Prognostic Value of Routine Biomarkers in the Early Stage of COVID-19
Abstract
:1. Introduction
2. Materials and Methods
Statistical Analysis
3. Results
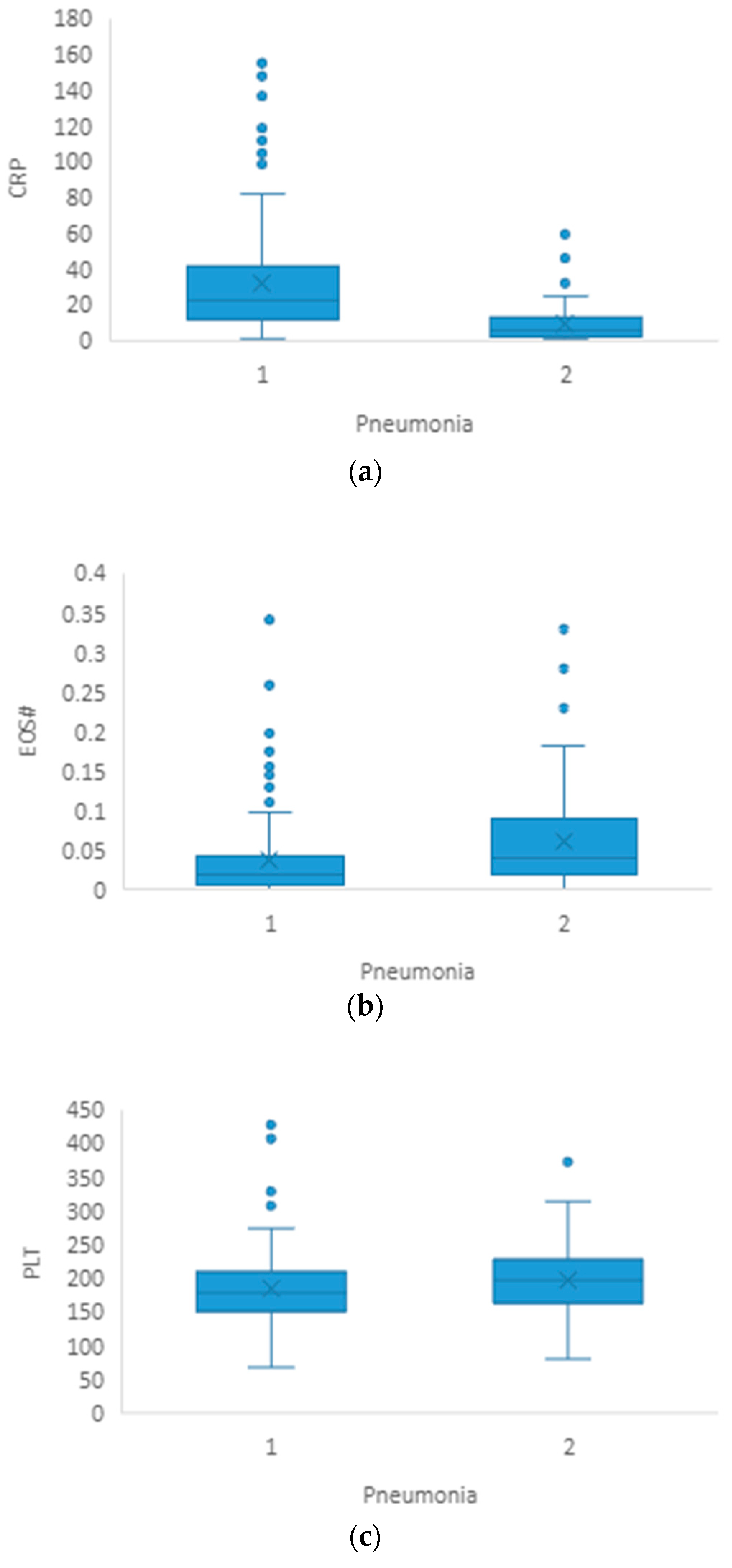
3.1. Pneumonia
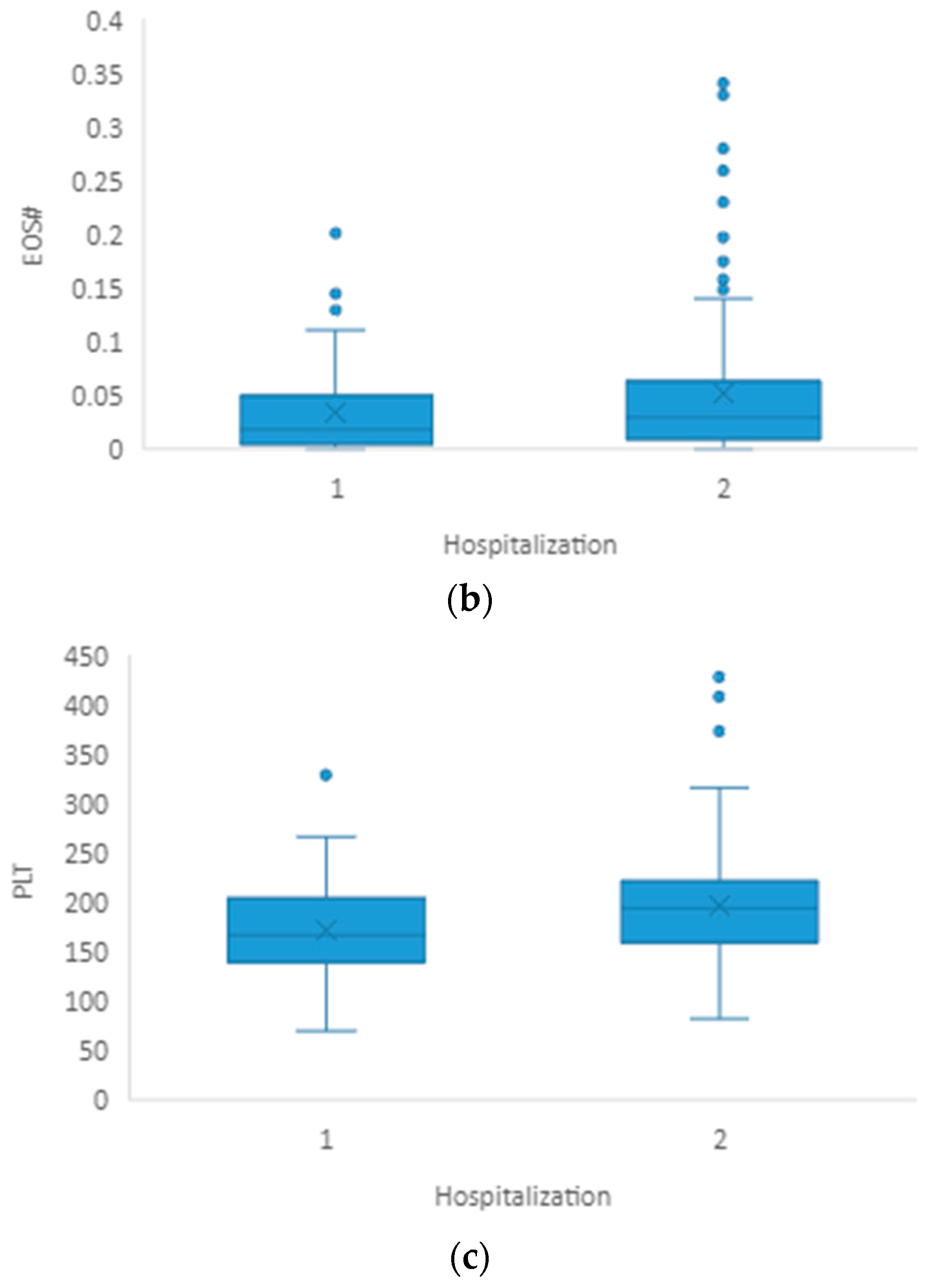
3.2. Hospitalization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, Y.; Bai, M.; You, Q. Associations between Serum Interleukins (IL-1β, IL-2, IL-4, IL-6, IL-8, and IL-10) and Disease Severity of COVID-19: A Systematic Review and Meta-Analysis. BioMed Res. Int. 2022, 2022, 2755246. [Google Scholar] [CrossRef]
- Ducastel, M.; Chenevier-Gobeaux, C.; Ballaa, Y.; Meritet, J.-F.; Brack, M.; Chapuis, N.; Pene, F.; Carlier, N.; Szwebel, T.-A.; Roche, N.; et al. Oxidative Stress and Inflammatory Biomarkers for the Prediction of Severity and ICU Admission in Unselected Patients Hospitalized with COVID-19. Int. J. Mol. Sci. 2021, 22, 7462. [Google Scholar] [CrossRef]
- Çakırca, G.; Damar Çakırca, T.; Üstünel, M.; Torun, A.; Koyuncu, İ. Thiol level and total oxidant/antioxidant status in patients with COVID-19 infection. Ir. J. Med.Sci. 2022, 191, 1925–1930. [Google Scholar] [CrossRef]
- Khodeir, M.M.; Shabana, H.A.; Alkhamiss, A.S.; Rasheed, Z.; Alsoghair, M.; Alsagaby, S.A.; Khan, M.I.; Fernández, N.; Al Abdulmonem, W. Early prediction keys for COVID-19 cases progression: A meta-analysis. J. Infect. Public Health 2021, 14, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Formica, V.; Minieri, M.; Bernardini, S.; Ciotti, M.; D’Agostini, C.; Roselli, M.; Andreoni, M.; Morelli, C.; Parisi, G.; Federici, M.; et al. Complete blood count might help to identify subjects with high probability of testing positive to SARS-CoV-2. Clin. Med. 2020, 20, e114–e119. [Google Scholar] [CrossRef]
- Henry, B.; de Oliveira, M.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem. Lab. Med. (CCLM) 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, A.; Zhang, Y.; Xuan, W.; Yan, T.; Pan, K.; Yu, W.; Zhang, J. Patients of COVID-19 may benefit from sustained Lopinavir-combined regimen and the increase of Eosinophil may predict the outcome of COVID-19 progression. Int. J. Infect. Dis. 2020, 95, 183–191. [Google Scholar] [CrossRef]
- Zahorec, R. Ratio of neutrophil to lymphocyte counts--rapid and simple parameter of systemic inflammation and stress in critically ill. Bratisl. Lek. Listy 2001, 102, 5–14. [Google Scholar]
- Meng, X.; Chang, Q.; Liu, Y.; Chen, L.; Wei, G.; Yang, J.; Zheng, P.; He, F.; Wang, W.; Ming, L. Determinant roles of gender and age on SII, PLR, NLR, LMR and MLR and their reference intervals defining in Henan, China: A posteriori and big-data-based. J. Clin. Lab Anal. 2018, 32, e22228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osadnik, T.; Bujak, K.; Osadnik, K.; Czarnecka, H.; Pawlas, N.; Reguła, R.; Fronczek, M.; Lejawa, M.; Gawlita, M.; Gonera, M.; et al. Novel inflammatory biomarkers may reflect subclinical inflammation in young healthy adults with obesity. Endokrynol. Pol. 2019, 70, 135–142. [Google Scholar] [CrossRef] [Green Version]
- Buonacera, A.; Stancanelli, B.; Colaci, M.; Malatino, L. Neutrophil to Lymphocyte Ratio: An Emerging Marker of the Relationships between the Immune System and Diseases. Int. J. Mol. Sci. 2022, 23, 3636. [Google Scholar] [CrossRef]
- Huang, Z.; Fu, Z.; Huang, W.; Huang, K. Prognostic value of neutrophil-to-lymphocyte ratio in sepsis: A meta-analysis. Am. J. Emerg. Med. 2020, 38, 641–647. [Google Scholar] [CrossRef]
- Zahorec, R. Neutrophil-to-lymphocyte ratio, past, present and future perspectives. Bratisl. Lek. Listy. 2021, 122, 474–488. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kalemaki, D.; Tzagkarakis, E.; Lydakis, C. Pitfalls in studies of eosinopenia and neutrophil-to-lymphocyte count ratio. Infect. Dis. 2018, 50, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Vafadar Moradi, E.; Teimouri, A.; Rezaee, R.; Morovatdar, N.; Foroughian, M.; Layegh, P.; Rezvani Kakhki, B.; Ahmadi Koupaei, S.R.; Ghorani, V. Increased age, neutrophil-to-lymphocyte ratio (NLR) and white blood cells count are associated with higher COVID-19 mortality. Am. J. Emerg. Med. 2021, 40, 11–14. [Google Scholar] [CrossRef]
- Sayed, A.A.; Allam, A.A.; Sayed, A.I.; Alraey, M.A.; Joseph, M.V. The use of neutrophil-to-lymphocyte ratio (NLR) as a marker for COVID-19 infection in Saudi Arabia: A case-control retrospective multicenter study. Saudi. Med. J. 2021, 42, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Saffar, A.S.; Ashdown, H.; Gounni, A.S. The molecular mechanisms of glucocorticoids-mediated neutrophil survival. Curr. Drug Targets 2011, 12, 556–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, M.; Terashima, T.; D’yachkova, Y.; Bondy, G.P.; Hogg, J.C.; van Eeden, S.F. Glucocorticoid-induced granulocytosis: Contribution of marrow release and demargination of intravascular granulocytes. Circulation 1998, 98, 2307–2313. [Google Scholar] [CrossRef] [Green Version]
- Dale, D.C.; Fauci, A.S.; Guerry, D.I.V.; Wolff, S.M. Comparison of agents producing a neutrophilic leukocytosis in man. Hydrocortisone, prednisone, endotoxin, and etiocholanolone. J. Clin. Investig. 1975, 56, 808–813. [Google Scholar] [CrossRef] [Green Version]
- Prazma, C.M.; Bel, E.H.; Price, R.G.; Bradford, E.S.; Albers, F.C.; Yancey, S.W. Oral corticosteroid dose changes and impact on peripheral blood eosinophil counts in patients with severe eosinophilic asthma: A post hoc analysis. Respir. Res. 2019, 20, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunsky, E.H.; Zweiman, B.; Fischler, E.; Levy, D.A. Early effects of corticosteroids on basophils, leukocyte histamine, and tissue histamine. J. Allergy Clin. Immunol. 1979, 63, 426–432. [Google Scholar] [CrossRef] [PubMed]
- Kournoutou, G.G.; Dinos, G. Azithromycin through the Lens of the COVID-19 Treatment. Antibiotics 2022, 11, 1063. [Google Scholar] [CrossRef]
- Zimmermann, P.; Ziesenitz, V.C.; Curtis, N.; Ritz, N. The Immunomodulatory Effects of Macrolides-A Systematic Review of the Underlying Mechanisms. Front. Immunol. 2018, 9, 302. [Google Scholar] [CrossRef] [Green Version]
- Bedel, C.; Korkut, M.; Armağan, H.H. Can NLR, PLR and LMR be used as prognostic indicators in patients with pulmonary embolism? A commentary. Bosn. J. Basic. Med. Sci. 2021, 21, 501. [Google Scholar] [CrossRef]
- Cai, J.; Li, H.; Zhang, C.; Chen, Z.; Liu, H.; Lei, F.; Qin, J.-J.; Liu, Y.-M.; Zhou, F.; Song, X.; et al. The Neutrophil-to-Lymphocyte Ratio Determines Clinical Efficacy of Corticosteroid Therapy in Patients with COVID-19. Cell Metab. 2021, 33, 258–269.e3. [Google Scholar] [CrossRef] [PubMed]
- Palladino, M. Complete blood count alterations in COVID-19 patients: A narrative review. Biochem. Med. 2021, 31, 030501. [Google Scholar] [CrossRef]
- Zong, X.; Gu, Y.; Yu, P.H.; Li, Z.; Wang, Y. Thrombocytopenia Is Associated with COVID-19 Severity and Outcome: An Updated Meta-Analysis of 5637 Patients with Multiple Outcomes. Lab. Med. 2021, 52, 10–15. [Google Scholar] [CrossRef]
- Seyit, M.; Avci, E.; Nar, R.; Senol, H.; Yilmaz, A.; Ozen, M.; Oskay, A.; Aybek, H. Neutrophil to lymphocyte ratio, lymphocyte to monocyte ratio and platelet to lymphocyte ratio to predict the severity of COVID-19. Am. J. Emerg. Med. 2021, 40, 110–114. [Google Scholar] [CrossRef]
- Liu, L.; Shao, Z.; Yu, H.; Zhang, W.; Wang, H.; Mei, Z. Is the platelet to lymphocyte ratio a promising biomarker to distinguish acute appendicitis? Evidence from a systematic review with meta-analysis. PLoS ONE 2020, 15, e0233470. [Google Scholar] [CrossRef]
- Dávila-Collado, R.; Jarquín-Durán, O.; Solís-Vallejo, A.; Nguyen, M.A.; Espinoza, J.L. Elevated Monocyte to Lymphocyte Ratio and Increased Mortality among Patients with Chronic Kidney Disease Hospitalized for COVID-19. J. Pers. Med. 2021, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, S.; Feng, Y.; Wu, W.; Chang, C.; Chen, S.; Zhen, G.; Yi, L. Decreased eosinophil counts and elevated lactate dehydrogenase predict severe COVID-19 in patients with underlying chronic airway diseases. Postgrad. Med. J. 2022, 98, 906–913. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Vieyra, R.; Gutiérrez-Castellanos, S.; Álvarez-Aguilar, C.; Baizabal-Aguirre, V.M.; Nuñez-Anita, R.E.; Rocha-López, A.G.; Gomez-Garcia, A. Behavior of Eosinophil Counts in Recovered and Deceased COVID-19 Patients over the Course of the Disease. Viruses 2021, 13, 1675. [Google Scholar] [CrossRef]
- Lourda, M.; Dzidic, M.; Hertwig, L.; Bergsten, H.; Medina, L.M.P.; Sinha, I.; Kvedaraite, E.; Chen, P.; Muvva, J.R.; Gorin, J.-B.; et al. High-dimensional profiling reveals phenotypic heterogeneity and disease-specific alterations of granulocytes in COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2109123118. [Google Scholar] [CrossRef] [PubMed]
- Public Health Ontario. An Overview of Cycle Threshold Values and Their Role in SARS-CoV-2 Real-Time PCR Test Interpretation [Internet]. 2020. Available online: https://www.publichealthontario.ca/-/media/documents/ncov/main/2020/09/cycle-threshold-values-sars-cov2-pcr.pdf?la=en (accessed on 12 July 2023).
- Biomerieux. ARGENE ® Real-Time PCR Assays [Internet]. 2020. Available online: https://www.biomerieux.cz/sites/clinic/files/argene_sars-cov-2_r-gene_ce_ivd.pdf (accessed on 12 July 2023).
- National Institutes of Health (NIH)- COVID 19 Treatment Guidelines Clinical Spectrum of SARS-CoV-2 Infection, 6 March 2023. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 14 July 2023).
- Mousavi-Nasab, S.D.; Mardani, R.; Azadani, H.N.; Vasmehjani, A.A.; Sabeti, S.; Darazam, I.A.; Ahmadi, N. Neutrophil to lymphocyte ratio and C-reactive protein level as prognostic markers in mild versus severe COVID-19 patients. Gastroenterol. Hepatol. Bed Bench 2020, 13, 361–366. [Google Scholar] [PubMed]
- Reusch, N.; De Domenico, E.; Bonaguro, L.; Schulte-Schrepping, J.; Baßler, K.; Schultze, J.L.; Aschenbrenner, A.C. Neutrophils in COVID-19. Front. Immunol. 2021, 12, 652470. [Google Scholar] [CrossRef]
- Tanni, F.; Akker, E.; Zaman, M.M.; Figueroa, N.; Tharian, B.; Hupart, K.H. Eosinopenia and COVID-19. J. Am. Osteopat. Assoc. 2020, 120, 504–508. [Google Scholar] [CrossRef]
- Sun, Y.; Dong, Y.; Wang, L.; Xie, H.; Li, B.; Chang, C.; Wang, F.-S. Characteristics and prognostic factors of disease severity in patients with COVID-19: The Beijing experience. J. Autoimmun. 2020, 112, 102473. [Google Scholar] [CrossRef]
- Roca, E.; Ventura, L.; Zattra, C.M.; Lombardi, C. EOSINOPENIA: An early, effective, and relevant COVID-19 biomarker? QJM 2020, 114, 68–69. [Google Scholar] [CrossRef]
- Li, Q.; Ding, X.; Xia, G.; Chen, H.-G.; Chen, F.; Geng, Z.; Xu, L.; Lei, S.; Pan, A.; Wang, L.; et al. Eosinopenia and elevated C-reactive protein facilitate triage of COVID-19 patients in fever clinic: A retrospective case-control study. EClinicalMedicine 2020, 23, 100375. [Google Scholar] [CrossRef]
- Du, Y.; Tu, L.; Zhu, P.; Mu, M.; Wang, R.; Yang, P.; Wang, X.; Hu, C.; Ping, R.; Hu, P.; et al. Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study. Am. J. Respir. Crit. Care Med. 2020, 201, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.P.; Soliman, A.; Al Masalamani, M.A.; De Sanctis, V.; Nashwan, A.J.; Sasi, S.; Ali, E.A.; Hassan, O.A.; Iqbal, F.M.; Yassin, M.A. Clinical Outcome of Eosinophilia in Patients with COVID-19: A Controlled Study. Acta Biomed. 2020, 91, e2020165. [Google Scholar] [PubMed]
- Ho, K.S.; Howell, D.; Rogers, L.; Narasimhan, B.; Verma, H.; Steiger, D. The Relationship between Asthma, Eosinophilia, and Outcomes in COVID-19 Infection. Ann. Allergy Asthma. Immunol. 2021, 127, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.M.; Costa, A.M.; Tuna, C.; Gonçalves, R.; Ferreira, S.; Belém, F.; Evangelista, M.C.; Ascensão, M. Eosinopenia as predictor of infection in patients admitted to an internal medicine ward: A cross-sectional study. Porto Biomed. J. 2020, 5, e084. [Google Scholar] [CrossRef]
- Ramirez, G.A.; Yacoub, M.-R.; Ripa, M.; Mannina, D.; Cariddi, A.; Saporiti, N.; Ciceri, F.; Castagna, A.; Colombo, G.; Dagna, L. Eosinophils from Physiology to Disease: A Comprehensive Review. BioMed Res. Int. 2018, 2018, 9095275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bass, D.A. Behavior of eosinophil leukocytes in acute inflammation II. Eosinophil dynamics during acute inflammation. J. Clin. Investig. 1975, 56, 870–879. [Google Scholar] [CrossRef] [Green Version]
- Bass, D.A.; Gonwa, T.A.; Szejda, P.; Cousart, M.S.; De Chatelet, L.R.; McCall, C.E. Eosinopenia of acute infection: Production of eosinopenia by chemotactic factors of acute inflammation. J. Clin. Investig. 1980, 65, 1265–1271. [Google Scholar] [CrossRef] [PubMed]
- Andreozzi, F.; Hermans, C.; Yombi, J.C. Eosinopenia and COVID-19 patients: So specific? EClinicalMedicine 2020, 24, 100439. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Moshensky, A.; Lam, M.T.; Odish, M.F.; Patel, A.; Xu, L.; Hansen, E.; Trescott, S.; Nguyen, C.; Kim, R.; et al. Increased Peripheral Blood Neutrophil Activation Phenotypes and Neutrophil Extracellular Trap Formation in Critically Ill Coronavirus Disease 2019 (COVID-19) Patients: A Case Series and Review of the Literature. Clin. Infect. Dis. 2022, 74, 479–489. [Google Scholar] [CrossRef]
- Harte, J.V.; Coleman-Vaughan, C.; Crowley, M.P.; Mykytiv, V. It’s in the blood: A review of the hematological system in SARS-CoV-2-associated COVID-19. Crit. Rev. Clin. Lab. Sci. 2023, 1–30. [Google Scholar] [CrossRef]
- López-Pereira, P.; Iturrate, I.; de La Cámara, R.; Cardeñoso, L.; Alegre, A.; Aguado, B. Can COVID-19 cause severe neutropenia? Clin. Case Rep. 2020, 8, 3349–3351. [Google Scholar] [CrossRef]
- Fu, W.; Chen, C.; Chen, X.; Wang, K.; Zuo, P.; Liu, Y.; Zhang, M.; Zhao, X.; Xie, S.; Zhang, H.; et al. A U-shaped association between baseline neutrophil count and COVID-19-related mortality: A retrospective cohort study. J. Med. Virol. 2021, 93, 4265–4272. [Google Scholar] [CrossRef] [PubMed]
- Illg, Z.; Muller, G.; Mueller, M.; Nippert, J.; Allen, B. Analysis of absolute lymphocyte count in patients with COVID-19. Am. J. Emerg. Med. 2021, 46, 16–19. [Google Scholar] [CrossRef] [PubMed]
- Huang, I.; Pranata, R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. J. Intensive Care 2020, 8, 36. [Google Scholar] [CrossRef]
- Toori, K.U.; Qureshi, M.A.; Chaudhry, A. Lymphopenia: A useful predictor of COVID-19 disease severity and mortality. Pak. J. Med. Sci. 2021, 37, 1984–1988. [Google Scholar] [CrossRef] [PubMed]
- Boccatonda, A.; D’Ardes, D.; Rossi, I.; Grignaschi, A.; Lanotte, A.; Cipollone, F.; Guagnano, M.T.; Giostra, F. Platelet Count in Patients with SARS-CoV-2 Infection: A Prognostic Factor in COVID-19. J. Clin. Med. 2022, 11, 4112. [Google Scholar] [CrossRef] [PubMed]
- Vanderschueren, S.; De Weerdt, A.; Malbrain, M.; Vankersschaever, D.; Frans, E.; Wilmer, A.; Bobbaers, H. Thrombocytopenia and prognosis in intensive care. Crit. Care Med. 2000, 28, 1871–1876. [Google Scholar] [CrossRef]
- Manne, B.K.; Denorme, F.; Middleton, E.A.; Portier, I.; Rowley, J.W.; Stubben, C.; Petrey, A.C.; Tolley, N.D.; Guo, L.; Cody, M.; et al. Platelet gene expression and function in patients with COVID-19. Blood 2020, 136, 1317–1329. [Google Scholar] [CrossRef]
- Raadsen, M.; Du Toit, J.; Langerak, T.; van Bussel, B.; van Gorp, E.; Goeijenbier, M. Thrombocytopenia in Virus Infections. J. Clin. Med. 2021, 10, 877. [Google Scholar] [CrossRef]
- Assinger, A. Platelets and infection—An emerging role of platelets in viral infection. Front. Immunol. 2014, 5, 649. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Zou, S.; Wang, C.; Tan, X.; Yu, M. Neutrophil-to-lymphocyte and platelet-to-lymphocyte ratio in Chinese Han population from Chaoshan region in South China. BMC Cardiovasc. Disord. 2019, 19, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azab, B.; Camacho-Rivera, M.; Taioli, E. Average values and racial differences of neutrophil lymphocyte ratio among a nationally representative sample of United States subjects. PLoS ONE 2014, 9, e112361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haghjooy Javanmard, S.; Vaseghi, G.; Manteghinejad, A.; Nasirian, M. Neutrophil-to-Lymphocyte ratio as a potential biomarker for disease severity in COVID-19 patients. J. Glob. Antimicrob. Resist. 2020, 22, 862–863. [Google Scholar] [CrossRef] [PubMed]
- Toori, K.U.; Qureshi, M.A.; Chaudhry, A.; Safdar, M.F. Neutrophil to lymphocyte ratio (NLR) in COVID-19: A cheap prognostic marker in a resource constraint setting. Pak. J. Med. Sci. 2021, 37, 1435–1439. [Google Scholar] [CrossRef]
- Ciccullo, A.; Borghetti, A.; Verme, L.Z.D.; Tosoni, A.; Lombardi, F.; Garcovich, M.; Biscetti, F.; Montalto, M.; Cauda, R.; Di Giambenedetto, S. Neutrophil-to-lymphocyte ratio and clinical outcome in COVID-19: A report from the Italian front line. Int. J. Antimicrob. Agents 2020, 56, 106017. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Xiang, P.; Pu, L.; Xiong, H.; Li, C.; Zhang, M.; Tan, J.; Xu, Y.; Song, R.; et al. Neutrophil-to-lymphocyte ratio predicts critical illness patients with 2019 coronavirus disease in the early stage. J. Transl. Med. 2020, 18, 206. [Google Scholar] [CrossRef]
- Sarkar, S.; Kannan, S.; Khanna, P.; Singh, A.K. Role of platelet-to-lymphocyte count ratio (PLR), as a prognostic indicator in COVID-19: A systematic review and meta-analysis. J. Med. Virol. 2022, 94, 211–221. [Google Scholar] [CrossRef]
- Balta, S.; Ozturk, C. The platelet-lymphocyte ratio: A simple, inexpensive and rapid prognostic marker for cardiovascular events. Platelets 2015, 26, 680–681. [Google Scholar] [CrossRef]
- Ravindra, R.; Ramamurthy, P.; Aslam, S.S.M.; Kulkarni, A.K.S.; Ramamurthy, P.S. Platelet Indices and Platelet to Lymphocyte Ratio (PLR) as Markers for Predicting COVID-19 Infection Severity. Cureus 2022, 14, e28206. [Google Scholar] [CrossRef]
- Simon, P.; Le Borgne, P.; Lefevbre, F.; Cipolat, L.; Remillon, A.; Dib, C.; Hoffmann, M.; Gardeur, I.; Sabah, J.; Kepka, S.; et al. Platelet-to-Lymphocyte Ratio (PLR) Is Not a Predicting Marker of Severity but of Mortality in COVID-19 Patients Admitted to the Emergency Department: A Retrospective Multicenter Study. J. Clin. Med. 2022, 11, 4903. [Google Scholar] [CrossRef]

| Without Pneumonia (n = 76) Median (IQR) | Pneumonia (n = 124) Median (IQR) | p-Value | |
|---|---|---|---|
| Age | 48.5 (35.5–64.0) | 58.0 (45.0–68.0) | 0.001 |
| CRP(mg/L) | 6.3 (2.2–13.0) | 22.5 (11.6–43.7) | <0.001 |
| WBC(×109/L) | 4.70 (4.05–5.80) | 5.15 (3.92–6.70) | 0.082 |
| NEU# | 2.87 (2.17–4.01) | 3.50 (2.34–4.80) | 0.031 |
| LY# | 1.38 (0.94–1.90) | 1.36 (1.08–1.81) | 0.691 |
| MONO# | 0.40 (0.31–0.60) | 0.38 (0.27–0.51) | 0.065 |
| EOS# | 0.04 (0.02–0.10) | 0.02 (0.007–0.04) | <0.001 |
| BASO# | 0.012 (0.006–0.020) | 0.010 (0.005–0.015) | 0.087 |
| PLT(×109/L) | 199 (164–227) | 179 (152–211) | 0.033 |
| NLR | 2.22 (1.22–3.48) | 2.42 (1.60–3.68) | 0.164 |
| PLR | 144 (98–208) | 129 (92–172) | 0.143 |
| LMR | 3.27 (2.10–5.12) | 3.80 (2.51–5.44) | 0.102 |
| p | Odds Ratio | 95% Confidence Interval for Odds Ratio | ||
|---|---|---|---|---|
| Lower | Upper | |||
| Age | 0.003 | 1.034 | 1.012 | 1.057 |
| PLT | 0.206 | 0.996 | 0.991 | 1.002 |
| NEU# | 0.413 | 1.105 | 0.871 | 1.402 |
| EOS# | 0.046 | 0.026 | 0.001 | 0.931 |
| CRP | 0.000 | 1.051 | 1.025 | 1.077 |
| Constant | 0.052 | 0.196 | ||
| Hosp (n = 52) Median (IQR) | NoHosp (n = 148) Median (IQR) | p-Value | |
|---|---|---|---|
| Age | 60 (45–69) | 52 (41–66) | 0.084 |
| CRP(mg/L) | 28.6 (15.3–57.7) | 10.4 (3.0–22.8) | <0.001 |
| WBC(×109/L) | 5.50 (4.25–7.05) | 4.90 (3.90–5.95) | 0.054 |
| NEU# | 3.62 (2.65–5.12) | 3.03 (2.25–4.26) | 0.051 |
| LY# | 1.33 (1.01–1.81) | 1.38 (1.08–1.83) | 0.905 |
| MONO# | 0.36 (0.29–0.53) | 0.39 (0.29–0.55) | 0.526 |
| EOS# | 0.018 (0.006–0.047) | 0.030 (0.011–0.067) | 0.032 |
| BASO# | 0.010 (0.005–0.179) | 0.011 (0.006–0.020) | 0.251 |
| PLT(×109/L) | 167 (141–204) | 195 (160–223) | 0.006 |
| NLR | 2.76 (1.61–3.73) | 2.21 (1.35–3.37) | 0.081 |
| PLR | 110.7 (86.8–173.0) | 139.5 (99.3–194.6) | 0.061 |
| LMR | 3.81 (2.43–5.28) | 3.48 (2.38–5.30) | 0.624 |
| p | Odds Ratio | 95% Confidence Interval for Odds Ratio | ||
|---|---|---|---|---|
| Lower | Upper | |||
| PLT | 0.005 | 0.989 | 0.981 | 0.997 |
| NEU# | 0.356 | 1.120 | 0.880 | 1.426 |
| EOS# | 0.162 | 0.003 | 0.000 | 10.771 |
| CRP | 0.001 | 1.021 | 1.008 | 1.033 |
| Constant | 0.736 | 1.265 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mihajlović, A.; Ivanov, D.; Tapavički, B.; Marković, M.; Vukas, D.; Miljković, A.; Bajić, D.; Semnic, I.; Bogdan, M.; Karaba Jakovljević, D.; et al. Prognostic Value of Routine Biomarkers in the Early Stage of COVID-19. Healthcare 2023, 11, 2137. https://doi.org/10.3390/healthcare11152137
Mihajlović A, Ivanov D, Tapavički B, Marković M, Vukas D, Miljković A, Bajić D, Semnic I, Bogdan M, Karaba Jakovljević D, et al. Prognostic Value of Routine Biomarkers in the Early Stage of COVID-19. Healthcare. 2023; 11(15):2137. https://doi.org/10.3390/healthcare11152137
Chicago/Turabian StyleMihajlović, Andrea, David Ivanov, Borislav Tapavički, Milica Marković, Dragana Vukas, Ana Miljković, Dejana Bajić, Isidora Semnic, Maja Bogdan, Dea Karaba Jakovljević, and et al. 2023. "Prognostic Value of Routine Biomarkers in the Early Stage of COVID-19" Healthcare 11, no. 15: 2137. https://doi.org/10.3390/healthcare11152137
APA StyleMihajlović, A., Ivanov, D., Tapavički, B., Marković, M., Vukas, D., Miljković, A., Bajić, D., Semnic, I., Bogdan, M., Karaba Jakovljević, D., Nikolić, S., Slavić, D., & Lendak, D. (2023). Prognostic Value of Routine Biomarkers in the Early Stage of COVID-19. Healthcare, 11(15), 2137. https://doi.org/10.3390/healthcare11152137








