Artificial Intelligence Applied to Pancreatic Imaging: A Narrative Review
Abstract
:1. Introduction
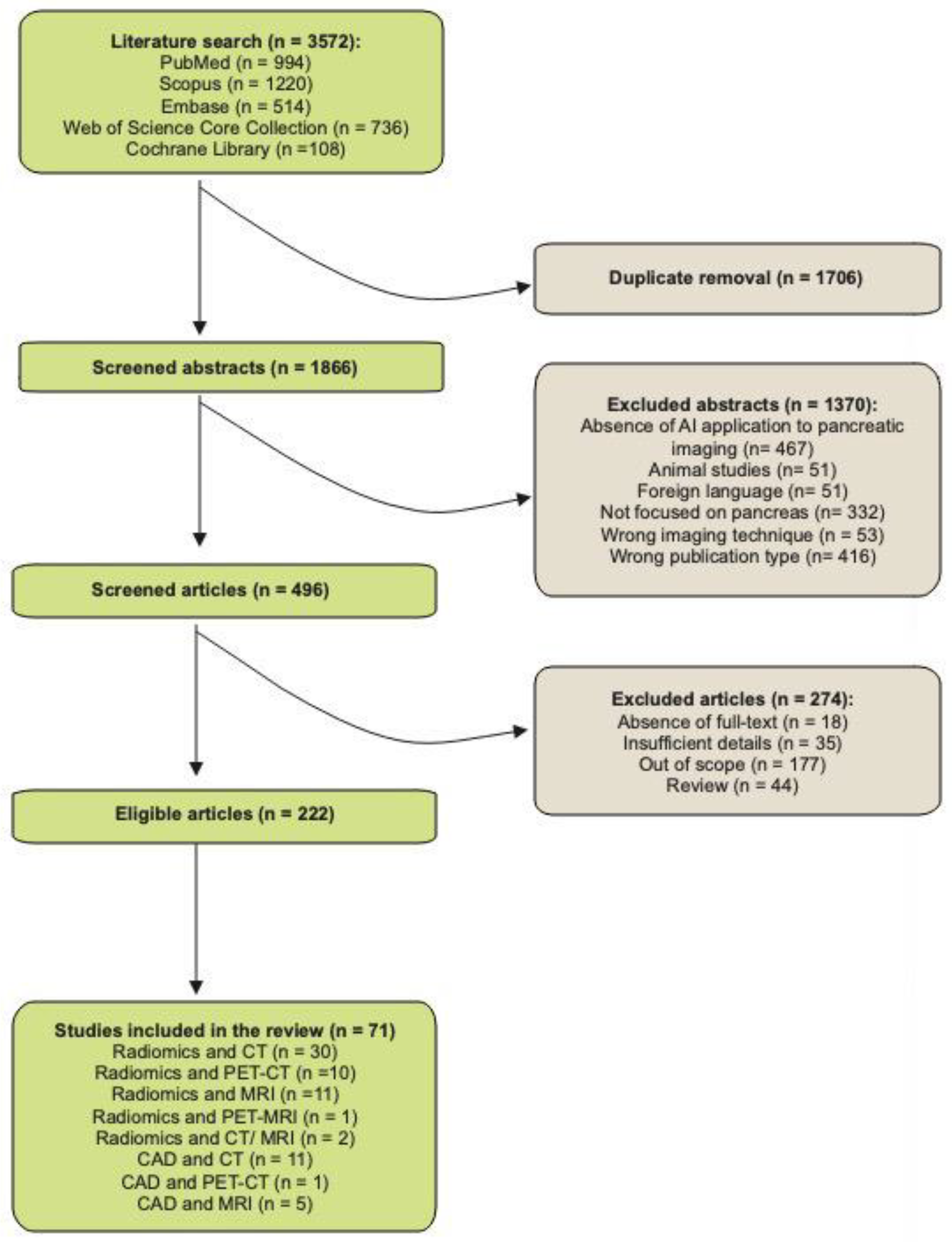
2. Materials and Methods
3. Insights on Radiomics Applied to Pancreatic Imaging
3.1. Radiomics and CT
3.1.1. Oncological Applications
Cystic Lesions
pNET
Adenocarcinoma
3.1.2. Non-Oncological Applications
Pancreatitis
3.2. Radiomics and PET-CT
3.2.1. Oncological Applications
pNET
Adenocarcinoma
3.3. Radiomics and MRI
3.3.1. Oncological Applications
Cystic Lesions
pNET
Adenocarcinoma
3.3.2. Non-Oncological Applications
Pancreatitis
3.4. Radiomics and PET-MRI
Oncological Applications
3.5. Radiomics in Combined CT and MRI Studies
Oncological Applications
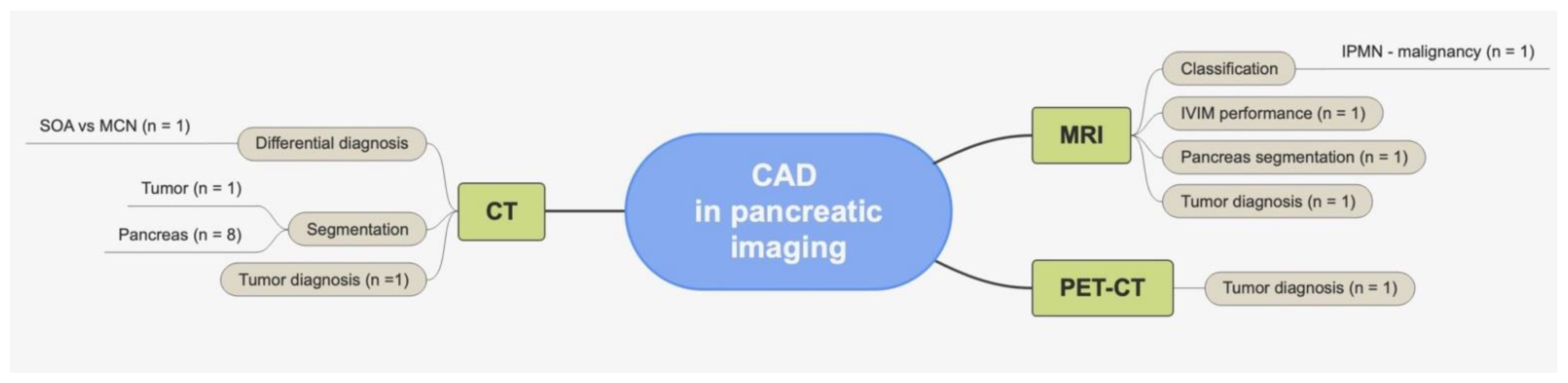
4. Insights on CAD Applied to Pancreatic Imaging
4.1. CAD and CT
4.1.1. Oncological Applications
Cystic Lesions
4.1.2. Non-Oncological Studies
4.2. CAD and PET-CT
4.3. CAD and MRI
4.3.1. Oncological Applications
Cystic Lesions
4.3.2. Non-Oncological Applications
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stevens, K.J.; Lisanti, C. Pancreas Imaging. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Chen, F.-M.; Ni, J.-M.; Zhang, Z.-Y.; Zhang, L.; Li, B.; Jiang, C.-J. Presurgical evaluation of pancreatic cancer: A comprehensive imaging comparison of CT versus MRI. AJR Am. J. Roentgenol. 2016, 206, 526–535. [Google Scholar] [CrossRef]
- Xing, H.; Hao, Z.; Zhu, W.; Sun, D.; Ding, J.; Zhang, H.; Liu, Y.; Huo, L. Preoperative prediction of pathological grade in pancreatic ductal adenocarcinoma based on 18F-FDG PET/CT radiomics. EJNMMI Res. 2021, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Belli, M.L.; Mori, M.; Broggi, S.; Cattaneo, G.M.; Bettinardi, V.; Dell’Oca, I.; Fallanca, F.; Passoni, P.; Vanoli, E.G.; Calandrino, R.; et al. Quantifying the robustness of [18F]FDG-PET/CT radiomic features with respect to tumor delineation in head and neck and pancreatic cancer patients. Phys. Med. 2018, 49, 105–111. [Google Scholar] [CrossRef]
- Santos, M.K.; Ferreira Júnior, J.R.; Wada, D.T.; Tenório, A.P.M.; Barbosa, M.H.N.; Marques, P.M.D.A. Artificial intelligence, machine learning, computer-aided diagnosis, and radiomics: Advances in imaging towards to precision medicine. Radiol. Bras. 2019, 52, 387–396. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Abunahel, B.M.; Pontre, B.; Kumar, H.; Petrov, M.S. Pancreas image mining: A systematic review of radiomics. Eur. Radiol. 2021, 31, 3447–3467. [Google Scholar] [CrossRef] [PubMed]
- Kumar, H.; DeSouza, S.V.; Petrov, M.S. Automated pancreas segmentation from computed tomography and magnetic resonance images: A systematic review. Comput. Methods Programs Biomed. 2019, 178, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, X.; Ou, X.; Zhang, W.; Ma, X. Discrimination of pancreatic serous cystadenomas from mucinous cystadenomas with CT textural features: Based on machine learning. Front. Oncol. 2019, 9, 494. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, X.; Zhang, H.; Zhang, W.; Song, J.; Xu, H.; Ma, X. Differential diagnosis of pancreatic serous cystadenoma and mucinous cystadenoma: Utility of textural features in combination with morphological characteristics. BMC Cancer 2019, 19, 1223. [Google Scholar] [CrossRef]
- Shen, X.; Yang, F.; Yang, P.; Yang, M.; Xu, L.; Zhuo, J.; Wang, J.; Lu, D.; Liu, Z.; Zheng, S.-S.; et al. A Contrast-Enhanced Computed Tomography Based Radiomics Approach for Preoperative Differentiation of Pancreatic Cystic Neoplasm Subtypes: A Feasibility Study. Front. Oncol. 2020, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Lin, K.; Yan, W.; Guo, Y.; Wang, Y.; Li, J.; Zhu, J. Computer-Aided Diagnosis of Pancreas Serous Cystic Neoplasms: A Radiomics Method on Preoperative MDCT Images. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Ma, S.; Guo, X.; Zhang, X.; Wang, X. Preoperative differentiation of pancreatic mucinous cystic neoplasm from macrocystic serous cystic adenoma using radiomics: Preliminary findings and comparison with radiological model. Eur. J. Radiol. 2020, 122, 108747. [Google Scholar] [CrossRef] [PubMed]
- Tobaly, D.; Santinha, J.; Sartoris, R.; Dioguardi Burgio, M.; Matos, C.; Cros, J.; Couvelard, A.; Rebours, V.; Sauvanet, A.; Ronot, M.; et al. CT-Based Radiomics Analysis to Predict Malignancy in Patients with Intraductal Papillary Mucinous Neoplasm (IPMN) of the Pancreas. Cancers 2020, 12, 3089. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ren, S.; Guo, K.; Daniels, M.J.; Wang, Z.; Chen, R. Preoperative differentiation of serous cystic neoplasms from mucin-producing pancreatic cystic neoplasms using a CT-based radiomics nomogram. Abdom. Radiol. 2021, 46, 2637–2646. [Google Scholar] [CrossRef]
- Hanania, A.N.; Bantis, L.E.; Feng, Z.; Wang, H.; Tamm, E.P.; Katz, M.H.; Maitra, A.; Koay, E.J. Quantitative imaging to evaluate malignant potential of IPMNs. Oncotarget 2016, 7, 85776–85784. [Google Scholar] [CrossRef] [PubMed]
- Permuth, J.B.; Choi, J.; Balarunathan, Y.; Kim, J.; Chen, D.-T.; Chen, L.; Orcutt, S.; Doepker, M.P.; Gage, K.; Zhang, G.; et al. Florida Pancreas Collaborative Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget 2016, 7, 85785–85797. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, M.A.; Chakraborty, J.; Doussot, A.; Langdon-Embry, L.; Mainarich, S.; Gönen, M.; Balachandran, V.P.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R.; et al. Survival prediction in pancreatic ductal adenocarcinoma by quantitative computed tomography image analysis. Ann. Surg. Oncol. 2018, 25, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Liu, Z.; Lin, Y.; Wan, J.; Li, J.; Xu, K.; Wang, Y.; Jin, Z.; Tian, J.; Xue, H. Differentiation of atypical non-functional pancreatic neuroendocrine tumor and pancreatic ductal adenocarcinoma using CT based radiomics. Eur. J. Radiol. 2019, 117, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, J.; Liang, P.; Li, A.; Hu, Y.; Shen, Y.; Hu, D.; Li, Z. Differentiation of atypical pancreatic neuroendocrine tumors from pancreatic ductal adenocarcinomas: Using whole-tumor CT texture analysis as quantitative biomarkers. Cancer Med. 2018, 7, 4924–4931. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Huang, Z.; Li, M.; Wei, Y.; Zhang, L.; Yang, C.; Zhang, Y.; Song, B. Differential diagnosis of nonhypervascular pancreatic neuroendocrine neoplasms from pancreatic ductal adenocarcinomas, based on computed tomography radiological features and texture analysis. Acad. Radiol. 2020, 27, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Canellas, R.; Burk, K.S.; Parakh, A.; Sahani, D.V. Prediction of pancreatic neuroendocrine tumor grade based on CT features and texture analysis. AJR Am. J. Roentgenol. 2018, 210, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Reinert, C.P.; Baumgartner, K.; Hepp, T.; Bitzer, M.; Horger, M. Complementary role of computed tomography texture analysis for differentiation of pancreatic ductal adenocarcinoma from pancreatic neuroendocrine tumors in the portal-venous enhancement phase. Abdom. Radiol. 2020, 45, 750–758. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, M.; Ciaravino, V.; Cardobi, N.; De Robertis, R.; Cingarlini, S.; Landoni, L.; Capelli, P.; Bassi, C.; Scarpa, A. CT enhancement and 3D texture analysis of pancreatic neuroendocrine neoplasms. Sci. Rep. 2019, 9, 2176. [Google Scholar] [CrossRef]
- Gu, D.; Hu, Y.; Ding, H.; Wei, J.; Chen, K.; Liu, H.; Zeng, M.; Tian, J. CT radiomics may predict the grade of pancreatic neuroendocrine tumors: A multicenter study. Eur. Radiol. 2019, 29, 6880–6890. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Yang, P.; Huang, R.; Xu, L.; Wang, J.; Liu, W.; Zhang, L.; Wan, D.; Huang, Q.; Lu, Y.; et al. A combined nomogram model to preoperatively predict histologic grade in pancreatic neuroendocrine tumors. Clin. Cancer Res. 2019, 25, 584–594. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhuge, X.; Wang, Z.; Wang, Q.; Sun, K.; Feng, Z.; Chen, X. Textural analysis on contrast-enhanced CT in pancreatic neuroendocrine neoplasms: Association with WHO grade. Abdom. Radiol. 2019, 44, 576–585. [Google Scholar] [CrossRef]
- Ren, S.; Zhao, R.; Cui, W.; Qiu, W.; Guo, K.; Cao, Y.; Duan, S.; Wang, Z.; Chen, R. Computed Tomography-Based Radiomics Signature for the Preoperative Differentiation of Pancreatic Adenosquamous Carcinoma from Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2020, 10, 1618. [Google Scholar] [CrossRef]
- Kaissis, G.A.; Ziegelmayer, S.; Lohöfer, F.K.; Harder, F.N.; Jungmann, F.; Sasse, D.; Muckenhuber, A.; Yen, H.-Y.; Steiger, K.; Siveke, J.; et al. Image-Based Molecular Phenotyping of Pancreatic Ductal Adenocarcinoma. J. Clin. Med. 2020, 9, 724. [Google Scholar] [CrossRef] [PubMed]
- Khalvati, F.; Zhang, Y.; Baig, S.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A. Prognostic value of CT radiomic features in resectable pancreatic ductal adenocarcinoma. Sci. Rep. 2019, 9, 5449. [Google Scholar] [CrossRef]
- Yun, G.; Kim, Y.H.; Lee, Y.J.; Kim, B.; Hwang, J.-H.; Choi, D.J. Tumor heterogeneity of pancreas head cancer assessed by CT texture analysis: Association with survival outcomes after curative resection. Sci. Rep. 2018, 8, 7226. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Wang, X.; Li, M.; Tong, T.; Yu, X.; Zhou, Z. Pancreatic ductal adenocarcinoma: A radiomics nomogram outperforms clinical model and TNM staging for survival estimation after curative resection. Eur. Radiol. 2020, 30, 2513–2524. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, Y.J.; Kim, K.G.; Park, J.S. Preoperative CT texture features predict prognosis after curative resection in pancreatic cancer. Sci. Rep. 2019, 9, 17389. [Google Scholar] [CrossRef] [PubMed]
- Eilaghi, A.; Baig, S.; Zhang, Y.; Zhang, J.; Karanicolas, P.; Gallinger, S.; Khalvati, F.; Haider, M.A. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma—A quantitative analysis. BMC Med. Imaging 2017, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.H.; Li, X.D.; Zhu, H.; Miao, F.; Qian, X.H.; Pan, Z.L.; Lin, X.Z. Resectable pancreatic ductal adenocarcinoma: Association between preoperative CT texture features and metastatic nodal involvement. Cancer Imaging 2020, 20, 17. [Google Scholar] [CrossRef]
- Li, K.; Yao, Q.; Xiao, J.; Li, M.; Yang, J.; Hou, W.; Du, M.; Chen, K.; Qu, Y.; Li, L.; et al. Contrast-enhanced CT radiomics for predicting lymph node metastasis in pancreatic ductal adenocarcinoma: A pilot study. Cancer Imaging 2020, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, T.-W.; Wu, C.-Q.; Lin, Q.; Hu, R.; Xie, C.-L.; Zuo, H.-D.; Wu, J.-L.; Mu, Q.-W.; Fu, Q.-S.; et al. Radiomics model of contrast-enhanced computed tomography for predicting the recurrence of acute pancreatitis. Eur. Radiol. 2019, 29, 4408–4417. [Google Scholar] [CrossRef] [PubMed]
- Mashayekhi, R.; Parekh, V.S.; Faghih, M.; Singh, V.K.; Jacobs, M.A.; Zaheer, A. Radiomic features of the pancreas on CT imaging accurately differentiate functional abdominal pain, recurrent acute pancreatitis, and chronic pancreatitis. Eur. J. Radiol. 2020, 123, 108778. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, C.; Liu, Z.; Wang, L.; Pan, G.; Sun, G.; Chang, Y.; Zuo, C.; Yang, X. Radiomics analysis for the differentiation of autoimmune pancreatitis and pancreatic ductal adenocarcinoma in 18 F-FDG PET/CT. Med. Phys. 2019, 46, 4520–4530. [Google Scholar] [CrossRef]
- Mapelli, P.; Partelli, S.; Salgarello, M.; Doraku, J.; Pasetto, S.; Rancoita, P.M.V.; Muffatti, F.; Bettinardi, V.; Presotto, L.; Andreasi, V.; et al. Dual tracer 68Ga-DOTATOC and 18F-FDG PET/computed tomography radiomics in pancreatic neuroendocrine neoplasms: An endearing tool for preoperative risk assessment. Nucl. Med. Commun. 2020, 41, 896–905. [Google Scholar] [CrossRef]
- Liberini, V.; Rampado, O.; Gallio, E.; De Santi, B.; Ceci, F.; Dionisi, B.; Thuillier, P.; Ciuffreda, L.; Piovesan, A.; Fioroni, F.; et al. 68Ga-DOTATOC PET/CT-Based Radiomic Analysis and PRRT Outcome: A Preliminary Evaluation Based on an Exploratory Radiomic Analysis on Two Patients. Front. Med. 2020, 7, 601853. [Google Scholar] [CrossRef]
- Lim, C.H.; Cho, Y.S.; Choi, J.Y.; Lee, K.-H.; Lee, J.K.; Min, J.H.; Hyun, S.H. Imaging phenotype using 18F-fluorodeoxyglucose positron emission tomography-based radiomics and genetic alterations of pancreatic ductal adenocarcinoma. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2113–2122. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Song, J.; Pollom, E.; Alagappan, M.; Shirato, H.; Chang, D.T.; Koong, A.C.; Li, R. Quantitative Analysis of (18)F-Fluorodeoxyglucose Positron Emission Tomography Identifies Novel Prognostic Imaging Biomarkers in Locally Advanced Pancreatic Cancer Patients Treated with Stereotactic Body Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 102–109. [Google Scholar] [CrossRef]
- Yue, Y.; Osipov, A.; Fraass, B.; Sandler, H.; Zhang, X.; Nissen, N.; Hendifar, A.; Tuli, R. Identifying prognostic intratumor heterogeneity using pre- and post-radiotherapy 18F-FDG PET images for pancreatic cancer patients. J. Gastrointest. Oncol. 2017, 8, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, M.; Zuo, C.; Yang, Z.; Yang, X.; Ren, S.; Peng, Y.; Sun, G.; Shen, J.; Cheng, C.; et al. Radiomics model of dual-time 2-[18F]FDG PET/CT imaging to distinguish between pancreatic ductal adenocarcinoma and autoimmune pancreatitis. Eur. Radiol. 2021, 31, 6983–6991. [Google Scholar] [CrossRef] [PubMed]
- Toyama, Y.; Hotta, M.; Motoi, F.; Takanami, K.; Minamimoto, R.; Takase, K. Prognostic value of FDG-PET radiomics with machine learning in pancreatic cancer. Sci. Rep. 2020, 10, 17024. [Google Scholar] [CrossRef]
- Costache, M.I.; Costache, C.A.; Dumitrescu, C.I.; Tica, A.A.; Popescu, M.; Baluta, E.A.; Anghel, A.C.; Saftoiu, A.; Dumitrescu, D. Which is the Best Imaging Method in Pancreatic Adenocarcinoma Diagnosis and Staging—CT, MRI or EUS? Curr. Health Sci. J. 2017, 43, 132–136. [Google Scholar] [CrossRef]
- Jeon, S.K.; Kim, J.H.; Yoo, J.; Kim, J.-E.; Park, S.J.; Han, J.K. Assessment of malignant potential in intraductal papillary mucinous neoplasms of the pancreas using MR findings and texture analysis. Eur. Radiol. 2021, 31, 3394–3404. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Tang, T.; Su, Q.; Wang, Y.; Shu, Z.; Yang, W.; Gong, X. Radiomic nomogram based on MRI to predict grade of branching type intraductal papillary mucinous neoplasms of the pancreas: A multicenter study. Cancer Imaging 2021, 21, 26. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.-G.; Ren, S.; Chen, X.; Wang, Q.-D.; Xiao, W.-B.; Zhang, J.-F.; Duan, S.-F.; Wang, Z.-Q. Pancreatic neuroendocrine tumor: Prediction of the tumor grade using magnetic resonance imaging findings and texture analysis with 3-T magnetic resonance. Cancer Manag. Res. 2019, 11, 1933–1944. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Zhang, Q.-W.; Duan, S.-F.; Bian, Y.; Hao, Q.; Xing, P.-Y.; Wang, T.-G.; Chen, L.-G.; Ma, C.; Lu, J.-P. MRI-based radiomics approach for differentiation of hypovascular non-functional pancreatic neuroendocrine tumors and solid pseudopapillary neoplasms of the pancreas. BMC Med. Imaging 2021, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, H.; Qian, X.; Chen, N.; Lin, X. MRI texture analysis for differentiating nonfunctional pancreatic neuroendocrine neoplasms from solid pseudopapillary neoplasms of the pancreas. Acad. Radiol. 2020, 27, 815–823. [Google Scholar] [CrossRef] [PubMed]
- Taffel, M.T.; Luk, L.; Ream, J.M.; Rosenkrantz, A.B. Exploratory study of apparent diffusion coefficient histogram metrics in assessing pancreatic malignancy. Can. Assoc. Radiol. J. 2019, 70, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Kaissis, G.; Ziegelmayer, S.; Lohöfer, F.; Algül, H.; Eiber, M.; Weichert, W.; Schmid, R.; Friess, H.; Rummeny, E.; Ankerst, D.; et al. A machine learning model for the prediction of survival and tumor subtype in pancreatic ductal adenocarcinoma from preoperative diffusion-weighted imaging. Eur. Radiol. Exp. 2019, 3, 41. [Google Scholar] [CrossRef] [PubMed]
- Kaissis, G.; Ziegelmayer, S.; Lohöfer, F.; Steiger, K.; Algül, H.; Muckenhuber, A.; Yen, H.-Y.; Rummeny, E.; Friess, H.; Schmid, R.; et al. A machine learning algorithm predicts molecular subtypes in pancreatic ductal adenocarcinoma with differential response to gemcitabine-based versus FOLFIRINOX chemotherapy. PLoS ONE 2019, 14, e0218642. [Google Scholar] [CrossRef]
- Becker, A.S.; Wagner, M.W.; Wurnig, M.C.; Boss, A. Diffusion-weighted imaging of the abdomen: Impact of b-values on texture analysis features. NMR Biomed. 2017, 30, e3669. [Google Scholar] [CrossRef]
- Lin, Q.; Ji, Y.-F.; Chen, Y.; Sun, H.; Yang, D.-D.; Chen, A.-L.; Chen, T.-W.; Zhang, X.M. Radiomics model of contrast-enhanced MRI for early prediction of acute pancreatitis severity. J. Magn. Reson. Imaging 2020, 51, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Frøkjær, J.B.; Lisitskaya, M.V.; Jørgensen, A.S.; Østergaard, L.R.; Hansen, T.M.; Drewes, A.M.; Olesen, S.S. Pancreatic magnetic resonance imaging texture analysis in chronic pancreatitis: A feasibility and validation study. Abdom. Radiol. 2020, 45, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Huang, X.; Meng, H.; Zhang, M.; Zhang, X.; Lin, X.; Li, B. Performance of multiparametric functional imaging and texture analysis in predicting synchronous metastatic disease in pancreatic ductal adenocarcinoma patients by hybrid PET/MR: Initial experience. Front. Oncol. 2020, 10, 198. [Google Scholar] [CrossRef] [PubMed]
- Azoulay, A.; Cros, J.; Vullierme, M.P.; de Mestier, L.; Couvelard, A.; Hentic, O.; Ruszniewski, P.; Sauvanet, A.; Vilgrain, V.; Ronot, M. Morphological imaging and CT histogram analysis to differentiate pancreatic neuroendocrine tumor grade 3 from neuroendocrine carcinoma. Diagn. Interv. Imaging 2020, 101, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Ohki, K.; Igarashi, T.; Ashida, H.; Takenaga, S.; Shiraishi, M.; Nozawa, Y.; Ojiri, H. Usefulness of texture analysis for grading pancreatic neuroendocrine tumors on contrast-enhanced computed tomography and apparent diffusion coefficient maps. Jpn. J. Radiol. 2021, 39, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Boers, T.G.W.; Hu, Y.; Gibson, E.; Barratt, D.C.; Bonmati, E.; Krdzalic, J.; van der Heijden, F.; Hermans, J.J.; Huisman, H.J. Interactive 3D U-net for the segmentation of the pancreas in computed tomography scans. Phys. Med. Biol. 2020, 65, 065002. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-L.; Wu, T.; Chen, P.-T.; Tsai, Y.M.; Roth, H.; Wu, M.-S.; Liao, W.-C.; Wang, W. Deep learning to distinguish pancreatic cancer tissue from non-cancerous pancreatic tissue: A retrospective study with cross-racial external validation. Lancet Digit. Health 2020, 2, e303–e313. [Google Scholar] [CrossRef]
- Li, C.; Lin, X.; Hui, C.; Lam, K.M.; Zhang, S. Computer-Aided Diagnosis for Distinguishing Pancreatic Mucinous Cystic Neoplasms from Serous Oligocystic Adenomas in Spectral CT Images. Technol. Cancer Res. Treat. 2016, 15, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.R.R.; Mala, G.A.; Sarika, C.; Shruthi, S.; Sripradha, S. Segmentation of pancreatic cysts and roi extraction from pancreatic ct images using machine learning. Eur. J. Mol. Clin. Med. 2020, 7, 2020. [Google Scholar]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic Multi-Organ Segmentation on Abdominal CT With Dense V-Networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; He, K.; Nie, D.; Adeli, E.; Shi, Z.; Lee, S.-W.; Zheng, Y.; Liu, X.; Li, D.; Shen, D. Cascaded MultiTask 3-D Fully Convolutional Networks for Pancreas Segmentation. IEEE Trans. Cybern. 2021, 51, 2153–2165. [Google Scholar] [CrossRef]
- Zheng, H.; Qian, L.; Qin, Y.; Gu, Y.; Yang, J. Improving the slice interaction of 2.5D CNN for automatic pancreas segmentation. Med. Phys. 2020, 47, 5543–5554. [Google Scholar] [CrossRef]
- Suman, G.; Panda, A.; Korfiatis, P.; Edwards, M.E.; Garg, S.; Blezek, D.J.; Chari, S.T.; Goenka, A.H. Development of a volumetric pancreas segmentation CT dataset for AI applications through trained technologists: A study during the COVID 19 containment phase. Abdom. Radiol. 2020, 45, 4302–4310. [Google Scholar] [CrossRef]
- Nishio, M.; Noguchi, S.; Fujimoto, K. Automatic Pancreas Segmentation Using Coarse-Scaled 2D Model of Deep Learning: Usefulness of Data Augmentation and Deep U-Net. Appl. Sci. 2020, 10, 3360. [Google Scholar] [CrossRef]
- Li, W.; Qin, S.; Li, F.; Wang, L. MAD-UNet: A deep U-shaped network combined with an attention mechanism for pancreas segmentation in CT images. Med. Phys. 2021, 48, 329–341. [Google Scholar] [CrossRef]
- Panda, A.; Korfiatis, P.; Suman, G.; Garg, S.K.; Polley, E.C.; Singh, D.P.; Chari, S.T.; Goenka, A.H. Two-stage deep learning model for fully automated pancreas segmentation on computed tomography: Comparison with intra-reader and inter-reader reliability at full and reduced radiation dose on an external dataset. Med. Phys. 2021, 48, 2468–2481. [Google Scholar] [CrossRef]
- Li, S.; Jiang, H.; Wang, Z.; Zhang, G.; Yao, Y.-D. An effective computer aided diagnosis model for pancreas cancer on PET/CT images. Comput. Methods Programs Biomed. 2018, 165, 205–214. [Google Scholar] [CrossRef]
- Balasubramanian, A.D.; Murugan, P.R.; Thiyagarajan, A.P. Analysis and classification of malignancy in pancreatic magnetic resonance images using neural network techniques. Int. J. Imaging Syst. Technol. 2019, 29, 399–418. [Google Scholar] [CrossRef]
- D’Onofrio, M.; Tedesco, G.; Cardobi, N.; De Robertis, R.; Sarno, A.; Capelli, P.; Martini, P.T.; Giannotti, G.; Beleù, A.; Marchegiani, G.; et al. Magnetic resonance (MR) for mural nodule detection studying Intraductal papillary mucinous neoplasms (IPMN) of pancreas: Imaging-pathologic correlation. Pancreatology 2021, 21, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, S.; Gurney-Champion, O.J.; Klaassen, R.; Thoeny, H.C. Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI. Magn. Reson. Med. 2020, 83, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ruan, D.; Xiao, J.; Wang, L.; Sun, B.; Saouaf, R.; Yang, W.; Li, D.; Fan, Z. Fully automated multiorgan segmentation in abdominal magnetic resonance imaging with deep neural networks. Med. Phys. 2020, 47, 4971–4982. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-B. Artificial intelligence in pancreatic disease. AIMI 2020, 1, 19–30. [Google Scholar] [CrossRef]

| Author | Year | Radiomics Analysis | Task | N Pts | Data Split | Ref Standard | CT Phase | Results |
|---|---|---|---|---|---|---|---|---|
| Yang | 2019 | LIFEx software | Differential diagnosis (MCN vs. SCN) | 78 (25 MCNs, 53 SCNs) | RW (TS:DS = 4:1) | Histopathology | AP, PVP | Radiomics features, 2 mm: AUC 0.66, Acc 74%, Sen 86%, Spe 71% Radiomics features, 5 mm: AUC 0.75, Acc 83%, Sen 85%, Spe 83% |
| Yang (1) | 2019 | LIFEx software | Differential diagnosis (MCN vs. SCN) | 91 (32 MCNs, 59 SCNs) | SW | Histopathology | PAP | Textural features: AUC 0.777 Textural features + morphological characteristics: AUC 0.893 |
| Xie | 2019 | In-house algorithm (MATLAB R2017a) | Differential diagnosis (MCN vs. SCN) | 57 (31 MCNs, 26 SCNs) | SW | Radiologist | AP, PVP, DP | Radiomics model: AUC 0.989, Acc 94.7%, Sen 93.6%, Spe 96.2% Combined model (radiomics + radiological features): AUC 0.994, Acc 98.2%, Sen 96.8%, Spe 100% |
| Chen | 2021 | Analysis Kit Software (v 3.0.0.R) | Differential diagnosis (PCN vs. SCN) | 89 (31 SCNs, 30 IPMNs, 28 MCNs) | RW (63 TS, 26 VS) | Radiologist | NECT, AP, PVP | Radiomics signature NECT + AP + PVP: AUC 0.817 |
| Wei | 2019 | NS | Differential diagnosis (PCN vs. SCN) | 260 (102 SCNs, 158 non-SCNs) | SW (200 TS, 60 VS) | Radiologist | AP, PVP | Radiomics method: AUC 0.837, Sen 66.7%, Spe 81.8% |
| Shen | 2020 | ANN, RF, SVM (MATLAB 2017b) | Differential diagnosis (PCN) | 164 (76 SCAs, 40 MCNs, 48 IPMNs) | SW (115 TS, 41 VS) | Histopathology | AP | Radiomics model (nine features) Acc 71.43% (SVM, ANN), 79.59% (RF) |
| He | 2019 | Pyradiomics | Differential diagnosis (PDAC vs. pNET) | 147 (80 PDACs, 67 pNETs) | SW (100 TS, 47 VS) | Radiologist | PAP, PVP | Radiomics signature: AUC 0.873, Acc 76.6%, Sen 92.3%, Spe 70.6% Integrated model (radiomics + clinical features): AUC 0.884, Acc 80.4%, Sen 80.0%, Spe 80.8% |
| Li | 2018 | FireVoxel Software | Differential diagnosis (PDAC vs. pNET) | 75 (50 PDACs, 25 pNETs) | SW | Radiologist | AP, PVP | Combined fifth + skewness as the best parameters: AUC 0.887, Sen 90%, Spe 80% |
| Reinert | 2020 | Pyradiomics | Differential diagnosis (PDAC vs. pNET) | 95 (53 PDACs, 42 pNETs) | SW | Radiologist | PVP | Significant discriminatory features: first-order features, i.e., median, total energy, energy, 10th percentile, 90th percentile, minimum, maximum; second-order feature, i.e., gray-level co-occurrence matrix informational measure of correlation (Sen 79%, Spe 71%) |
| Yu | 2020 | Analysis Kit Software | Differential diagnosis (PDAC vs. pNET) | 120 (80 PDACs, 40 pNETs) | RW | Radiologist | AP, PVP | AP texture model: AUC 0.855 PVP texture model: AUC 0.929 |
| Ren | 2020 | Analysis Kit Software (v 3.0.0.R) | Differential diagnosis (PDAC vs. PASC) | 112 (81 PDACs, 31 PASCs) | RW (TS:DS = 2:1) | Histopathology | PAP, PVP | Acc 94.5%, Sen 98.3%, Spe 90.1%, PPV 91.9%, NPV 97.8% |
| Tobaly | 2020 | Pyradiomics (v 2.2.0) | IPMN grading | 408 (181 benign, 227 malignant) | SW (296 TS, 112 VS) | Histopathology | PAP, PVP | Benign vs. malignant IPMN radiomics model: AUC 0.71, Acc 64%, Sen 69%, Spe 57% Radiomics + surgical indication: AUC 0.75, Acc 67%, Sen 69%, Spe 65% |
| Hanania | 2016 | IBEX | Prediction of IPMN malignancy | 53 (34 high-grade, 19 low-grade) | SW(TS:DS = 7:3) | Histopathology | AP | Radiomics panel (10 features): AUC 0.96, Sen 97%, Spe 88% |
| Permuth | 2016 | In-house algorithm (Definiens Platform) | Prediction of IPMN malignancy | 38 (20 benign, 18 malignant) | SW(TS:DS = 9:1) | Histopathology | AP, PVP | Radiomics signature (14 features): AUC 0.77, Sen 83%, Spe 74% Integrated model 1 (radiomics + genomic data): AUC 0.92, Sen 83%, Spe 89% Integrated model 2 (radiomics + standard imaging + genomic data): AUC 0.93, Sen 89%, Spe 89% |
| Canellas | 2018 | TexRAD (v 3.1) | pNET grading | 101 (63 grade 1, 35 grade 2, 3 grade 3) | SW | Histopathology | PVP | Entropy as an independent predictor: OR 3.7, AUC 0.65, values > 4.65 with differences in DFS (G1 vs. G2/G3) |
| Gu | 2019 | Pyradiomics (v 1.3.0) | pNET grading (G1 vs. G2/G3) | 138 (57 grade 1, 69 grade 2, 12 grade 3) | RW (104 TS, 34 VS) | Histopathology | AP, PVP | Nomogram (radiomics features + clinical risk factor tumor margin): AUC 0.902 |
| Guo | 2019 | MATLAB R2014a | pNET grading (G1/G2 vs. G3) | 37 (13 grade 1, 11 grade 2, 13 grade 3) | RW | Histopathology | NECT, AP, PVP | Texture features AUC 0.93, Sen 91.7%, Spe 84.6% Size/margin + texture features AUC 0.958, Sen 91.6%, Spe 87.5% |
| Liang | 2019 | In-house algorithm (MATLAB R2016a) | pNET grading (G1 vs. G2/G3) | 137 (70 grade 1, 67 grade 2/3) | RW (86 TS, 51 VS) | Histopathology | AP | Nomogram (eight radiomics features + clinical stage): AUC 0.891 |
| D’Onofrio | 2019 | MaZda Software (v 4.6) | pNET grading | 100 (31 grade 1, 52 grade 2, 17 grade 3) | RW | Radiologist | AP, PVP | Kurtosis is different among three G groups: AUC 0.924, Sen 82%, Spe 85% for G3 diagnosis Entropy different between G1 and G3 and G2 and G3 groups: AUC 0.732, Sen 82%, Spe 64% for G3 diagnosis |
| Kaissis | 2020 | Pyradiomics | PDAC classification | 207 (45 QM, 136 non-QM, 26 unclassifiable) | SW (181 TS, 26 VS) | Histopathology | PVP | AUC 0.93, Sen 0.84, Spe 0.92 |
| Attiyeh | 2018 | MATLAB R2015a | PDAC prognosis | 161 | SW (113 TS, 48 VS) | Radiologist | PVP | Model A, preoperative CA19-9 and image features: c-index 0.69 Model B, preoperative CA19-9, Brennan score (postresection pathological variables), and image features: c-index 0.74 |
| Khalvati | 2019 | Pyradiomics | PDAC prognosis | 98 | SW (30 TS, 68 VS) | Radiologist | PAP, PVP | Radiomics signature: HR 1.35 (Reader 2), 1.56 (Reader 1) |
| Yun | 2018 | NS | PDAC prognosis | 88 (70 recurrence, 18 non-recurrence) | SW | Radiologist | PAP, PVP | Correlation of recurrence with texture features Average: AUC 0.736, standard deviation: AUC 0.709, contrast: AUC 0.692, correlation: AUC 0.698 Survival analysis nodal metastasis: HR 2.0375, average: HR 0.5599, standard deviation HR 0.5745 |
| Xie | 2020 | NS | PDAC prognosis | 220 | SW (147 TS, 73 VS) | NS | PAP | Rad-score: low-RS correlated with better prognosis (AUC 0.715), HR 2.556 for DFS, HR 3.741 for OS |
| Kim | 2019 | NS | PDAC prognosis | 116 | SW | Radiologist | AP | GLN135: higher levels correlated with shorter DFS (HR 6.030) |
| Eilaghi | 2017 | MATLAB R2015a | PDAC prognosis | 30 | SW | Radiologist | PAP, PVP | Prediction of OS Tumor dissimilarity: AUC 0.716 Inverse difference normalized: AUC 0.716 |
| Fang | 2020 | MaZda Software (v 4.6) | Prediction of LN metastasis | 155 (73 nodal matastases, 82 without nodal metastases) | RW | Histopathology | AP, PVP | Ten texture features with significance in ROC analysis: biggest AUC 0.630 for wavelet-based feature WavEnLH_s-2 |
| Li | 2020 | Pyradiomics | Prediction of LN metastasis | 159 (59 nodal matastases, 100 without nodal metastases) | SW (118 TS, 41 VS) | Histopathology | AP, PVP | Radiomics signature (15 features): AUC 0.912 |
| Chen | 2019 | IBEX | AcP prognosis | 389 (181 recurrent AcP) | RW (271 TS, 118 VS) | Radiologist | AP, PVP | Recurrence prediction: AUC 0.929, Acc 89.0% |
| Mashayekhi | 2020 | In-house algorithm (MATLAB) | Differential diagnosis (recurrent AcP vs. CP) | 56 (20 recurrent AcP, 19 functional abdominal pain, 17 CP) | SW | Radiologist | PVP | Acc 82.1%; recurrent AP: AUC 0.88, Sen 95%, Spe 78%; CP: AUC 0.90, Sen 71%, Spe 95% |
| Author | Year | Radiomics Analysis | Task | N Pts | Data Split | Reference Standard | Radiotracer | CT Phase | Results |
|---|---|---|---|---|---|---|---|---|---|
| Liu | 2021 | SVM (MATLAB R2018a) | Differential diagnosis (PDAC vs. autoimmune pancreatitis) | 112 (64 PDACs, 48 autoimmune pancreatitis) | RW | Radiologist | FDG | NECT | AUC 0.9668, Acc 89.91%, Sen 85.31%, Spe 96.04% |
| Zhang | 2019 | SVM (MATLAB R2017a) | Differential diagnosis (PDAC vs. autoimmune pancreatitis) | 111 (66 PDACs, 45 autoimmune pancreatitis) | RW | Radiologist | FDG | NECT | AUC 0.93, Acc 85%, Sen 86%, Spe 84% |
| Lim | 2020 | MIM (v 6.4) | PDAC classification | 48 | SW | Radiologist | FDG | NECT | KRAS gene mutation: significant association with long-run emphasis (AUC 0.806), zone emphasis (AUC 0.794), large-zone emphasis (AUC 0.829); SMAD4 gene mutation: significant association with standardized uptake value skewness (AUC 0.727), long-run emphasis (AUC 0.692), high-intensity textural features such as run emphasis (AUC 0.775), short-run emphasis (AUC 0.736), zone emphasis (AUC 0.750), and short-zone emphasis (AUC 0.725) |
| 2021 | Pyradiomics | PDAC grading | 149 | RW (99 TS, 50 VS) | Nuclear medicine physician | FDG | NECT | Prediction model (12 features): AUC 0.921 for G1 vs. G2/3 | |
| Mapelli | 2020 | Chang-Gung Image Texture Analysis software package (v 1.3) | pNET prognosis | 61 | RW | NS | DOTADOC, FDG | NECT | DOTATOC PET: SZV, entropy, intensity variability, and SRD were predictive of tumor dimension; FDG PET: intensity variability, SZV, homogeneity, SUVmax, and MTV were predictive for tumor dimension |
| Liberini | 2020 | LIFEx software (v 5.10) | pNET prognosis | 2 | SW | NS | DOTADOC | NECT | A significant difference of 28 radiomics features in pre- and post-treatment studies |
| Toyama | 2020 | LIFEx software | PDAC prognosis | 161 | SW | Histopathology | FDG | NECT | GLZLM GLNU as an independent predictor factor for poor prognosis (HR 2.0) |
| Cui | 2016 | MITK software (v 3.1.0.A) | PDAC prognosis | 139 | SW (90 TS, 49 VS) | NS | FDG | NECT | Prognostic signature (seven features): HR 3.72 |
| Yue | 2017 | 3D kernel-based approach | PDAC prognosis | 26 | SW | NS | FDG | NECT | Low-risk group: higher texture variation (>30%) and longer mean OS (29.3 months); high-risk group: lower texture variation (<15%) and shorter mean OS (17.7 months) |
| Belli | 2018 | CGITA software (v 1.4) | Tumor segmentation | 25 | SW | Radiologist | FDG | NECT | DSC 0.73 |
| Author | Year | Radiomics Analysis | Task | N Pts | Data Split | Reference Standard | MRI Phase | Results |
|---|---|---|---|---|---|---|---|---|
| Song | 2021 | Pyradiomics | Differential diagnosis (NF-pNET vs. SPN) | 79 (22 NF-pNETs, 57 SPNs) | RW (TS:DS = 7:3) | Histopathology | T2WI, DWI, T1WI, CE-T1WI | Precontrast T1WI: AUC 0.853 AP: AUC 0.907 PVP: AUC 0.773 DP: AUC 0.773 Clinic-radiomics nomogram: AUC 0.920, Acc 90.0%, Sen 100.0%, Spec 71.4% |
| Li | 2019 | MaZda (v 4.6) | Differential diagnosis (NF-pNET vs. SPN) | 119 (61 NF-pNETs, 58 SPNs) | RW (101 TS, 18 DS) | Histopathology | T2WI, DWI, T1WI, CE-T1WI | AP: AUC 0.925 DP: AUC 0.950 |
| Cui | 2021 | MITK Software (v 3.1.0.A) | IPMN grading | 202 (152 low-grade, 50 high-grade) | RW (103 TS, 48 VS1, 51 VS2) | Histopathology | T2WI, T1WI, CE-T1WI | SET 1 Radiomics signature: AUC 0.811; Nomogram: AUC 0.884, Sen 90.0%, Spe 79.0% SET 2 Radiomics signature: AUC 0.822; Nomogram: AUC 0.876, Sen 85.7%, Spe 83.7% |
| Jeon | 2021 | MEDIP | Prediction of IPMN malignancy | 248 (142 Benign, 106 Malignant) | SW | Histopathology | MRCP | AUC 0.85 (Greater entropy and smaller compactness as independent predictors) |
| Guo | 2019 | Omni-Kinetics software (v 2.0.10) | pNET grading | 77 (31 grade 1, 29 grade 2, 17 grade 3) | RW | Histopathology | T2WI, DWI, T1WI, CE-T1WI | Independent predictors of T2WI: inverse difference moment for G1 vs. G2 (AUC 0.833), energy+correlation+difference entropy for G1 vs. G3 (AUC 0.989), difference entropy for G2 vs. G3 (AUC 0.813); Independent predictors of DWI: correlation+contrast+inverse difference moment for G1 vs. G2 (AUC 0.841), maxintensity+entropy+inverse difference moment for G1 vs. G3 (AUC 0.962), maxintensity for G2 vs. G3 (AUC 0.703) |
| Kaissis | 2019 | Pyradiomics | PDAC prognosis | 132 | SW (100 TS, 32 VS) | Histopathology | T2WI, DWI, T1WI, CE-T1WI | AUC 0.90, Sen 87%, Spe 80% |
| Kaissis (1) | 2019 | Pyradiomics | PDAC classification | 55 (27 KRT81+, 28 KRT81-) | SW | Histopathology | T2WI, DWI, T1WI, CE-T1WI | AUC 0.93, Sen 90%, Spe 92% |
| Taffel | 2019 | In-house software FireVoxel | Tumor diagnosis | 42 (36 PDACs, 6 pNETs) | SW | Histopathology | T2WI, DWI, T1WI, CE-T1WI | ADC histogram differentiation NET-PDAC: AUC 0.88-0.92, Sen 94–97%, Spe 83–88%; Differentiation nodal status: AUC 0.80–0.82, Sen 87%, Spe 67–83% |
| Becker | 2017 | In-house algorithm (MATLAB R2015b) | Impact of b-values | 8 controls | RW | Radiologist | DWI | Significant positive correlations with b-value: skewness, contrast, correlation, energy, LRE, GLN, RP; Significant negative correlations with b-value: kurtosis, entropy, homogeneity, LGRE, SRLGE, LRLGE |
| Lin | 2019 | IBEX | AcP classification | 259 (142 mild AcP, 117 severe AcP) | SW (180 TS, 79 VS) | Radiologist | CE-T1WI | AUC 0.848, Acc 81.0%, Sen 75.0%, Spe 86.0% |
| Frokjaer | 2020 | SlicerRadiomics extension (v 4.10.1) | CP classification | 99 (77 CP, 22 controls) | SW | Radiologist | T2WI, DWI, MRCP | Acc 98%, Sen 97%, Spe 100% |
| Author | Year | Radiomics Analysis | Task | N Pts | Data Split | Reference Standard | Radiotracer | MRI Phase | Results |
|---|---|---|---|---|---|---|---|---|---|
| Gao | 2020 | LIFEx software | Prediction of metastatic disease | 17 (11 metastatic PDACs, 6 non-metastatic PDACs) | RW | Radiologist and nuclear medicine physician | FDG | T2W HASTE, DWI, T1WI DIXON | SUV: AUC 0.818, Sen 72.7%, Spe 100%MTV: AUC 0.818, Sen 63.6%, Spe 100%TLG: AUC 0.848, Sen 72.7%, Spe 100% |
| Author | Year | Radiomics Analysis | Task | N Pts | Data Split | Reference Standard | CT/MRI Phase | Results |
|---|---|---|---|---|---|---|---|---|
| Azoulay | 2019 | TexRAD | Differential diagnosis (G3-pNET vs. NEC) | 37 (14 G3-pNETs, 23 NECs) | RW | Radiologist | CT: NECT, AP, PVP MRI: T1WI, T2WI, DWI, AP, PVP | CT histogram analysis AP skewness filter 4: AUC 0.736 AP skewness filter 5: AUC 0.758 PVP mean filter 0:AUC 0.712 PVP MPP filter 0: AUC 0.712 PVP entropy filter 0: AUC 0.719 |
| Ohki | 2021 | NS | pNET Grading (G1 vs. G2–G3) | 33 (22 grade 1, 11 grade 2/3) | RW | Radiologist | CT: AP, PVP MRI: ADC map | AP log-sigma 1.0 joint-energy: AUC 0.855 PVP log-sigma 1.5 kurtosis: AUC 0.860 ADC log-sigma 1.0 correlation: AUC 0.847 |
| Author | Year | AI Model | Task | N Pts | Data Split | Reference Standard | CT Phase | Results |
|---|---|---|---|---|---|---|---|---|
| Li | 2016 | SVM | Differential diagnosis (SOA vs. MCN) | 42 (23 SOAs, 19 MCNs) | RW | Radiologist | NECT, AP, PVP | Acc 93.2% |
| Liu | 2020 | CNN | Tumor diagnosis | 690 local set 1(370 cases, 320 controls), 189 local set 2 (101 cases, 88 controls), 363 US test set (281 cases, 82 controls) | SW (412 TS, 139 VS, 139 test set 1, 189 test set 2) | Pathology | PVP | Local set 1: AUC 0.997, Acc 98.6%, Sen 97.3%, Spe 100% Local set 2: AUC 0.999, Acc 98.9%, Sen 99.0%, Spe 98.9% US set: AUC 0.920, Acc 83.2%, Sen 79.0%, Spe 97.6% |
| Roy | 2020 | ANN | Tumor segmentation | NS | NS | NS | NS | NS |
| Gibson | 2018 | Dense V-Network FCN | Pancreas segmentation | 90 (43 public dataset 1, 47 public dataset 2) | SW | Radiologist | CECT | DSC 78% |
| Xue | 2021 | 3D FCN | Pancreas segmentation | 59 | SW | Radiologist | CECT | DSC 86.9% JC 77.3% |
| Zheng | 2020 | VNet | Pancreas segmentation | 82 | RW | Radiologist | CECT | DSC 86.21% Sen 87.49% Spe 85.11% |
| Boers | 2020 | Interactive 3D UNet | Pancreas segmentation | 100 | RW (90 TS, 10 VS) | Radiologist | PVP | DSC 78.1%, average automated baseline performance 78%, semiautomatic segmentation performance in 8 min 86% |
| Suman | 2021 | NVIDIA | Pancreas segmentation | 188 first batch, 159 second batch | SW | Radiologist | PVP | DSC 63%, JC 48%, FP 21%, FN 43% |
| Nishio | 2020 | Deep UNet | Pancreas segmentation | 80 | RW | Radiologist | CECT | DSC 70.3–78.9%, JC 0.563–0.658, Sen 64.5–76.2%, Spe 100% |
| Panda | 2021 | 3D CNN | Pancreas segmentation | 1917 internal dataset, 41 external dataset 1, 80 external dataset 2 | RW (1380 TS, 248 VS, 289 internal test set, 50 external test set 1, 82 external test set 2) | Radiologist | PVP | Internal dataset: DSC 91% External dataset 1: DSC 83–84% External dataset 2: DSC 89% |
| Li | 2021 | MAD-UNet | Pancreas segmentation | 363 (82 public dataset 1, 281 public dataset 2) | RW | UNet, VNet, Attention UNet, SegNet | CECT | DSC 86.10% JC 75.55% Sen 86.43% Spe 84.97% |
| Author | Year | AI Model | Task | N Pts | Data Split | Reference Standard | Radiotracer | CT Phase | Results |
|---|---|---|---|---|---|---|---|---|---|
| Li | 2018 | HFB-SVM-RF | Tumor Diagnosis | 80 (40 cancer patients, 40 controls) | RW | Radiologist | FDG | NECT | Acc 96.47%, Sen 95.23%, Spe 97.51% |
| Author | Year | AI Model | Task | N Pts | Data Split | Reference Standard | MRI Phase | Results |
|---|---|---|---|---|---|---|---|---|
| D’Onofrio | 2021 | NS | Prediction of IPMN malignancy | 91 | SW | Histopathology | T2WI, T1WI, DWI, MRCP | ADC map: entropy = 10.32, J Youden index 0.48, AUC 0.7288, Sen 68.75%, Spe 79.25% |
| Balasubramanian | 2019 | ANN, SVM | Tumor diagnosis | 168 (68 with lesion, 100 controls) | RW (TS:VS = 7:3) | NS | NS | ANN BP 2 features (HOMO, CP): Acc 98%, Sen 100%, Spe 95% |
| Barbieri | 2020 | DNN | Evaluation of IVIM performance | 10 | SW | Radiologist | DWI | Dt: ICC 94–97% Fp: ICC 66% Dp: 50–51% |
| Chen | 2020 | UNet-based ALAMO | Pancreas segmentation | 102 | SW (66 TS, 16 VS, 20 test set) | Radiologist | T1WI-VIBE | Single slice: DSC 0.871 20 slices: DSC 0.880 40 slices: DSC 0.871 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laino, M.E.; Ammirabile, A.; Lofino, L.; Mannelli, L.; Fiz, F.; Francone, M.; Chiti, A.; Saba, L.; Orlandi, M.A.; Savevski, V. Artificial Intelligence Applied to Pancreatic Imaging: A Narrative Review. Healthcare 2022, 10, 1511. https://doi.org/10.3390/healthcare10081511
Laino ME, Ammirabile A, Lofino L, Mannelli L, Fiz F, Francone M, Chiti A, Saba L, Orlandi MA, Savevski V. Artificial Intelligence Applied to Pancreatic Imaging: A Narrative Review. Healthcare. 2022; 10(8):1511. https://doi.org/10.3390/healthcare10081511
Chicago/Turabian StyleLaino, Maria Elena, Angela Ammirabile, Ludovica Lofino, Lorenzo Mannelli, Francesco Fiz, Marco Francone, Arturo Chiti, Luca Saba, Matteo Agostino Orlandi, and Victor Savevski. 2022. "Artificial Intelligence Applied to Pancreatic Imaging: A Narrative Review" Healthcare 10, no. 8: 1511. https://doi.org/10.3390/healthcare10081511
APA StyleLaino, M. E., Ammirabile, A., Lofino, L., Mannelli, L., Fiz, F., Francone, M., Chiti, A., Saba, L., Orlandi, M. A., & Savevski, V. (2022). Artificial Intelligence Applied to Pancreatic Imaging: A Narrative Review. Healthcare, 10(8), 1511. https://doi.org/10.3390/healthcare10081511








