | N | Y | Y | Y | Y | 2 | 4 | 5 | 8 |
| 0.080 | 0.062 | N | N | N | N | Y | 0 | 0 | 3 | 6 |
| 0.054 | 0.038 | N | N | N | Y | Y | 1 | 4 | 5 | 6 |
| 0.096 | 0.003 | Y | Y | Y | Y | Y | 0 | 0 | 2 | 4 |
| 0.130 | 0.027 | Y | N | N | Y | Y | 0 | 0 | 1 | 1 |
| 0.063 | 0.047 | N | N | N | Y | Y | 0 | 2 | 5 | 6 |
| 0.097 | 0.112 | N | N | N | N | N | 1 | 1 | 3 | 5 |
| 0.215 | 0.115 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.103 | 0.087 | N | N | N | N | Y | 2 | 2 | 2 | 5 |
| 0.053 | 0.010 | N | N | Y | Y | Y | 2 | 4 | 4 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.060 | 0.035 | N | N | N | Y | Y | 1 | 2 | 3 | 7 |
| 0.060 | 0.019 | N | N | Y | Y | Y | 1 | 1 | 3 | 7 |
| 0.060 | 0.001 | Y | Y | Y | Y | Y | 0 | 1 | 3 | 7 |
| 0.072 | 0.103 | N | N | N | N | N | 1 | 1 | 3 | 7 |
| 0.060 | 0.091 | N | N | N | N | Y | 1 | 2 | 4 | 7 |
| 0.060 | 0.105 | N | N | N | N | N | 1 | 2 | 4 | 8 |
| 0.074 | 0.081 | N | N | N | N | Y | 1 | 1 | 3 | 7 |
| 0.060 | 0.049 | N | N | N | Y | Y | 1 | 2 | 3 | 7 |
| 0.060 | 0.061 | N | N | N | N | Y | 1 | 2 | 4 | 7 |
| 0.060 | 0.116 | N | N | N | N | N | 1 | 2 | 4 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.053 | 0.108 | N | N | N | N | N | 0 | 5 | 6 | 8 |
| 0.109 | 0.122 | N | N | N | N | N | 0 | 0 | 1 | 6 |
| 0.053 | 0.071 | N | N | N | N | Y | 1 | 5 | 6 | 7 |
| 0.091 | 0.111 | N | N | N | N | N | 0 | 0 | 1 | 5 |
| 0.168 | 0.110 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.048 | 0.146 | N | N | N | N | N | 1 | 5 | 7 | 9 |
| 0.038 | 0.013 | N | N | Y | Y | Y | 2 | 4 | 6 | 8 |
| 0.261 | 0.244 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.140 | 0.112 | N | N | N | N | N | 0 | 0 | 0 | 2 |
| 0.060 | 0.116 | N | N | N | N | N | 1 | 2 | 4 | 8 |
Appendix B.2.5. Trials 41–50
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.589 | 0.562 | 0.051 | 0.066 | 0.037 |
| 0.569 | 0.582 | 0.045 | 0.061 | 0.033 |
| 0.644 | 0.487 | 0.163 | 0.194 | 0.135 |
| 0.496 | 0.573 | 0.077 | 0.099 | 0.064 |
| 0.565 | 0.591 | 0.048 | 0.065 | 0.035 |
| 0.565 | 0.512 | 0.072 | 0.094 | 0.057 |
| 0.516 | 0.509 | 0.047 | 0.070 | 0.043 |
| 0.782 | 0.512 | 0.270 | 0.302 | 0.241 |
| 0.538 | 0.591 | 0.064 | 0.079 | 0.051 |
| 0.550 | 0.487 | 0.088 | 0.118 | 0.065 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.550 | 0.580 | 0.050 | 0.082 | 0.037 |
| 0.550 | 0.605 | 0.066 | 0.102 | 0.056 |
| 0.644 | 0.487 | 0.163 | 0.194 | 0.135 |
| 0.550 | 0.541 | 0.044 | 0.070 | 0.037 |
| 0.499 | 0.616 | 0.117 | 0.152 | 0.099 |
| 0.549 | 0.518 | 0.056 | 0.080 | 0.051 |
| 0.550 | 0.514 | 0.061 | 0.085 | 0.053 |
| 0.782 | 0.482 | 0.299 | 0.326 | 0.272 |
| 0.526 | 0.616 | 0.090 | 0.128 | 0.083 |
| 0.550 | 0.487 | 0.088 | 0.118 | 0.065 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.550 | 0.562 | 0.038 | 0.053 | 0.027 |
| 0.550 | 0.582 | 0.048 | 0.063 | 0.035 |
| 0.550 | 0.487 | 0.088 | 0.118 | 0.065 |
| 0.550 | 0.530 | 0.047 | 0.065 | 0.032 |
| 0.550 | 0.591 | 0.056 | 0.072 | 0.041 |
| 0.500 | 0.482 | 0.060 | 0.086 | 0.051 |
| 0.550 | 0.509 | 0.064 | 0.087 | 0.052 |
| 0.550 | 0.487 | 0.088 | 0.118 | 0.065 |
| 0.550 | 0.591 | 0.056 | 0.072 | 0.041 |
| 0.550 | 0.487 | 0.088 | 0.118 | 0.065 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.549 | 0.562 | 0.038 | 0.054 | 0.027 |
| 0.549 | 0.582 | 0.049 | 0.063 | 0.036 |
| 0.637 | 0.487 | 0.157 | 0.188 | 0.129 |
| 0.547 | 0.571 | 0.047 | 0.067 | 0.042 |
| 0.500 | 0.591 | 0.094 | 0.109 | 0.087 |
| 0.550 | 0.512 | 0.060 | 0.082 | 0.050 |
| 0.509 | 0.509 | 0.047 | 0.068 | 0.042 |
| 0.803 | 0.487 | 0.316 | 0.347 | 0.288 |
| 0.528 | 0.591 | 0.071 | 0.086 | 0.060 |
| 0.550 | 0.487 | 0.088 | 0.118 | 0.065 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.575 | 0.014 | 0.011 | 0.018 |
| 0.594 | 0.025 | 0.013 | 0.032 |
| 0.673 | 0.029 | 0.004 | 0.049 |
| 0.506 | 0.010 | 0.001 | 0.024 |
| 0.534 | 0.030 | 0.017 | 0.044 |
| 0.540 | 0.025 | 0.039 | 0.074 |
| 0.543 | 0.027 | 0.014 | 0.044 |
| 0.498 | 0.283 | 0.275 | 0.301 |
| 0.565 | 0.026 | 0.027 | 0.027 |
| 0.536 | 0.014 | 0.000 | 0.099 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.545 | 0.005 | 0.000 | 0.011 |
| 0.600 | 0.049 | 0.031 | 0.107 |
| 0.673 | 0.029 | 0.004 | 0.049 |
| 0.566 | 0.016 | 0.008 | 0.026 |
| 0.526 | 0.027 | 0.052 | 0.073 |
| 0.531 | 0.018 | 0.046 | 0.068 |
| 0.548 | 0.002 | 0.015 | 0.017 |
| 0.498 | 0.283 | 0.275 | 0.301 |
| 0.559 | 0.033 | 0.018 | 0.039 |
| 0.536 | 0.014 | 0.000 | 0.099 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.575 | 0.025 | 0.020 | 0.028 |
| 0.594 | 0.044 | 0.033 | 0.051 |
| 0.673 | 0.123 | 0.098 | 0.143 |
| 0.480 | 0.070 | 0.060 | 0.077 |
| 0.534 | 0.016 | 0.003 | 0.030 |
| 0.479 | 0.021 | 0.067 | 0.086 |
| 0.543 | 0.007 | 0.010 | 0.020 |
| 0.498 | 0.052 | 0.043 | 0.069 |
| 0.565 | 0.014 | 0.015 | 0.015 |
| 0.536 | 0.014 | 0.000 | 0.099 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.575 | 0.026 | 0.022 | 0.029 |
| 0.594 | 0.045 | 0.034 | 0.052 |
| 0.673 | 0.037 | 0.012 | 0.056 |
| 0.506 | 0.041 | 0.027 | 0.050 |
| 0.534 | 0.035 | 0.021 | 0.048 |
| 0.540 | 0.010 | 0.055 | 0.059 |
| 0.543 | 0.034 | 0.022 | 0.052 |
| 0.498 | 0.305 | 0.296 | 0.322 |
| 0.565 | 0.036 | 0.037 | 0.037 |
| 0.536 | 0.014 | 0.000 | 0.099 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 27.30% | 28.71% | 27.41% |
| 55.21% | 94.38% | 21.98% |
| 17.96% | 36.01% | 2.07% |
| 13.42% | 37.52% | 0.87% |
| 62.55% | 67.74% | 50.36% |
| 34.99% | 78.68% | 69.65% |
| 56.47% | 102.73% | 19.78% |
| 105.16% | 113.71% | 99.59% |
| 41.69% | 51.61% | 33.52% |
| 16.01% | 83.19% | 0.00% |
| 43.07% | 69.43% | 32.52% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 10.41% | 13.53% | 0.29% |
| 75.29% | 190.81% | 30.38% |
| 17.96% | 36.01% | 2.07% |
| 34.88% | 69.08% | 12.10% |
| 23.29% | 73.12% | 34.19% |
| 32.50% | 91.73% | 85.41% |
| 2.99% | 29.09% | 19.47% |
| 94.78% | 100.94% | 92.22% |
| 36.60% | 47.27% | 14.01% |
| 16.01% | 83.19% | 0.00% |
| 34.47% | 73.48% | 29.01% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 64.87% | 103.48% | 38.40% |
| 91.43% | 144.92% | 51.67% |
| 140.26% | 220.25% | 82.77% |
| 148.90% | 186.00% | 118.98% |
| 27.75% | 41.06% | 7.40% |
| 34.77% | 132.02% | 99.25% |
| 11.36% | 23.01% | 19.72% |
| 59.08% | 66.46% | 58.28% |
| 25.73% | 35.76% | 20.18% |
| 16.01% | 83.19% | 0.00% |
| 62.02% | 103.62% | 49.67% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 67.44% | 107.65% | 40.35% |
| 92.44% | 145.30% | 52.95% |
| 23.42% | 43.51% | 6.13% |
| 86.39% | 74.60% | 63.97% |
| 37.23% | 54.47% | 19.27% |
| 16.69% | 110.22% | 71.78% |
| 73.21% | 124.07% | 31.51% |
| 96.42% | 102.62% | 92.70% |
| 51.53% | 60.63% | 42.34% |
| 16.01% | 83.19% | 0.00% |
| 56.08% | 90.63% | 42.10% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.051 | 0.014 | Y | N | Y | Y | Y | 0 | 1 | 4 | 9 |
| 0.045 | 0.025 | N | N | Y | Y | Y | 2 | 4 | 5 | 8 |
| 0.163 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 0 |
| 0.077 | 0.010 | N | N | Y | Y | Y | 1 | 1 | 2 | 7 |
| 0.048 | 0.030 | N | N | N | Y | Y | 3 | 4 | 5 | 8 |
| 0.072 | 0.025 | N | N | N | Y | Y | 0 | 1 | 3 | 7 |
| 0.047 | 0.027 | N | N | N | Y | Y | 2 | 4 | 6 | 8 |
| 0.270 | 0.283 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.064 | 0.026 | N | N | N | Y | Y | 1 | 2 | 4 | 7 |
| 0.088 | 0.014 | N | N | Y | Y | Y | 0 | 1 | 1 | 5 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.050 | 0.005 | Y | Y | Y | Y | Y | 0 | 2 | 6 | 7 |
| 0.066 | 0.049 | N | N | N | Y | Y | 1 | 2 | 6 | 6 |
| 0.163 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 0 |
| 0.044 | 0.016 | N | N | Y | Y | Y | 2 | 4 | 5 | 8 |
| 0.117 | 0.027 | Y | N | N | Y | Y | 0 | 0 | 1 | 4 |
| 0.056 | 0.018 | N | N | Y | Y | Y | 2 | 2 | 5 | 7 |
| 0.061 | 0.002 | Y | Y | Y | Y | Y | 1 | 1 | 3 | 7 |
| 0.299 | 0.283 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.090 | 0.033 | N | N | N | Y | Y | 2 | 2 | 2 | 6 |
| 0.088 | 0.014 | N | N | Y | Y | Y | 0 | 1 | 1 | 5 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.038 | 0.025 | N | N | Y | Y | Y | 2 | 4 | 7 | 8 |
| 0.048 | 0.044 | N | N | N | Y | Y | 2 | 5 | 5 | 8 |
| 0.088 | 0.123 | N | N | N | N | N | 0 | 2 | 2 | 6 |
| 0.047 | 0.070 | N | N | N | N | Y | 1 | 2 | 5 | 8 |
| 0.056 | 0.016 | N | N | Y | Y | Y | 0 | 3 | 4 | 7 |
| 0.060 | 0.021 | N | N | Y | Y | Y | 2 | 2 | 5 | 7 |
| 0.064 | 0.007 | Y | Y | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.088 | 0.052 | N | N | N | N | Y | 0 | 2 | 2 | 5 |
| 0.056 | 0.014 | N | N | Y | Y | Y | 0 | 3 | 4 | 7 |
| 0.088 | 0.014 | N | N | Y | Y | Y | 0 | 1 | 1 | 5 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.038 | 0.026 | N | N | N | Y | Y | 2 | 4 | 7 | 8 |
| 0.049 | 0.045 | N | N | N | Y | Y | 2 | 5 | 5 | 8 |
| 0.157 | 0.037 | Y | N | N | Y | Y | 0 | 0 | 0 | 1 |
| 0.047 | 0.041 | N | N | N | Y | Y | 3 | 5 | 6 | 7 |
| 0.094 | 0.035 | N | N | N | Y | Y | 0 | 1 | 1 | 5 |
| 0.060 | 0.010 | N | N | Y | Y | Y | 1 | 1 | 3 | 7 |
| 0.047 | 0.034 | N | N | N | Y | Y | 1 | 4 | 6 | 8 |
| 0.316 | 0.305 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.071 | 0.036 | N | N | N | Y | Y | 1 | 1 | 5 | 7 |
| 0.088 | 0.014 | N | N | Y | Y | Y | 0 | 1 | 1 | 5 |
Appendix B.3
Appendix B.3.1. Trials 1–10
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.398 | 0.453 | 0.056 | 0.071 | 0.041 |
| 0.435 | 0.452 | 0.026 | 0.032 | 0.017 |
| 0.720 | 0.606 | 0.133 | 0.156 | 0.110 |
| 0.516 | 0.606 | 0.090 | 0.112 | 0.081 |
| 0.472 | 0.453 | 0.028 | 0.043 | 0.029 |
| 0.685 | 0.603 | 0.112 | 0.138 | 0.086 |
| 0.664 | 0.603 | 0.100 | 0.125 | 0.074 |
| 0.501 | 0.584 | 0.085 | 0.106 | 0.076 |
| 0.530 | 0.603 | 0.082 | 0.105 | 0.074 |
| 0.592 | 0.603 | 0.069 | 0.094 | 0.052 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.370 | 0.487 | 0.117 | 0.153 | 0.088 |
| 0.439 | 0.487 | 0.061 | 0.097 | 0.044 |
| 0.720 | 0.603 | 0.136 | 0.161 | 0.110 |
| 0.516 | 0.603 | 0.092 | 0.116 | 0.083 |
| 0.431 | 0.487 | 0.067 | 0.103 | 0.045 |
| 0.685 | 0.603 | 0.112 | 0.138 | 0.086 |
| 0.664 | 0.603 | 0.100 | 0.125 | 0.074 |
| 0.499 | 0.588 | 0.091 | 0.112 | 0.083 |
| 0.530 | 0.603 | 0.082 | 0.105 | 0.074 |
| 0.592 | 0.603 | 0.069 | 0.094 | 0.052 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.453 | 0.047 | 0.063 | 0.046 |
| 0.500 | 0.453 | 0.047 | 0.063 | 0.046 |
| 0.500 | 0.603 | 0.104 | 0.128 | 0.094 |
| 0.500 | 0.603 | 0.104 | 0.128 | 0.094 |
| 0.500 | 0.453 | 0.047 | 0.063 | 0.046 |
| 0.500 | 0.603 | 0.104 | 0.128 | 0.094 |
| 0.500 | 0.603 | 0.104 | 0.128 | 0.094 |
| 0.500 | 0.584 | 0.086 | 0.107 | 0.077 |
| 0.500 | 0.603 | 0.104 | 0.128 | 0.094 |
| 0.500 | 0.603 | 0.104 | 0.128 | 0.094 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.371 | 0.453 | 0.082 | 0.096 | 0.066 |
| 0.437 | 0.453 | 0.026 | 0.041 | 0.016 |
| 0.712 | 0.603 | 0.130 | 0.155 | 0.105 |
| 0.511 | 0.603 | 0.096 | 0.119 | 0.086 |
| 0.432 | 0.453 | 0.029 | 0.044 | 0.017 |
| 0.685 | 0.603 | 0.112 | 0.138 | 0.086 |
| 0.642 | 0.603 | 0.087 | 0.112 | 0.065 |
| 0.517 | 0.584 | 0.074 | 0.094 | 0.066 |
| 0.544 | 0.603 | 0.074 | 0.098 | 0.064 |
| 0.533 | 0.603 | 0.080 | 0.103 | 0.072 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.393 | 0.005 | 0.000 | 0.019 |
| 0.446 | 0.011 | 0.000 | 0.037 |
| 0.816 | 0.096 | 0.081 | 0.102 |
| 0.523 | 0.007 | 0.005 | 0.026 |
| 0.472 | 0.000 | 0.000 | 0.000 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.641 | 0.023 | 0.001 | 0.048 |
| 0.550 | 0.048 | 0.024 | 0.069 |
| 0.554 | 0.025 | 0.016 | 0.031 |
| 0.540 | 0.052 | 0.024 | 0.135 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.376 | 0.007 | 0.011 | 0.016 |
| 0.473 | 0.034 | 0.004 | 0.121 |
| 0.816 | 0.096 | 0.081 | 0.102 |
| 0.523 | 0.007 | 0.005 | 0.026 |
| 0.488 | 0.057 | 0.008 | 0.095 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.641 | 0.023 | 0.001 | 0.048 |
| 0.547 | 0.048 | 0.023 | 0.070 |
| 0.554 | 0.025 | 0.016 | 0.031 |
| 0.540 | 0.052 | 0.024 | 0.135 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.393 | 0.107 | 0.102 | 0.121 |
| 0.446 | 0.054 | 0.028 | 0.065 |
| 0.816 | 0.316 | 0.300 | 0.322 |
| 0.523 | 0.023 | 0.010 | 0.042 |
| 0.472 | 0.028 | 0.028 | 0.028 |
| 0.707 | 0.207 | 0.194 | 0.222 |
| 0.641 | 0.141 | 0.117 | 0.164 |
| 0.550 | 0.050 | 0.025 | 0.071 |
| 0.554 | 0.054 | 0.045 | 0.060 |
| 0.540 | 0.040 | 0.044 | 0.115 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.393 | 0.022 | 0.009 | 0.027 |
| 0.446 | 0.009 | 0.002 | 0.035 |
| 0.816 | 0.104 | 0.088 | 0.110 |
| 0.523 | 0.012 | 0.001 | 0.031 |
| 0.472 | 0.040 | 0.040 | 0.040 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.641 | 0.000 | 0.022 | 0.025 |
| 0.550 | 0.033 | 0.008 | 0.054 |
| 0.554 | 0.010 | 0.001 | 0.016 |
| 0.540 | 0.006 | 0.077 | 0.082 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 9.43% | 25.95% | 0.00% |
| 40.78% | 222.22% | 0.00% |
| 72.33% | 92.43% | 51.70% |
| 8.23% | 32.28% | 4.90% |
| 0.00% | 0.00% | 0.00% |
| 19.69% | 42.95% | 6.90% |
| 22.60% | 37.88% | 0.68% |
| 57.03% | 91.52% | 22.33% |
| 29.85% | 41.27% | 14.74% |
| 74.94% | 143.69% | 45.19% |
| 33.49% | 73.02% | 14.64% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 5.72% | 18.29% | 6.86% |
| 55.62% | 275.94% | 4.11% |
| 70.67% | 92.81% | 49.98% |
| 8.08% | 31.31% | 4.76% |
| 85.89% | 209.77% | 7.75% |
| 19.69% | 42.95% | 6.90% |
| 22.60% | 37.88% | 0.68% |
| 52.99% | 84.18% | 20.22% |
| 29.85% | 41.27% | 14.74% |
| 74.94% | 143.69% | 45.19% |
| 42.61% | 97.81% | 16.12% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 228.34% | 223.93% | 190.82% |
| 115.84% | 102.93% | 61.47% |
| 302.48% | 341.66% | 233.92% |
| 21.97% | 44.10% | 7.80% |
| 59.59% | 61.47% | 44.34% |
| 198.03% | 235.39% | 151.27% |
| 135.46% | 173.75% | 90.84% |
| 57.93% | 92.22% | 23.42% |
| 51.75% | 63.76% | 35.09% |
| 37.85% | 122.21% | 33.92% |
| 120.92% | 146.14% | 87.29% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 26.48% | 41.00% | 8.81% |
| 32.88% | 218.75% | 4.84% |
| 79.69% | 103.89% | 56.76% |
| 12.48% | 35.57% | 0.84% |
| 138.55% | 240.96% | 90.70% |
| 19.69% | 42.95% | 6.90% |
| 0.16% | 33.98% | 22.34% |
| 44.89% | 81.74% | 8.76% |
| 13.47% | 25.02% | 1.02% |
| 8.13% | 114.37% | 74.24% |
| 37.64% | 93.82% | 27.52% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.117 | 0.007 | Y | Y | Y | Y | Y | 0 | 0 | 0 | 3 |
| 0.061 | 0.034 | N | N | N | Y | Y | 1 | 2 | 5 | 8 |
| 0.136 | 0.096 | N | N | N | N | Y | 0 | 1 | 1 | 2 |
| 0.092 | 0.007 | Y | Y | Y | Y | Y | 0 | 2 | 4 | 6 |
| 0.067 | 0.057 | N | N | N | N | Y | 1 | 1 | 4 | 7 |
| 0.112 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.100 | 0.023 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.091 | 0.048 | N | N | N | Y | Y | 1 | 1 | 4 | 6 |
| 0.082 | 0.025 | N | N | Y | Y | Y | 1 | 2 | 4 | 6 |
| 0.069 | 0.052 | N | N | N | N | Y | 2 | 2 | 5 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.047 | 0.107 | N | N | N | N | N | 2 | 3 | 5 | 9 |
| 0.047 | 0.054 | N | N | N | N | Y | 2 | 3 | 5 | 8 |
| 0.104 | 0.316 | N | N | N | N | N | 1 | 2 | 3 | 6 |
| 0.104 | 0.023 | N | N | Y | Y | Y | 1 | 1 | 2 | 5 |
| 0.047 | 0.028 | N | N | N | Y | Y | 2 | 3 | 4 | 8 |
| 0.104 | 0.207 | N | N | N | N | N | 1 | 2 | 3 | 6 |
| 0.104 | 0.141 | N | N | N | N | N | 1 | 2 | 3 | 6 |
| 0.086 | 0.050 | N | N | N | Y | Y | 0 | 2 | 4 | 6 |
| 0.104 | 0.054 | N | N | N | N | Y | 1 | 2 | 3 | 5 |
| 0.104 | 0.040 | N | N | N | Y | Y | 1 | 2 | 2 | 5 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.082 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 0 | 5 |
| 0.026 | 0.009 | N | Y | Y | Y | Y | 3 | 4 | 7 | 9 |
| 0.130 | 0.104 | N | N | N | N | N | 1 | 1 | 1 | 2 |
| 0.096 | 0.012 | Y | N | Y | Y | Y | 0 | 1 | 4 | 6 |
| 0.029 | 0.040 | N | N | N | Y | Y | 3 | 5 | 7 | 9 |
| 0.112 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.087 | 0.000 | Y | Y | Y | Y | Y | 0 | 0 | 2 | 5 |
| 0.074 | 0.033 | N | N | N | Y | Y | 1 | 3 | 4 | 7 |
| 0.074 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.080 | 0.006 | Y | Y | Y | Y | Y | 0 | 2 | 4 | 6 |
Appendix B.3.2. Trials 11–20
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.970 | 0.916 | 0.056 | 0.092 | 0.027 |
| 0.924 | 0.916 | 0.054 | 0.089 | 0.035 |
| 0.916 | 0.916 | 0.058 | 0.093 | 0.039 |
| 0.924 | 0.944 | 0.030 | 0.057 | 0.025 |
| 0.984 | 0.916 | 0.068 | 0.106 | 0.029 |
| 0.925 | 0.916 | 0.054 | 0.089 | 0.035 |
| 1.000 | 0.916 | 0.084 | 0.122 | 0.040 |
| 0.695 | 0.916 | 0.227 | 0.266 | 0.208 |
| 0.940 | 0.916 | 0.049 | 0.084 | 0.030 |
| 0.862 | 0.916 | 0.093 | 0.132 | 0.059 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.970 | 0.916 | 0.056 | 0.092 | 0.027 |
| 0.924 | 0.916 | 0.054 | 0.089 | 0.035 |
| 0.916 | 0.916 | 0.058 | 0.093 | 0.039 |
| 0.516 | 0.603 | 0.092 | 0.116 | 0.083 |
| 0.984 | 0.916 | 0.068 | 0.106 | 0.029 |
| 0.925 | 0.916 | 0.054 | 0.089 | 0.035 |
| 0.925 | 0.916 | 0.054 | 0.089 | 0.035 |
| 1.000 | 0.916 | 0.084 | 0.122 | 0.040 |
| 0.940 | 0.916 | 0.049 | 0.084 | 0.030 |
| 0.862 | 0.916 | 0.093 | 0.132 | 0.059 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.500 | 0.896 | 0.396 | 0.456 | 0.342 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| 0.500 | 0.916 | 0.416 | 0.460 | 0.378 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.669 | 0.916 | 0.248 | 0.291 | 0.229 |
| 0.757 | 0.916 | 0.177 | 0.216 | 0.146 |
| 0.883 | 0.916 | 0.077 | 0.115 | 0.050 |
| 0.722 | 0.916 | 0.206 | 0.244 | 0.181 |
| 0.744 | 0.916 | 0.188 | 0.226 | 0.159 |
| 0.685 | 0.916 | 0.235 | 0.276 | 0.216 |
| 0.826 | 0.916 | 0.123 | 0.161 | 0.081 |
| 0.566 | 0.916 | 0.350 | 0.395 | 0.313 |
| 0.765 | 0.916 | 0.171 | 0.209 | 0.138 |
| 0.804 | 0.916 | 0.140 | 0.178 | 0.099 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.972 | 0.002 | 0.009 | 0.011 |
| 0.915 | 0.009 | 0.039 | 0.059 |
| 0.977 | 0.062 | 0.040 | 0.069 |
| 0.964 | 0.040 | 0.017 | 0.076 |
| 0.880 | 0.104 | 0.006 | 0.167 |
| 0.952 | 0.027 | 0.017 | 0.041 |
| 0.937 | 0.063 | 0.034 | 0.101 |
| 0.953 | 0.258 | 0.209 | 0.299 |
| 0.943 | 0.004 | 0.012 | 0.014 |
| 0.664 | 0.198 | 0.045 | 0.290 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.972 | 0.002 | 0.009 | 0.011 |
| 0.915 | 0.009 | 0.039 | 0.059 |
| 0.977 | 0.062 | 0.040 | 0.069 |
| 0.523 | 0.007 | 0.005 | 0.026 |
| 0.880 | 0.104 | 0.006 | 0.167 |
| 0.952 | 0.027 | 0.017 | 0.041 |
| 0.937 | 0.012 | 0.026 | 0.042 |
| 0.953 | 0.047 | 0.007 | 0.097 |
| 0.943 | 0.004 | 0.012 | 0.014 |
| 0.664 | 0.198 | 0.045 | 0.290 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.857 | 0.357 | 0.276 | 0.436 |
| 0.915 | 0.415 | 0.365 | 0.463 |
| 0.977 | 0.477 | 0.456 | 0.484 |
| 0.964 | 0.464 | 0.441 | 0.500 |
| 0.880 | 0.380 | 0.318 | 0.478 |
| 0.952 | 0.452 | 0.442 | 0.466 |
| 0.937 | 0.437 | 0.399 | 0.467 |
| 0.953 | 0.453 | 0.404 | 0.493 |
| 0.943 | 0.443 | 0.428 | 0.454 |
| 0.664 | 0.164 | 0.072 | 0.317 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.972 | 0.303 | 0.292 | 0.312 |
| 0.915 | 0.158 | 0.108 | 0.206 |
| 0.977 | 0.095 | 0.073 | 0.102 |
| 0.964 | 0.243 | 0.220 | 0.279 |
| 0.880 | 0.136 | 0.074 | 0.234 |
| 0.952 | 0.268 | 0.258 | 0.281 |
| 0.937 | 0.112 | 0.074 | 0.141 |
| 0.953 | 0.387 | 0.338 | 0.428 |
| 0.943 | 0.178 | 0.163 | 0.189 |
| 0.664 | 0.140 | 0.014 | 0.232 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 3.84% | 41.51% | 9.28% |
| 17.07% | 110.48% | 66.11% |
| 106.67% | 177.00% | 43.17% |
| 132.34% | 304.00% | 29.82% |
| 151.92% | 157.75% | 20.76% |
| 50.67% | 116.05% | 19.13% |
| 74.55% | 83.96% | 83.09% |
| 113.53% | 143.20% | 78.69% |
| 7.93% | 46.59% | 13.63% |
| 212.52% | 220.36% | 76.47% |
| 87.11% | 140.09% | 44.02% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 3.84% | 41.51% | 9.28% |
| 17.07% | 110.48% | 66.11% |
| 106.67% | 177.00% | 43.17% |
| 8.08% | 31.31% | 4.76% |
| 151.92% | 157.75% | 20.76% |
| 50.67% | 116.05% | 19.13% |
| 22.49% | 118.91% | 29.26% |
| 56.17% | 79.39% | 17.54% |
| 7.93% | 46.59% | 13.63% |
| 212.52% | 220.36% | 76.47% |
| 63.74% | 109.94% | 30.01% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 90.21% | 127.43% | 60.47% |
| 99.77% | 122.34% | 79.33% |
| 114.78% | 127.89% | 99.00% |
| 111.64% | 132.12% | 95.85% |
| 91.47% | 126.30% | 69.01% |
| 108.82% | 123.00% | 96.07% |
| 105.16% | 123.27% | 86.72% |
| 108.89% | 130.27% | 87.70% |
| 106.66% | 119.83% | 93.02% |
| 39.35% | 83.76% | 15.65% |
| 97.68% | 121.62% | 78.28% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 122.15% | 136.12% | 100.31% |
| 88.92% | 141.14% | 50.09% |
| 122.23% | 202.80% | 63.37% |
| 117.88% | 153.49% | 89.96% |
| 72.54% | 147.22% | 32.52% |
| 113.80% | 129.82% | 93.43% |
| 91.08% | 174.61% | 45.71% |
| 110.55% | 136.60% | 85.66% |
| 104.32% | 136.64% | 77.92% |
| 99.83% | 129.76% | 13.57% |
| 104.33% | 148.82% | 65.25% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.056 | 0.002 | Y | Y | Y | Y | Y | 2 | 4 | 6 | 8 |
| 0.054 | 0.009 | Y | Y | Y | Y | Y | 0 | 2 | 7 | 8 |
| 0.058 | 0.062 | N | N | N | N | Y | 1 | 2 | 7 | 8 |
| 0.030 | 0.040 | N | N | N | Y | Y | 1 | 4 | 8 | 9 |
| 0.068 | 0.104 | N | N | N | N | N | 1 | 3 | 7 | 8 |
| 0.054 | 0.027 | N | N | N | Y | Y | 0 | 3 | 7 | 8 |
| 0.084 | 0.063 | N | N | N | N | Y | 0 | 1 | 5 | 7 |
| 0.227 | 0.258 | N | N | N | N | N | 0 | 0 | 1 | 1 |
| 0.049 | 0.004 | N | Y | Y | Y | Y | 1 | 5 | 7 | 8 |
| 0.093 | 0.198 | N | N | N | N | N | 0 | 1 | 1 | 6 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.248 | 0.303 | N | N | N | N | N | 1 | 1 | 1 | 1 |
| 0.177 | 0.158 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.077 | 0.095 | N | N | N | N | Y | 1 | 1 | 2 | 8 |
| 0.206 | 0.243 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.188 | 0.136 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.235 | 0.268 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.123 | 0.112 | N | N | N | N | N | 0 | 0 | 0 | 2 |
| 0.350 | 0.387 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.171 | 0.178 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.140 | 0.140 | N | N | N | N | N | 0 | 0 | 0 | 1 |
Appendix B.3.3. Trials 21–30
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.448 | 0.507 | 0.060 | 0.076 | 0.041 |
| 0.485 | 0.512 | 0.035 | 0.060 | 0.017 |
| 0.541 | 0.507 | 0.042 | 0.054 | 0.044 |
| 0.484 | 0.545 | 0.061 | 0.080 | 0.044 |
| 0.522 | 0.507 | 0.032 | 0.047 | 0.030 |
| 0.685 | 0.604 | 0.111 | 0.138 | 0.085 |
| 0.448 | 0.507 | 0.060 | 0.076 | 0.041 |
| 0.634 | 0.555 | 0.084 | 0.101 | 0.070 |
| 0.504 | 0.507 | 0.029 | 0.043 | 0.026 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.370 | 0.502 | 0.132 | 0.170 | 0.091 |
| 0.439 | 0.502 | 0.075 | 0.114 | 0.045 |
| 0.560 | 0.502 | 0.082 | 0.123 | 0.053 |
| 0.600 | 0.538 | 0.071 | 0.101 | 0.049 |
| 0.431 | 0.502 | 0.082 | 0.120 | 0.048 |
| 0.685 | 0.604 | 0.111 | 0.138 | 0.085 |
| 0.583 | 0.502 | 0.097 | 0.141 | 0.063 |
| 0.599 | 0.553 | 0.061 | 0.085 | 0.047 |
| 0.466 | 0.502 | 0.054 | 0.093 | 0.053 |
| 0.600 | 0.502 | 0.111 | 0.155 | 0.073 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| 0.600 | 0.541 | 0.062 | 0.078 | 0.054 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| 0.600 | 0.604 | 0.069 | 0.095 | 0.053 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| 0.600 | 0.555 | 0.060 | 0.077 | 0.047 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| 0.600 | 0.507 | 0.093 | 0.113 | 0.081 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.371 | 0.507 | 0.136 | 0.151 | 0.116 |
| 0.437 | 0.507 | 0.070 | 0.085 | 0.051 |
| 0.552 | 0.507 | 0.051 | 0.065 | 0.052 |
| 0.599 | 0.541 | 0.060 | 0.077 | 0.054 |
| 0.432 | 0.453 | 0.029 | 0.044 | 0.017 |
| 0.685 | 0.604 | 0.111 | 0.138 | 0.085 |
| 0.471 | 0.507 | 0.042 | 0.058 | 0.024 |
| 0.606 | 0.555 | 0.064 | 0.081 | 0.051 |
| 0.469 | 0.507 | 0.044 | 0.060 | 0.025 |
| 0.338 | 0.507 | 0.169 | 0.184 | 0.149 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.443 | 0.005 | 0.000 | 0.019 |
| 0.496 | 0.011 | 0.000 | 0.037 |
| 0.506 | 0.034 | 0.019 | 0.056 |
| 0.498 | 0.015 | 0.010 | 0.021 |
| 0.522 | 0.000 | 0.000 | 0.000 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.485 | 0.037 | 0.037 | 0.037 |
| 0.549 | 0.085 | 0.073 | 0.099 |
| 0.485 | 0.019 | 0.019 | 0.019 |
| 0.584 | 0.016 | 0.000 | 0.115 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.376 | 0.007 | 0.011 | 0.016 |
| 0.480 | 0.041 | 0.009 | 0.121 |
| 0.531 | 0.029 | 0.019 | 0.047 |
| 0.615 | 0.015 | 0.010 | 0.021 |
| 0.488 | 0.057 | 0.008 | 0.095 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.489 | 0.094 | 0.025 | 0.131 |
| 0.513 | 0.086 | 0.071 | 0.103 |
| 0.472 | 0.006 | 0.019 | 0.027 |
| 0.577 | 0.023 | 0.000 | 0.161 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.443 | 0.157 | 0.152 | 0.171 |
| 0.496 | 0.104 | 0.078 | 0.115 |
| 0.506 | 0.094 | 0.078 | 0.115 |
| 0.498 | 0.102 | 0.095 | 0.106 |
| 0.522 | 0.078 | 0.078 | 0.078 |
| 0.707 | 0.107 | 0.094 | 0.122 |
| 0.485 | 0.115 | 0.115 | 0.115 |
| 0.549 | 0.051 | 0.039 | 0.065 |
| 0.485 | 0.115 | 0.115 | 0.115 |
| 0.584 | 0.016 | 0.000 | 0.115 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.443 | 0.072 | 0.059 | 0.077 |
| 0.496 | 0.059 | 0.048 | 0.085 |
| 0.506 | 0.046 | 0.030 | 0.067 |
| 0.498 | 0.100 | 0.094 | 0.105 |
| 0.472 | 0.040 | 0.040 | 0.040 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.485 | 0.015 | 0.015 | 0.015 |
| 0.549 | 0.057 | 0.045 | 0.071 |
| 0.485 | 0.016 | 0.017 | 0.017 |
| 0.584 | 0.246 | 0.147 | 0.262 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 8.76% | 24.25% | 0.00% |
| 30.20% | 222.22% | 0.00% |
| 82.17% | 102.59% | 41.67% |
| 23.79% | 47.75% | 12.59% |
| 0.00% | 0.00% | 0.00% |
| 19.94% | 43.30% | 6.90% |
| 61.30% | 90.91% | 48.49% |
| 101.06% | 104.85% | 98.55% |
| 64.59% | 71.43% | 43.02% |
| 17.72% | 101.63% | 0.00% |
| 40.95% | 80.89% | 25.12% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 5.07% | 17.62% | 6.17% |
| 54.97% | 268.00% | 7.46% |
| 35.11% | 38.30% | 35.00% |
| 20.47% | 43.25% | 9.96% |
| 70.15% | 195.45% | 6.64% |
| 19.94% | 43.30% | 6.90% |
| 96.40% | 93.11% | 39.04% |
| 141.50% | 151.07% | 121.04% |
| 10.87% | 35.55% | 28.97% |
| 20.72% | 104.07% | 0.00% |
| 47.52% | 98.97% | 26.12% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 169.67% | 188.70% | 150.68% |
| 112.65% | 101.63% | 96.83% |
| 101.25% | 101.63% | 96.83% |
| 164.91% | 175.62% | 136.38% |
| 84.14% | 96.83% | 68.94% |
| 155.25% | 227.95% | 98.90% |
| 124.06% | 142.77% | 101.63% |
| 84.60% | 83.88% | 82.04% |
| 124.06% | 142.77% | 101.63% |
| 17.72% | 101.63% | 0.00% |
| 113.83% | 136.34% | 93.39% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 52.62% | 66.47% | 38.63% |
| 83.32% | 165.53% | 56.17% |
| 89.89% | 102.84% | 57.97% |
| 165.71% | 174.77% | 136.56% |
| 138.55% | 240.96% | 90.70% |
| 19.94% | 43.30% | 6.90% |
| 34.23% | 61.70% | 24.87% |
| 89.38% | 88.20% | 87.68% |
| 37.54% | 66.80% | 27.55% |
| 145.05% | 176.02% | 79.70% |
| 85.62% | 118.66% | 60.67% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.060 | 0.005 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 8 |
| 0.035 | 0.011 | N | N | Y | Y | Y | 2 | 3 | 7 | 9 |
| 0.042 | 0.034 | N | N | N | Y | Y | 1 | 3 | 5 | 9 |
| 0.061 | 0.015 | N | N | Y | Y | Y | 0 | 1 | 3 | 7 |
| 0.032 | 0.000 | Y | Y | Y | Y | Y | 1 | 3 | 7 | 9 |
| 0.111 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.060 | 0.037 | N | N | N | Y | Y | 1 | 1 | 4 | 8 |
| 0.084 | 0.085 | N | N | N | N | Y | 0 | 2 | 2 | 5 |
| 0.029 | 0.019 | N | N | Y | Y | Y | 2 | 6 | 7 | 9 |
| 0.093 | 0.016 | Y | N | Y | Y | Y | 0 | 0 | 0 | 4 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.093 | 0.157 | N | N | N | N | N | 0 | 1 | 1 | 5 |
| 0.093 | 0.104 | N | N | N | N | N | 0 | 1 | 1 | 5 |
| 0.093 | 0.094 | N | N | N | N | Y | 0 | 1 | 1 | 4 |
| 0.062 | 0.102 | N | N | N | N | N | 1 | 2 | 3 | 8 |
| 0.093 | 0.078 | N | N | N | N | Y | 0 | 1 | 1 | 4 |
| 0.069 | 0.107 | N | N | N | N | N | 2 | 2 | 6 | 7 |
| 0.093 | 0.115 | N | N | N | N | N | 0 | 1 | 1 | 5 |
| 0.060 | 0.051 | N | N | N | N | Y | 0 | 1 | 3 | 9 |
| 0.093 | 0.115 | N | N | N | N | N | 0 | 1 | 1 | 5 |
| 0.093 | 0.016 | Y | N | Y | Y | Y | 0 | 0 | 0 | 4 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.136 | 0.072 | Y | N | N | N | Y | 0 | 0 | 0 | 0 |
| 0.070 | 0.059 | N | N | N | N | Y | 1 | 1 | 4 | 7 |
| 0.051 | 0.046 | N | N | N | Y | Y | 0 | 2 | 4 | 8 |
| 0.060 | 0.100 | N | N | N | N | N | 1 | 2 | 3 | 8 |
| 0.029 | 0.040 | N | N | N | Y | Y | 3 | 5 | 7 | 9 |
| 0.111 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.042 | 0.015 | Y | N | Y | Y | Y | 0 | 2 | 5 | 8 |
| 0.064 | 0.057 | N | N | N | N | Y | 1 | 1 | 3 | 8 |
| 0.044 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 5 | 8 |
| 0.169 | 0.246 | N | N | N | N | N | 0 | 0 | 0 | 0 |
Appendix B.3.4. Trials 31–40
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.498 | 0.562 | 0.065 | 0.081 | 0.041 |
| 0.535 | 0.562 | 0.035 | 0.052 | 0.015 |
| 0.591 | 0.562 | 0.046 | 0.059 | 0.044 |
| 0.517 | 0.565 | 0.053 | 0.087 | 0.017 |
| 0.572 | 0.562 | 0.036 | 0.052 | 0.030 |
| 0.628 | 0.564 | 0.074 | 0.091 | 0.069 |
| 0.498 | 0.562 | 0.065 | 0.081 | 0.041 |
| 0.806 | 0.564 | 0.243 | 0.265 | 0.226 |
| 0.554 | 0.562 | 0.033 | 0.048 | 0.026 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.370 | 0.517 | 0.147 | 0.190 | 0.091 |
| 0.439 | 0.516 | 0.089 | 0.134 | 0.045 |
| 0.560 | 0.517 | 0.085 | 0.125 | 0.061 |
| 0.646 | 0.517 | 0.136 | 0.192 | 0.101 |
| 0.431 | 0.517 | 0.097 | 0.140 | 0.048 |
| 0.487 | 0.517 | 0.058 | 0.100 | 0.062 |
| 0.583 | 0.517 | 0.098 | 0.141 | 0.072 |
| 0.700 | 0.527 | 0.173 | 0.224 | 0.133 |
| 0.466 | 0.517 | 0.069 | 0.112 | 0.054 |
| 0.700 | 0.517 | 0.183 | 0.242 | 0.140 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.567 | 0.133 | 0.156 | 0.116 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| 0.700 | 0.562 | 0.138 | 0.163 | 0.123 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.371 | 0.562 | 0.191 | 0.206 | 0.166 |
| 0.437 | 0.562 | 0.125 | 0.140 | 0.100 |
| 0.552 | 0.562 | 0.033 | 0.048 | 0.025 |
| 0.646 | 0.562 | 0.091 | 0.109 | 0.083 |
| 0.432 | 0.562 | 0.130 | 0.145 | 0.105 |
| 0.490 | 0.562 | 0.072 | 0.088 | 0.049 |
| 0.471 | 0.562 | 0.091 | 0.107 | 0.066 |
| 0.696 | 0.567 | 0.130 | 0.152 | 0.113 |
| 0.469 | 0.562 | 0.093 | 0.109 | 0.068 |
| 0.338 | 0.562 | 0.224 | 0.239 | 0.199 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.493 | 0.005 | 0.000 | 0.019 |
| 0.546 | 0.011 | 0.000 | 0.037 |
| 0.556 | 0.034 | 0.019 | 0.056 |
| 0.535 | 0.019 | 0.019 | 0.019 |
| 0.572 | 0.000 | 0.000 | 0.000 |
| 0.578 | 0.051 | 0.037 | 0.112 |
| 0.535 | 0.037 | 0.037 | 0.037 |
| 0.587 | 0.220 | 0.217 | 0.223 |
| 0.535 | 0.019 | 0.019 | 0.019 |
| 0.676 | 0.024 | 0.000 | 0.165 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.376 | 0.007 | 0.011 | 0.016 |
| 0.487 | 0.048 | 0.009 | 0.121 |
| 0.531 | 0.029 | 0.019 | 0.047 |
| 0.665 | 0.019 | 0.019 | 0.019 |
| 0.488 | 0.057 | 0.008 | 0.095 |
| 0.477 | 0.011 | 0.095 | 0.141 |
| 0.496 | 0.086 | 0.025 | 0.131 |
| 0.478 | 0.222 | 0.196 | 0.231 |
| 0.508 | 0.042 | 0.027 | 0.069 |
| 0.663 | 0.037 | 0.000 | 0.261 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.493 | 0.207 | 0.202 | 0.221 |
| 0.546 | 0.154 | 0.128 | 0.165 |
| 0.556 | 0.144 | 0.128 | 0.165 |
| 0.535 | 0.165 | 0.165 | 0.165 |
| 0.572 | 0.128 | 0.128 | 0.128 |
| 0.578 | 0.123 | 0.035 | 0.184 |
| 0.535 | 0.165 | 0.165 | 0.165 |
| 0.587 | 0.113 | 0.110 | 0.116 |
| 0.535 | 0.165 | 0.165 | 0.165 |
| 0.676 | 0.024 | 0.000 | 0.165 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.493 | 0.122 | 0.109 | 0.127 |
| 0.546 | 0.109 | 0.098 | 0.135 |
| 0.556 | 0.004 | 0.017 | 0.020 |
| 0.535 | 0.111 | 0.111 | 0.111 |
| 0.572 | 0.140 | 0.140 | 0.140 |
| 0.578 | 0.088 | 0.027 | 0.176 |
| 0.535 | 0.065 | 0.065 | 0.065 |
| 0.587 | 0.109 | 0.107 | 0.113 |
| 0.535 | 0.067 | 0.067 | 0.067 |
| 0.676 | 0.338 | 0.197 | 0.362 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 8.18% | 22.76% | 0.00% |
| 30.17% | 250.00% | 0.00% |
| 74.53% | 93.91% | 41.67% |
| 35.04% | 111.11% | 21.28% |
| 0.00% | 0.00% | 0.00% |
| 68.26% | 122.33% | 53.90% |
| 57.24% | 90.91% | 45.51% |
| 90.44% | 95.88% | 84.04% |
| 56.18% | 71.43% | 38.54% |
| 17.03% | 101.13% | 0.00% |
| 43.71% | 95.95% | 28.49% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 4.55% | 17.62% | 5.52% |
| 54.18% | 268.00% | 6.34% |
| 34.22% | 37.72% | 30.35% |
| 13.59% | 18.34% | 9.66% |
| 59.29% | 195.45% | 5.70% |
| 18.10% | 227.05% | 94.67% |
| 88.09% | 93.11% | 34.10% |
| 128.37% | 146.80% | 103.19% |
| 60.37% | 127.19% | 24.02% |
| 20.36% | 107.94% | 0.00% |
| 48.11% | 123.92% | 31.36% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 149.76% | 164.83% | 135.15% |
| 111.57% | 104.45% | 101.13% |
| 103.93% | 104.45% | 101.13% |
| 119.21% | 134.64% | 101.13% |
| 92.48% | 104.45% | 78.46% |
| 88.50% | 112.47% | 28.56% |
| 119.21% | 134.64% | 101.13% |
| 84.80% | 94.86% | 74.60% |
| 119.21% | 134.64% | 101.13% |
| 17.03% | 101.13% | 0.00% |
| 100.57% | 119.06% | 82.24% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 63.86% | 76.58% | 52.56% |
| 87.15% | 135.20% | 69.78% |
| 12.58% | 80.00% | 35.20% |
| 122.65% | 133.49% | 101.69% |
| 108.04% | 133.52% | 96.25% |
| 122.08% | 356.71% | 30.65% |
| 70.81% | 97.21% | 60.31% |
| 84.36% | 94.59% | 73.97% |
| 71.44% | 97.29% | 61.04% |
| 151.36% | 182.05% | 82.27% |
| 89.43% | 138.66% | 66.37% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.065 | 0.005 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 8 |
| 0.035 | 0.011 | N | N | Y | Y | Y | 3 | 4 | 7 | 8 |
| 0.046 | 0.034 | N | N | N | Y | Y | 1 | 3 | 4 | 9 |
| 0.053 | 0.019 | Y | N | Y | Y | Y | 0 | 2 | 4 | 8 |
| 0.036 | 0.000 | Y | Y | Y | Y | Y | 1 | 3 | 7 | 8 |
| 0.074 | 0.051 | N | N | N | N | Y | 0 | 0 | 2 | 8 |
| 0.065 | 0.037 | N | N | N | Y | Y | 1 | 1 | 4 | 8 |
| 0.243 | 0.220 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.033 | 0.019 | N | N | Y | Y | Y | 2 | 6 | 7 | 8 |
| 0.138 | 0.024 | Y | N | Y | Y | Y | 0 | 0 | 0 | 0 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.147 | 0.007 | Y | Y | Y | Y | Y | 0 | 0 | 0 | 1 |
| 0.089 | 0.048 | N | N | N | Y | Y | 0 | 0 | 3 | 7 |
| 0.085 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 6 |
| 0.136 | 0.019 | N | N | Y | Y | Y | 0 | 1 | 1 | 1 |
| 0.097 | 0.057 | N | N | N | N | Y | 0 | 0 | 2 | 6 |
| 0.058 | 0.011 | N | N | Y | Y | Y | 2 | 5 | 6 | 6 |
| 0.098 | 0.086 | N | N | N | N | Y | 0 | 0 | 0 | 6 |
| 0.173 | 0.222 | N | N | N | N | N | 0 | 0 | 1 | 2 |
| 0.069 | 0.042 | N | N | N | Y | Y | 1 | 3 | 5 | 7 |
| 0.183 | 0.037 | N | N | N | Y | Y | 0 | 0 | 1 | 1 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.138 | 0.207 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.154 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.144 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.165 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.128 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.123 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.165 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.133 | 0.113 | N | N | N | N | N | 0 | 0 | 1 | 1 |
| 0.138 | 0.165 | N | N | N | N | N | 0 | 1 | 1 | 1 |
| 0.138 | 0.024 | Y | N | Y | Y | Y | 0 | 0 | 0 | 0 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.191 | 0.122 | Y | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.125 | 0.109 | N | N | N | N | N | 0 | 0 | 0 | 4 |
| 0.033 | 0.004 | Y | Y | Y | Y | Y | 1 | 5 | 7 | 8 |
| 0.091 | 0.111 | N | N | N | N | N | 0 | 0 | 1 | 5 |
| 0.130 | 0.140 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| 0.072 | 0.088 | N | N | N | N | Y | 1 | 1 | 4 | 7 |
| 0.091 | 0.065 | N | N | N | N | Y | 0 | 1 | 1 | 5 |
| 0.130 | 0.109 | N | N | N | N | N | 0 | 0 | 1 | 1 |
| 0.093 | 0.067 | N | N | N | N | Y | 0 | 1 | 1 | 5 |
| 0.224 | 0.338 | N | N | N | N | N | 0 | 0 | 0 | 0 |
Appendix B.3.5. Trials 41–50
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.423 | 0.480 | 0.058 | 0.074 | 0.041 |
| 0.460 | 0.480 | 0.029 | 0.044 | 0.015 |
| 0.720 | 0.603 | 0.135 | 0.161 | 0.110 |
| 0.496 | 0.571 | 0.075 | 0.095 | 0.064 |
| 0.497 | 0.480 | 0.030 | 0.044 | 0.030 |
| 0.685 | 0.603 | 0.112 | 0.138 | 0.086 |
| 0.664 | 0.603 | 0.099 | 0.125 | 0.074 |
| 0.563 | 0.565 | 0.048 | 0.067 | 0.039 |
| 0.479 | 0.480 | 0.027 | 0.041 | 0.026 |
| 0.550 | 0.480 | 0.070 | 0.088 | 0.063 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.370 | 0.494 | 0.125 | 0.160 | 0.091 |
| 0.439 | 0.494 | 0.068 | 0.104 | 0.045 |
| 0.720 | 0.603 | 0.135 | 0.161 | 0.110 |
| 0.550 | 0.575 | 0.048 | 0.070 | 0.039 |
| 0.431 | 0.494 | 0.074 | 0.110 | 0.048 |
| 0.685 | 0.603 | 0.112 | 0.138 | 0.086 |
| 0.664 | 0.603 | 0.099 | 0.125 | 0.074 |
| 0.549 | 0.569 | 0.047 | 0.069 | 0.038 |
| 0.466 | 0.494 | 0.049 | 0.086 | 0.052 |
| 0.550 | 0.494 | 0.078 | 0.115 | 0.047 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.550 | 0.480 | 0.070 | 0.088 | 0.063 |
| 0.550 | 0.480 | 0.070 | 0.088 | 0.063 |
| 0.550 | 0.603 | 0.071 | 0.096 | 0.062 |
| 0.550 | 0.571 | 0.047 | 0.067 | 0.042 |
| 0.550 | 0.480 | 0.070 | 0.088 | 0.063 |
| 0.550 | 0.603 | 0.071 | 0.096 | 0.062 |
| 0.550 | 0.603 | 0.071 | 0.096 | 0.062 |
| 0.550 | 0.565 | 0.045 | 0.064 | 0.039 |
| 0.550 | 0.480 | 0.070 | 0.088 | 0.063 |
| 0.550 | 0.480 | 0.070 | 0.088 | 0.063 |
| Actual | Average | Average Error | Max Error | Min Error |
|---|---|---|---|---|
| 0.371 | 0.480 | 0.109 | 0.124 | 0.091 |
| 0.437 | 0.480 | 0.047 | 0.063 | 0.030 |
| 0.712 | 0.603 | 0.129 | 0.155 | 0.105 |
| 0.547 | 0.571 | 0.047 | 0.067 | 0.042 |
| 0.432 | 0.480 | 0.051 | 0.067 | 0.034 |
| 0.685 | 0.603 | 0.112 | 0.138 | 0.086 |
| 0.642 | 0.603 | 0.086 | 0.112 | 0.065 |
| 0.561 | 0.565 | 0.047 | 0.067 | 0.039 |
| 0.469 | 0.480 | 0.027 | 0.043 | 0.020 |
| 0.338 | 0.480 | 0.142 | 0.157 | 0.124 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.418 | 0.005 | 0.000 | 0.019 |
| 0.471 | 0.011 | 0.000 | 0.037 |
| 0.816 | 0.096 | 0.081 | 0.102 |
| 0.506 | 0.010 | 0.001 | 0.024 |
| 0.497 | 0.000 | 0.000 | 0.000 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.641 | 0.023 | 0.001 | 0.048 |
| 0.545 | 0.018 | 0.002 | 0.038 |
| 0.460 | 0.019 | 0.019 | 0.019 |
| 0.537 | 0.013 | 0.000 | 0.090 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.376 | 0.007 | 0.011 | 0.016 |
| 0.477 | 0.038 | 0.009 | 0.121 |
| 0.816 | 0.096 | 0.081 | 0.102 |
| 0.561 | 0.010 | 0.001 | 0.024 |
| 0.488 | 0.057 | 0.008 | 0.095 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.641 | 0.023 | 0.001 | 0.048 |
| 0.530 | 0.019 | 0.000 | 0.040 |
| 0.454 | 0.012 | 0.006 | 0.027 |
| 0.534 | 0.016 | 0.000 | 0.111 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.418 | 0.132 | 0.127 | 0.146 |
| 0.471 | 0.079 | 0.053 | 0.090 |
| 0.816 | 0.266 | 0.250 | 0.272 |
| 0.506 | 0.044 | 0.030 | 0.053 |
| 0.497 | 0.053 | 0.053 | 0.053 |
| 0.707 | 0.157 | 0.144 | 0.172 |
| 0.641 | 0.091 | 0.067 | 0.114 |
| 0.545 | 0.005 | 0.011 | 0.025 |
| 0.460 | 0.090 | 0.090 | 0.090 |
| 0.537 | 0.013 | 0.000 | 0.090 |
| Test Avg | Avg Err | Min | Max |
|---|---|---|---|
| 0.418 | 0.047 | 0.034 | 0.052 |
| 0.471 | 0.034 | 0.023 | 0.060 |
| 0.816 | 0.104 | 0.088 | 0.110 |
| 0.506 | 0.041 | 0.027 | 0.050 |
| 0.497 | 0.065 | 0.065 | 0.065 |
| 0.707 | 0.022 | 0.009 | 0.037 |
| 0.641 | 0.000 | 0.022 | 0.025 |
| 0.545 | 0.017 | 0.000 | 0.036 |
| 0.460 | 0.009 | 0.009 | 0.009 |
| 0.537 | 0.199 | 0.122 | 0.212 |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 9.08% | 25.07% | 0.00% |
| 36.94% | 250.00% | 0.00% |
| 71.04% | 92.81% | 49.98% |
| 13.74% | 37.74% | 0.90% |
| 0.00% | 0.00% | 0.00% |
| 19.81% | 42.95% | 6.90% |
| 22.76% | 37.88% | 0.68% |
| 37.99% | 56.12% | 4.37% |
| 69.81% | 71.43% | 45.68% |
| 18.41% | 102.10% | 0.00% |
| 29.96% | 71.61% | 10.85% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 5.38% | 17.62% | 6.56% |
| 55.50% | 268.00% | 8.17% |
| 71.04% | 92.81% | 49.98% |
| 21.71% | 61.68% | 1.23% |
| 77.23% | 195.45% | 7.25% |
| 19.81% | 42.95% | 6.90% |
| 22.76% | 37.88% | 0.68% |
| 40.77% | 57.91% | 1.20% |
| 24.59% | 31.51% | 11.51% |
| 20.20% | 96.77% | 0.00% |
| 35.90% | 90.26% | 9.35% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 189.40% | 201.43% | 165.06% |
| 113.72% | 102.10% | 84.06% |
| 373.36% | 436.85% | 261.23% |
| 93.01% | 79.21% | 72.00% |
| 75.88% | 84.06% | 60.12% |
| 220.16% | 275.95% | 150.47% |
| 128.40% | 182.62% | 69.49% |
| 11.54% | 38.25% | 29.07% |
| 128.86% | 142.74% | 102.10% |
| 18.41% | 102.10% | 0.00% |
| 135.27% | 164.53% | 99.36% |
| % Err Avg | % Max Avg | % Min Avg |
|---|---|---|
| 42.80% | 57.24% | 27.03% |
| 71.41% | 203.39% | 36.74% |
| 80.13% | 103.89% | 56.76% |
| 86.39% | 74.60% | 63.97% |
| 127.42% | 194.03% | 97.60% |
| 19.81% | 42.95% | 6.90% |
| 0.17% | 33.98% | 22.34% |
| 34.99% | 54.09% | 0.10% |
| 31.58% | 42.71% | 20.00% |
| 140.09% | 171.17% | 77.73% |
| 63.48% | 97.81% | 40.92% |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.125 | 0.007 | Y | Y | Y | Y | Y | 0 | 0 | 0 | 3 |
| 0.068 | 0.038 | N | N | N | Y | Y | 0 | 1 | 5 | 8 |
| 0.135 | 0.096 | N | N | N | N | Y | 0 | 1 | 1 | 2 |
| 0.048 | 0.010 | N | N | Y | Y | Y | 1 | 5 | 6 | 8 |
| 0.074 | 0.057 | N | N | N | N | Y | 0 | 1 | 4 | 6 |
| 0.112 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.099 | 0.023 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.047 | 0.019 | N | N | Y | Y | Y | 1 | 4 | 6 | 8 |
| 0.049 | 0.012 | N | N | Y | Y | Y | 2 | 5 | 5 | 8 |
| 0.078 | 0.016 | Y | N | Y | Y | Y | 0 | 1 | 1 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.070 | 0.132 | N | N | N | N | N | 0 | 1 | 3 | 9 |
| 0.070 | 0.079 | N | N | N | N | Y | 0 | 1 | 3 | 8 |
| 0.071 | 0.266 | N | N | N | N | N | 3 | 3 | 5 | 8 |
| 0.047 | 0.044 | N | N | N | Y | Y | 3 | 5 | 6 | 7 |
| 0.070 | 0.053 | N | N | N | N | Y | 0 | 1 | 3 | 8 |
| 0.071 | 0.157 | N | N | N | N | N | 3 | 3 | 5 | 8 |
| 0.071 | 0.091 | N | N | N | N | Y | 3 | 3 | 5 | 7 |
| 0.045 | 0.005 | Y | Y | Y | Y | Y | 3 | 3 | 6 | 8 |
| 0.070 | 0.090 | N | N | N | N | Y | 0 | 1 | 3 | 8 |
| 0.070 | 0.013 | Y | N | Y | Y | Y | 0 | 0 | 2 | 8 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.109 | 0.047 | Y | N | N | Y | Y | 0 | 0 | 0 | 4 |
| 0.047 | 0.034 | N | N | N | Y | Y | 0 | 4 | 5 | 8 |
| 0.129 | 0.104 | N | N | N | N | N | 1 | 1 | 1 | 2 |
| 0.047 | 0.041 | N | N | N | Y | Y | 3 | 5 | 6 | 7 |
| 0.051 | 0.065 | N | N | N | N | Y | 0 | 1 | 6 | 8 |
| 0.112 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.086 | 0.000 | Y | Y | Y | Y | Y | 0 | 0 | 2 | 6 |
| 0.047 | 0.017 | N | N | Y | Y | Y | 1 | 4 | 5 | 8 |
| 0.027 | 0.009 | N | Y | Y | Y | Y | 3 | 4 | 7 | 9 |
| 0.142 | 0.199 | N | N | N | N | N | 0 | 0 | 0 | 1 |
References
- James, J.T. A New, Evidence-based Estimate of Patient Harms Associated with Hospital Care. J. Patient Saf. 2013, 9, 122–128. [Google Scholar] [CrossRef]
- Ponemon Institute. The Real Cost of Patient Misidentification; Ponemon Institute: Traverse City, MI, USA, 2016. [Google Scholar]
- Ferguson, C.; Hickman, L.; Macbean, C.; Jackson, D. The wicked problem of patient misidentification: How could the technological revolution help address patient safety? J. Clin. Nurs. 2018. [Google Scholar] [CrossRef] [PubMed]
- Kulju, S.; Morrish, W.; King, L.; Bender, J.; Gunnar, W. Patient Misidentification Events in the Veterans Health Administration: A Comprehensive Review in the Context of High-Reliability Health Care. J. Patient Saf. 2022, 18, E290–E296. [Google Scholar] [CrossRef] [PubMed]
- Mannos, D. NCPS Patient Misidentification Study: A Summary of Root Cause Analyses. Top. Patient Saf. 2003, 3. [Google Scholar] [CrossRef]
- Dawood, M.; Matthews, W. Patient misidentification and its ramifications. Divers. Equal. Health Care 2014, 11, 161–162. [Google Scholar]
- O’Neill, K.A.; Shinn, D.; Starr, K.T.; Kelley, J. Patient misidentification in a pediatric emergency department: Patient safety and legal perspectives. Pediatr. Emerg. Care 2004, 20, 487–492. [Google Scholar] [CrossRef]
- Ortiz, J.; Amatucci, C. A case of mistaken identity: Staff input on patient id errors. Nurs. Manag. 2009, 40, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Dunn, E.J.; Moga, P.J. Patient Misidentification in Laboratory Medicine: A Qualitative Analysis of 227 Root Cause Analysis Reports in the Veterans Health Administration. Arch. Pathol. Lab. Med. 2010, 134, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E.; Suresh, G.; Ursprung, R.; Edwards, W.H.; Nickerson, J.; Shiono, P.H.; Plsek, P.; Goldmann, D.A.; Horbar, J. Patient Misidentification in the Neonatal Intensive Care Unit: Quantification of Risk. Pediatrics 2006, 117, e43–e47. [Google Scholar] [CrossRef]
- Bittle, M.J.; Charache, P.; Wassilchalk, D.M. Registration-Associated Patient Misidentification in an Academic Medical Center: Causes and Corrections. Jt. Comm. J. Qual. Patient Saf. 2007, 33, 25–33. [Google Scholar] [CrossRef]
- Levin, H.I.; Levin, J.E.; Docimo, S.G. “I meant that med for Baylee not Bailey!”: A mixed method study to identify incidence and risk factors for CPOE patient misidentification. AMIA Annu. Symp. Proc. 2012, 2012, 1294. [Google Scholar]
- Tase, T.H.; Quadrado, E.R.S.; Tronchin, D.M.R. Evaluation of the risk of misidentification of women in a public maternity hospital. Rev. Bras. Enferm. 2018, 71, 120–125. [Google Scholar] [CrossRef]
- Abraham, P.; Augey, L.; Duclos, A.; Michel, P.; Piriou, V. Descriptive Analysis of Patient Misidentification from Incident Report System Data in a Large Academic Hospital Federation. J. Patient Saf. 2021, 17, e615–e621. [Google Scholar] [CrossRef]
- Spruill, A.; Eron, B.; Coghill, A.; Talbert, G. Decreasing Patient Misidentification Before Chemotherapy Administration. Clin. J. Oncol. Nurs. 2009, 13, 716–717. [Google Scholar] [CrossRef][Green Version]
- Ursprung, R.; Gray, J.E.; Edwards, W.H.; Horbar, J.D.; Nickerson, J.; Plsek, P.; Shiono, P.H.; Suresh, G.K.; Goldmann, D.A. Real time patient safety audits: Improving safety every day. BMJ Qual. Saf. 2005, 14, 284–289. [Google Scholar] [CrossRef]
- Fukami, T.; Uemura, M.; Terai, M.; Umemura, T.; Maeda, M.; Ichikawa, M.; Sawai, N.; Kitano, F.; Nagao, Y. Intervention efficacy for eliminating patient misidentification using step-by-step problem-solving procedures to improve patient safety. Nagoya J. Med. Sci. 2020, 82, 315. [Google Scholar] [CrossRef]
- Sandhu, P.; Bandyopadhyay, K.; Ernst, D.J.; Hunt, W.; Thomas, H.; Taylor, J.; Birch, R.; Krolak, J.; Geaghan, S. Effectiveness of Laboratory Practices to Reducing Patient Misidentification Due to Specimen Labeling Errors at the Time of Specimen Collection in Healthcare Settings: LMBPTM Systematic Review. J. Appl. Lab. Med. 2017, 2, 244. [Google Scholar] [CrossRef]
- Lutoschkin, A.; Peters, A.; Grosswiler, R.; Siegrist, R.; Käser, Y.; Kubik-Huch, R.A.; Niemann, T. Improved Patient Safety in Computed Tomography through Avoidance of Patient Misidentification by Implementation of New Identification Wristbands. Available online: https://epos.myesr.org/poster/esr/ecr2019/C-0771 (accessed on 23 July 2022).
- Kawauchi, K.; Hirata, K.; Katoh, C.; Ichikawa, S.; Manabe, O.; Kobayashi, K.; Watanabe, S.; Furuya, S.; Shiga, T. A convolutional neural network-based system to prevent patient misidentification in FDG-PET examinations. Sci. Rep. 2019, 9, 7192. [Google Scholar] [CrossRef]
- Aguilar, A.; van der Putten, W.; Maguire, G. Positive Patient Identification using RFID and Wireless Networks. In Proceedings of the HISI 11th Annual Conference and Scientific Symposium, Dublin, Ireland, November 2006. [Google Scholar]
- Natarajan, S.; Wottawa, C.R.; Dutson, E.P. Minimization of Patient Misidentification through Proximity-Based Medical Record Retrieval. In Proceedings of the 2009 ICME International Conference on Complex Medical Engineering, Tempe, AZ, USA, 9–11 April 2009. [Google Scholar] [CrossRef]
- Malhotra, D.K.; Malhotra, K.; Malhotra, R. Evaluating Consumer Loans Using Machine Learning Techniques; Emerald Publishing Limited: Bingley, UK, 2020; pp. 59–69. [Google Scholar]
- Gillespie, T. Content moderation, AI, and the question of scale. Big Data Soc. 2020, 7, 205395172094323. [Google Scholar] [CrossRef]
- Coppersmith, G.; Leary, R.; Crutchley, P.; Fine, A. Natural Language Processing of Social Media as Screening for Suicide Risk. Biomed. Inform. Insights 2018, 10, 1178222618792860. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Liang, W. CNN-RNN Based Intelligent Recommendation for Online Medical Pre-Diagnosis Support. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 18, 912–921. [Google Scholar] [CrossRef]
- Barlow, H.B. Unsupervised Learning. Neural. Comput. 1989, 1, 295–311. [Google Scholar] [CrossRef]
- Goldberg, X. Introduction to semi-supervised learning. Synth. Lect. Artif. Intell. Mach. Learn. 2009, 6, 1–116. [Google Scholar] [CrossRef]
- Caruana, R.; Niculescu-Mizil, A. An Empirical Comparison of Supervised Learning Algorithms. In Proceedings of the ACM International Conference Proceeding Series; ACM Press: New York, NY, USA, 2006; Volume 148, pp. 161–168. [Google Scholar]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2016, arXiv:1609.04747. [Google Scholar]
- Rojas, R. The Backpropagation Algorithm. In Neural Networks; Springer: Berlin/Heidelberg, Germany, 1996; pp. 149–182. [Google Scholar]
- Zwass, V. Expert System. Available online: https://www.britannica.com/technology/expert-system (accessed on 24 February 2021).
- Lindsay, R.K.; Buchanan, B.G.; Feigenbaum, E.A.; Lederberg, J. DENDRAL: A case study of the first expert system for scientific hypothesis formation. Artif. Intell. 1993, 61, 209–261. [Google Scholar] [CrossRef]
- Waterman, D. A Guide to Expert Systems; Addison-Wesley Pub. Co.: Reading, MA, USA, 1986. [Google Scholar]
- Renders, J.M.; Themlin, J.M. Optimization of Fuzzy Expert Systems Using Genetic Algorithms and Neural Networks. IEEE Trans. Fuzzy Syst. 1995, 3, 300–312. [Google Scholar] [CrossRef]
- Sahin, S.; Tolun, M.R.; Hassanpour, R. Hybrid expert systems: A survey of current approaches and applications. Expert Syst. Appl. 2012, 39, 4609–4617. [Google Scholar] [CrossRef]
- Zadeh, L.A. Fuzzy sets. Inf. Control 1965, 8, 338–353. [Google Scholar] [CrossRef]
- Mitra, S.; Pal, S.K. Neuro-fuzzy expert systems: Relevance, features and methodologies. IETE J. Res. 1996, 42, 335–347. [Google Scholar] [CrossRef]
- Barredo Arrieta, A.; Díaz-Rodríguez, N.; del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Straub, J. Expert system gradient descent style training: Development of a defensible artificial intelligence technique. Knowledge-Based Syst. 2021, 228, 107275. [Google Scholar] [CrossRef]
- Straub, J. Impact of techniques to reduce error in high error rule-based expert system gradient descent networks. J. Intell. Inf. Syst. 2021, 2021, 481–512. [Google Scholar] [CrossRef]
- Straub, J. Assessment of Gradient Descent Trained Rule-Fact Network Expert System Multi-Path Training Technique Performance. Computers 2021, 10, 103. [Google Scholar] [CrossRef]
- Straub, J. Automating the Design and Development of Gradient Descent Trained Expert System Networks. Knowl.-Based Syst. 2022, 254, 109465. [Google Scholar] [CrossRef]
- Liang, X.S.; Straub, J. Deceptive Online Content Detection Using Only Message Characteristics and a Machine Learning Trained Expert System. Sensors 2021, 21, 7083. [Google Scholar] [CrossRef] [PubMed]
- Straub, J. Gradient descent training expert system. Softw. Impacts 2021, 10, 100121. [Google Scholar] [CrossRef]
- Liu, D.; Görges, M.; Jenkins, S.A. University of Queensland vital signs dataset: Development of an accessible repository of anesthesia patient monitoring data for research. Anesth. Analg. 2012, 114, 584–589. [Google Scholar] [CrossRef]
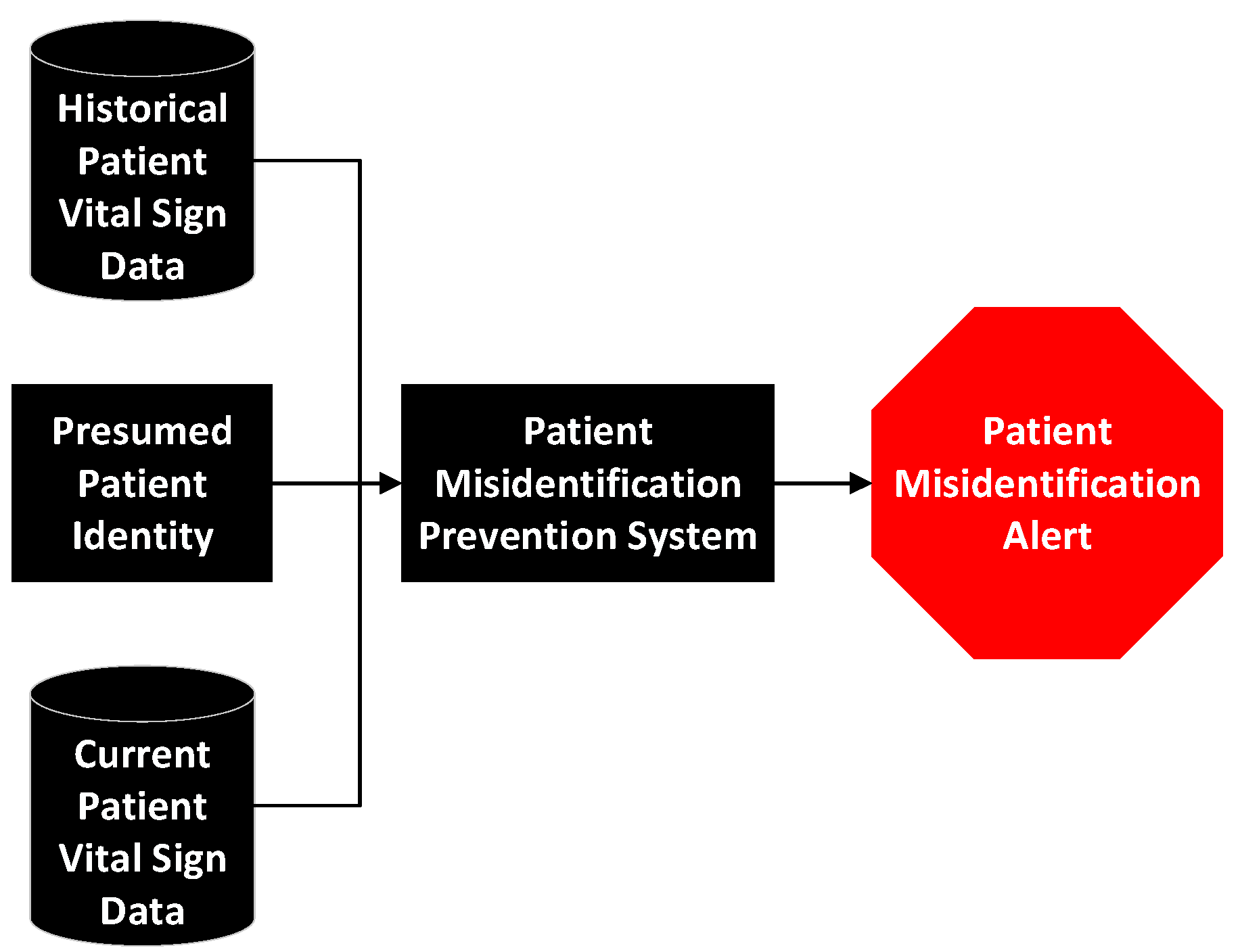

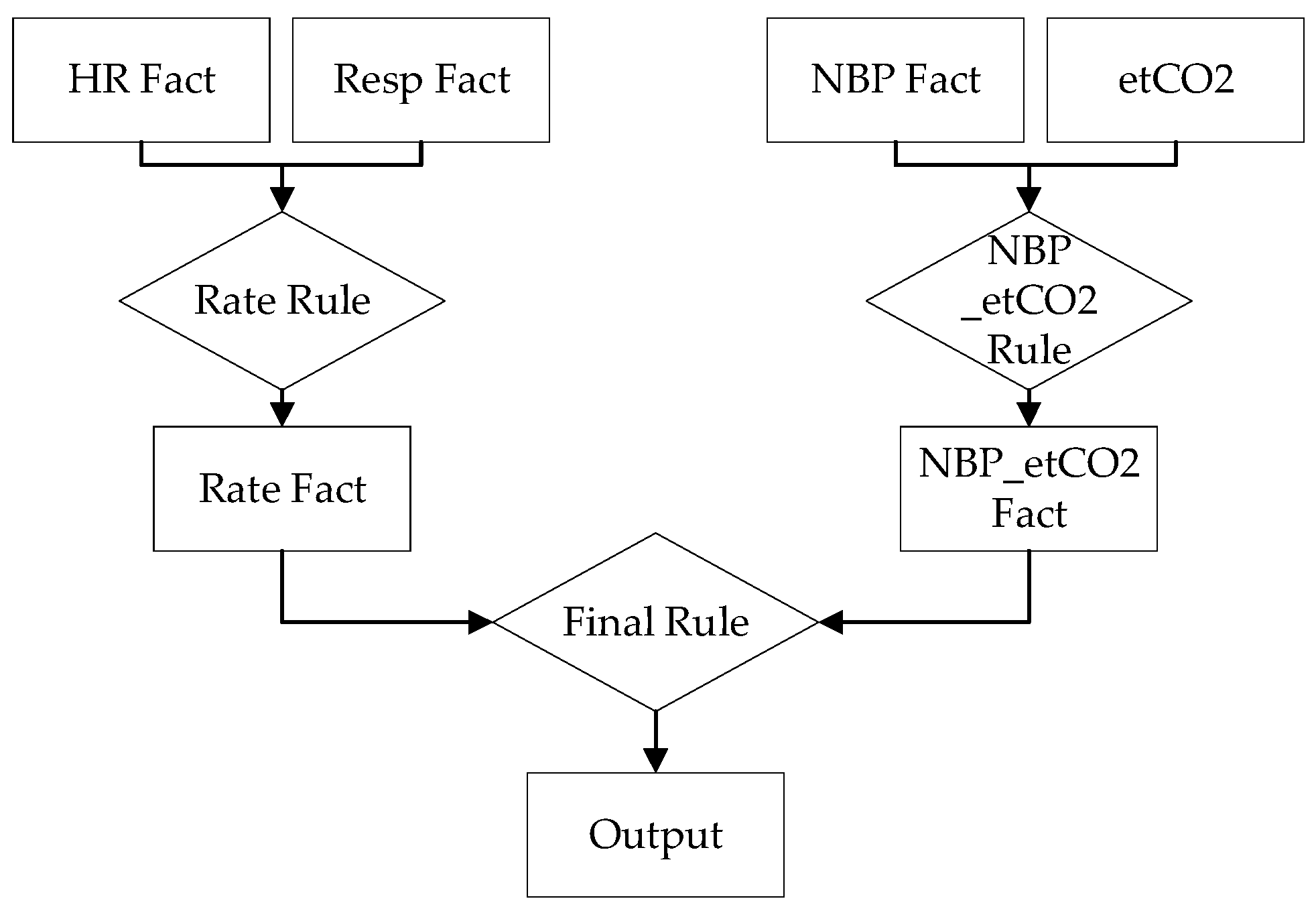
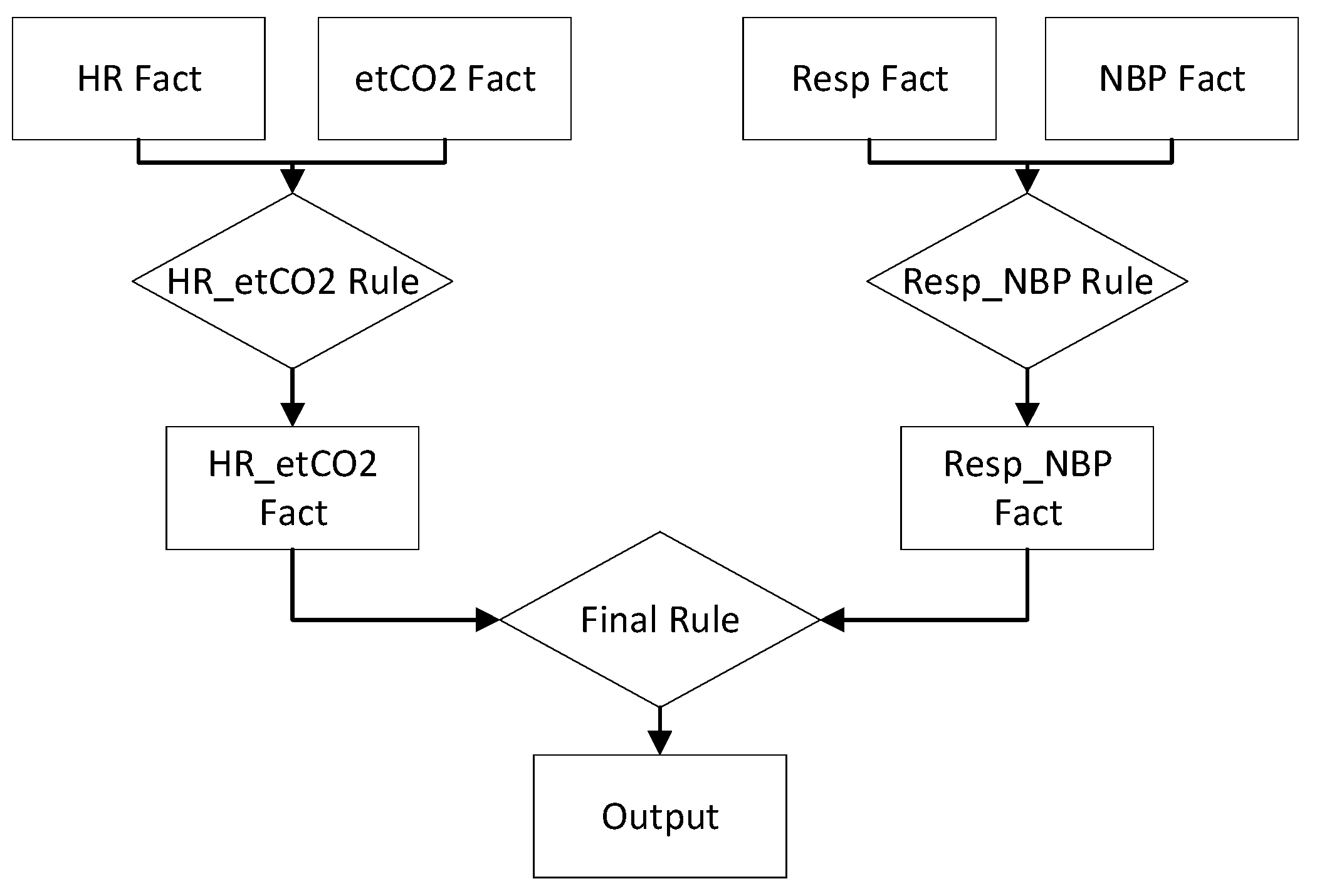
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.037 | 0.012 | N | N | Y | Y | Y | 0 | 5 | 7 | 8 |
| 0.044 | 0.035 | N | N | N | Y | Y | 1 | 5 | 7 | 8 |
| 0.129 | 0.064 | N | N | N | N | Y | 0 | 0 | 0 | 2 |
| 0.064 | 0.010 | N | N | Y | Y | Y | 1 | 2 | 6 | 6 |
| 0.034 | 0.033 | N | N | N | Y | Y | 3 | 5 | 7 | 8 |
| 0.045 | 0.004 | Y | Y | Y | Y | Y | 0 | 2 | 6 | 8 |
| 0.059 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.056 | 0.024 | N | N | Y | Y | Y | 0 | 2 | 5 | 8 |
| 0.050 | 0.002 | Y | Y | Y | Y | Y | 2 | 3 | 5 | 8 |
| 0.153 | 0.100 | N | N | N | N | Y | 0 | 0 | 1 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.199 | 0.165 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.193 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.243 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.218 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.168 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.222 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.216 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.226 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.208 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.204 | 0.111 | Y | N | N | N | N | 0 | 0 | 0 | 0 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.053 | 0.024 | N | N | Y | Y | Y | 1 | 3 | 4 | 8 |
| 0.053 | 0.035 | N | N | N | Y | Y | 0 | 4 | 6 | 8 |
| 0.055 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 4 | 8 |
| 0.076 | 0.016 | Y | N | Y | Y | Y | 0 | 2 | 3 | 7 |
| 0.049 | 0.030 | N | N | N | Y | Y | 0 | 2 | 7 | 8 |
| 0.061 | 0.002 | Y | Y | Y | Y | Y | 0 | 1 | 3 | 7 |
| 0.068 | 0.010 | N | N | Y | Y | Y | 1 | 3 | 3 | 7 |
| 0.062 | 0.085 | N | N | N | N | Y | 2 | 2 | 3 | 7 |
| 0.065 | 0.008 | N | Y | Y | Y | Y | 2 | 3 | 3 | 7 |
| 0.297 | 0.173 | N | N | N | N | N | 0 | 0 | 0 | 1 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.040 | 0.008 | N | Y | Y | Y | Y | 1 | 4 | 7 | 7 |
| 0.038 | 0.022 | N | N | Y | Y | Y | 2 | 5 | 7 | 8 |
| 0.161 | 0.037 | Y | N | N | Y | Y | 0 | 0 | 0 | 1 |
| 0.096 | 0.012 | Y | N | Y | Y | Y | 0 | 1 | 4 | 6 |
| 0.074 | 0.010 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 7 |
| 0.069 | 0.036 | N | N | N | Y | Y | 0 | 3 | 3 | 7 |
| 0.053 | 0.061 | N | N | N | N | Y | 2 | 5 | 6 | 7 |
| 0.321 | 0.305 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.038 | 0.010 | N | Y | Y | Y | Y | 1 | 5 | 7 | 8 |
| 0.060 | 0.007 | N | Y | Y | Y | Y | 1 | 2 | 5 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.402 | 0.407 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.384 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.457 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.468 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.418 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.334 | Y | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.466 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.476 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.458 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.423 | 0.361 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.050 | 0.006 | N | Y | Y | Y | Y | 2 | 3 | 4 | 8 |
| 0.072 | 0.055 | N | N | N | N | Y | 0 | 1 | 5 | 6 |
| 0.159 | 0.029 | Y | N | N | Y | Y | 0 | 0 | 0 | 1 |
| 0.055 | 0.011 | N | N | Y | Y | Y | 2 | 2 | 4 | 7 |
| 0.121 | 0.027 | Y | N | N | Y | Y | 0 | 0 | 1 | 3 |
| 0.053 | 0.009 | N | Y | Y | Y | Y | 1 | 1 | 4 | 8 |
| 0.071 | 0.040 | N | N | N | Y | Y | 0 | 1 | 3 | 7 |
| 0.206 | 0.115 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.042 | 0.002 | N | Y | Y | Y | Y | 1 | 4 | 6 | 8 |
| 0.124 | 0.021 | Y | N | Y | Y | Y | 0 | 0 | 0 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.056 | 0.005 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 9 |
| 0.026 | 0.011 | N | N | Y | Y | Y | 3 | 4 | 7 | 9 |
| 0.133 | 0.096 | N | N | N | N | Y | 0 | 1 | 1 | 2 |
| 0.090 | 0.007 | Y | Y | Y | Y | Y | 1 | 2 | 4 | 6 |
| 0.028 | 0.000 | Y | Y | Y | Y | Y | 1 | 4 | 8 | 9 |
| 0.112 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.100 | 0.023 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.085 | 0.048 | N | N | N | Y | Y | 0 | 2 | 4 | 6 |
| 0.082 | 0.025 | N | N | Y | Y | Y | 1 | 2 | 4 | 6 |
| 0.069 | 0.052 | N | N | N | N | Y | 2 | 2 | 5 | 7 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.056 | 0.002 | Y | Y | Y | Y | Y | 2 | 4 | 6 | 8 |
| 0.054 | 0.009 | Y | Y | Y | Y | Y | 0 | 2 | 7 | 8 |
| 0.058 | 0.062 | N | N | N | N | Y | 1 | 2 | 7 | 8 |
| 0.092 | 0.007 | Y | Y | Y | Y | Y | 0 | 2 | 4 | 6 |
| 0.068 | 0.104 | N | N | N | N | N | 1 | 3 | 7 | 8 |
| 0.054 | 0.027 | N | N | N | Y | Y | 0 | 3 | 7 | 8 |
| 0.054 | 0.012 | N | N | Y | Y | Y | 0 | 2 | 7 | 8 |
| 0.084 | 0.047 | N | N | N | Y | Y | 0 | 1 | 4 | 7 |
| 0.049 | 0.004 | N | Y | Y | Y | Y | 1 | 5 | 7 | 8 |
| 0.093 | 0.198 | N | N | N | N | N | 0 | 1 | 1 | 6 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.396 | 0.357 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.415 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.477 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.464 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.380 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.452 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.437 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.453 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.443 | N | N | N | N | N | 0 | 0 | 0 | 0 |
| 0.416 | 0.164 | Y | N | N | N | N | 0 | 0 | 0 | 0 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.132 | 0.007 | Y | Y | Y | Y | Y | 0 | 0 | 0 | 2 |
| 0.075 | 0.041 | N | N | N | Y | Y | 0 | 0 | 4 | 7 |
| 0.082 | 0.029 | N | N | N | Y | Y | 0 | 1 | 1 | 7 |
| 0.071 | 0.015 | Y | N | Y | Y | Y | 0 | 0 | 3 | 6 |
| 0.082 | 0.057 | N | N | N | N | Y | 0 | 0 | 3 | 6 |
| 0.111 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.097 | 0.094 | N | N | N | N | Y | 1 | 1 | 1 | 5 |
| 0.061 | 0.086 | N | N | N | N | Y | 1 | 2 | 4 | 7 |
| 0.054 | 0.006 | N | Y | Y | Y | Y | 2 | 5 | 5 | 7 |
| 0.111 | 0.023 | Y | N | Y | Y | Y | 0 | 0 | 0 | 2 |
| Average Error | Avg Err | Lowest | Correct | False | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| At 0.01 | At 0.025 | At 0.05 | At 0.1 | At 0.01 | At 0.025 | At 0.05 | At 0.10 | |||
| 0.058 | 0.005 | Y | Y | Y | Y | Y | 0 | 0 | 4 | 8 |
| 0.029 | 0.011 | N | N | Y | Y | Y | 3 | 4 | 7 | 9 |
| 0.135 | 0.096 | N | N | N | N | Y | 0 | 1 | 1 | 2 |
| 0.075 | 0.010 | N | N | Y | Y | Y | 1 | 1 | 3 | 7 |
| 0.030 | 0.000 | Y | Y | Y | Y | Y | 1 | 3 | 8 | 9 |
| 0.112 | 0.022 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.099 | 0.023 | Y | N | Y | Y | Y | 0 | 0 | 1 | 3 |
| 0.048 | 0.018 | N | N | Y | Y | Y | 1 | 4 | 5 | 8 |
| 0.027 | 0.019 | N | N | Y | Y | Y | 2 | 6 | 7 | 9 |
| 0.070 | 0.013 | Y | N | Y | Y | Y | 0 | 0 | 2 | 8 |
| Model (Set) | 1 | 2 | 3 | 3 | 3 | 3 |
|---|---|---|---|---|---|---|
| Default Fact Value | 000.600 | 000.600 | 000.500 | 000.500 | 000.600 | 000.550 |
| PR Approach | Actual PR value | Actual PR value | Single 0.5 PR value | Actual PR value | Actual PR value | Single 0.5 PR value |
| Best Error Margin | 0.01 | 0.01 | 0.025 | 0.01 | 0.025 | 0.025 |
| Correct Patients | 3 | 3 | 7 | 4 | 5 | 9 |
| Avg Incorrect Patients | 0.6 | 0.5 | 1.7 | 0.5 | 0.9 | 1.9 |
| Correct-to-Incorrect Ratio | 5 | 5 | 4.12 | 8 | 5.56 | 4.74 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maul, J.; Straub, J. Assessment of the Use of Patient Vital Sign Data for Preventing Misidentification and Medical Errors. Healthcare 2022, 10, 2440. https://doi.org/10.3390/healthcare10122440
Maul J, Straub J. Assessment of the Use of Patient Vital Sign Data for Preventing Misidentification and Medical Errors. Healthcare. 2022; 10(12):2440. https://doi.org/10.3390/healthcare10122440
Chicago/Turabian StyleMaul, Jared, and Jeremy Straub. 2022. "Assessment of the Use of Patient Vital Sign Data for Preventing Misidentification and Medical Errors" Healthcare 10, no. 12: 2440. https://doi.org/10.3390/healthcare10122440
APA StyleMaul, J., & Straub, J. (2022). Assessment of the Use of Patient Vital Sign Data for Preventing Misidentification and Medical Errors. Healthcare, 10(12), 2440. https://doi.org/10.3390/healthcare10122440






