1. Introduction
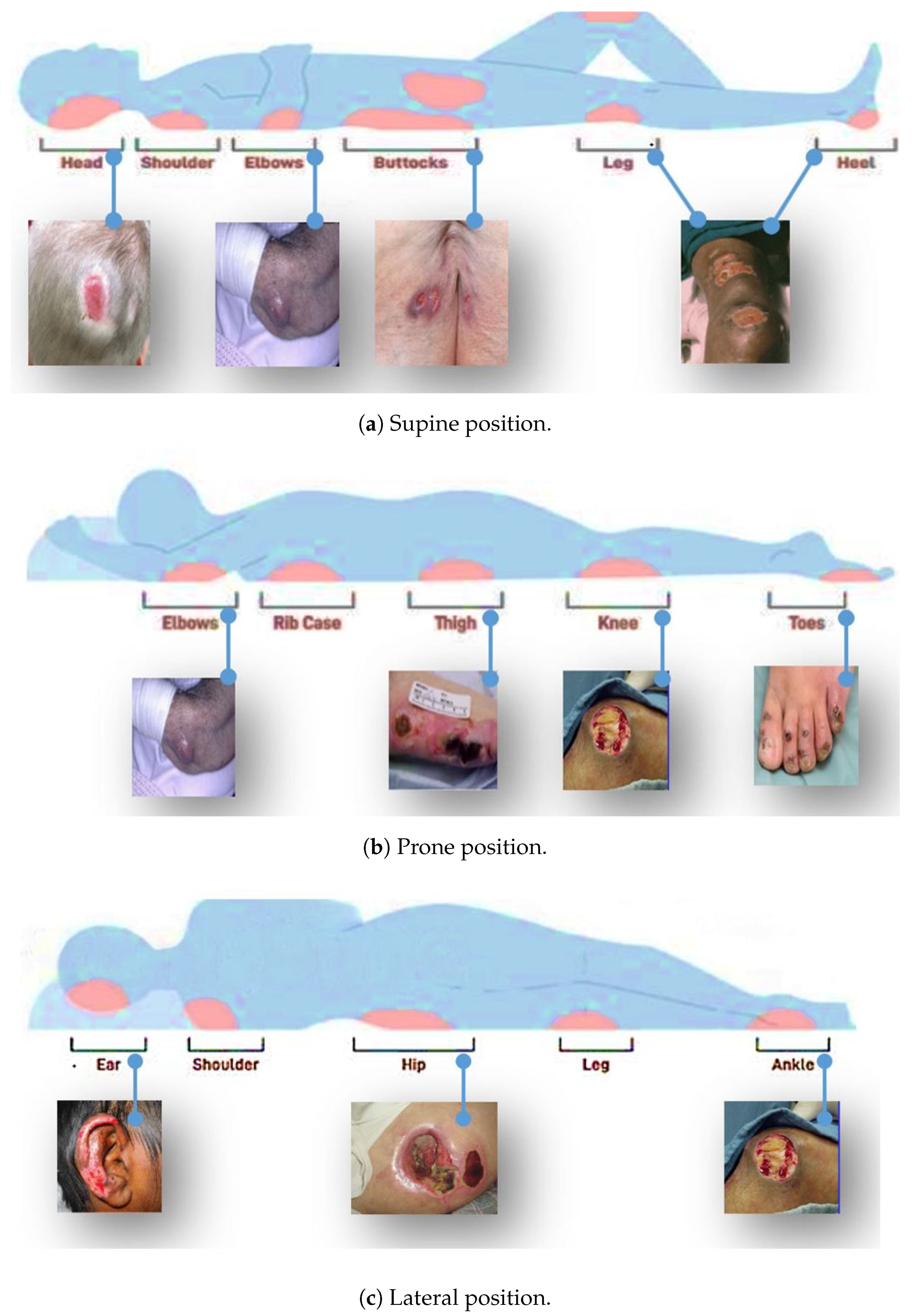
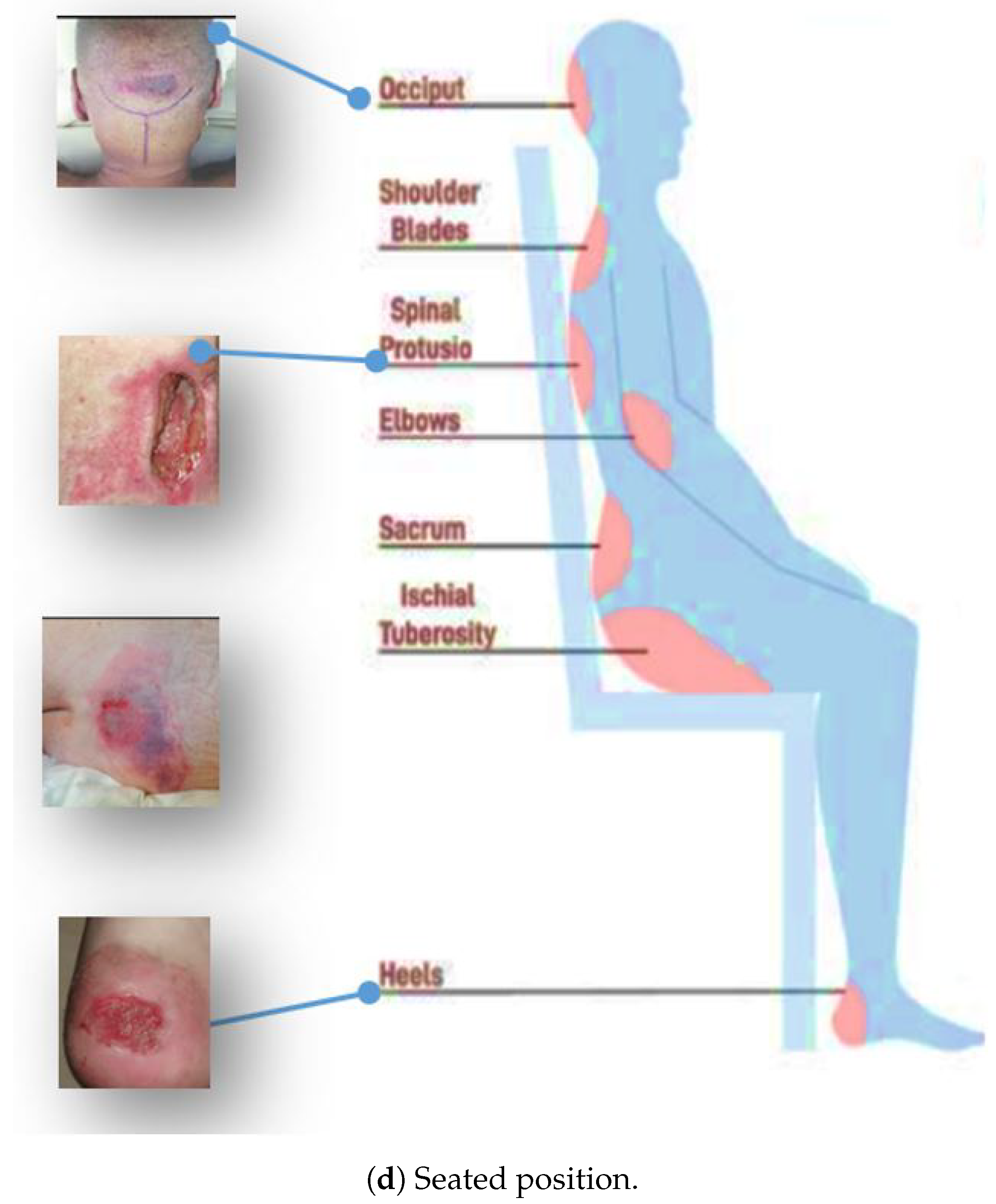
Pressure ulcers, sometimes known as bed sores, are small, localized wounds to the skin and underlying tissues brought on by persistent pressure or friction against the skin [
1]. They are serious health concerns, especially for older individuals and those with disabilities who spend a lot of time in bed or sitting down. However, newborn and pediatric children are also susceptible to pressure ulcers, albeit at a lower level [
2]. Pressure ulcers are becoming more prevalent, particularly among elderly individuals who are more susceptible due to decreased mobility, sensory perception, and skin integrity [
3,
4]. Additionally, extended hospital stays in chronic illnesses, neurological conditions, complex organ failure, cancer, radiation treatments, and prolonged stays in the intensive care unit (ICU) [
5], as well as long hospital stays during pandemics, such as COVID-19 [
6,
7,
8], increase the risk of developing pressure ulcers. Moreover, the prevalence of chronic diseases, such as obesity, diabetes, and cardiovascular disease, is rising. Many of these diseases also increase the risk of developing pressure ulcers [
9,
10]. Sedentary lifestyles have led to more pressure ulcers, especially among older adults and those with disabilities [
11,
12,
13,
14]. The most commonly affected areas include the buttocks, head, shoulders, sacrum, coccyx, elbows, heels, hips, and ears.
Figure 1 shows some of the areas where pressure ulcers may likely develop.
Patients who develop pressure ulcers may experience considerable effects, including pain, discomfort, and reduced mobility. In severe cases, they can lead to serious infections, hospitalization, and even death [
15]. For the effective care and treatment of pressure ulcers, early and correct identification is essential [
16]. However, classifying pressure ulcers into different stages can be challenging, especially for inexperienced healthcare providers [
17,
18]. As a result, there is a need for a more efficient and accurate method for classifying pressure ulcers that can improve patient outcomes and reduce healthcare costs. The current methods for classifying pressure ulcers into stages typically involve a visual assessment of the wound and a consideration of its depth and extent. There are several different classification systems in use, but the most widely used is the National Pressure Ulcer Advisory Panel (NPUAP) staging system [
1].
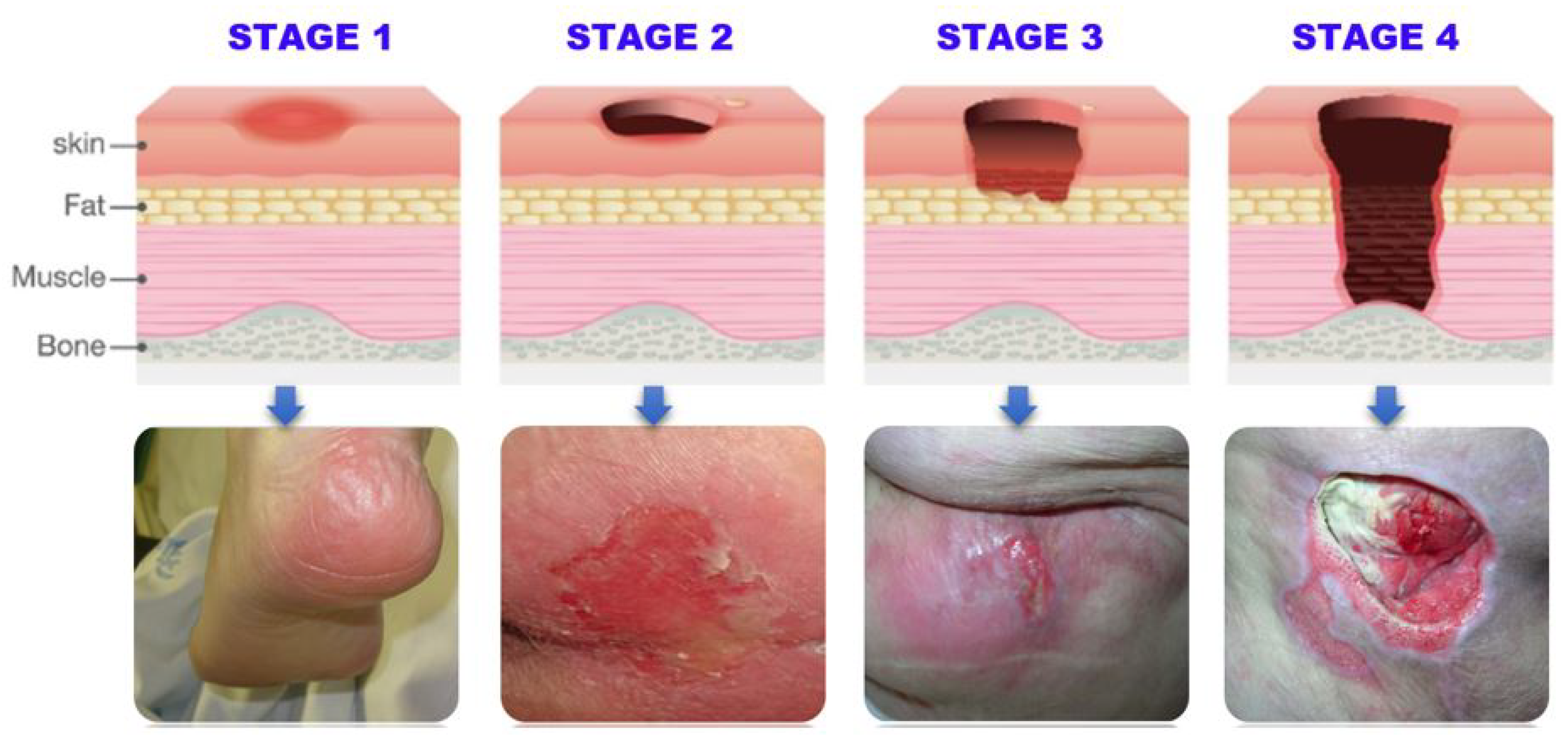
Table 1 summarizes the NPUAP staging system classification of pressure ulcers into four stages based on the severity of tissue damage, ranging from stage I (least severe) to stage IV (most severe).
Figure 2 provides a visual representation of the different stages.
Doctors and nurses typically classify pressure ulcers through visual inspection and manual palpation. During the assessment, they examine the wound site, look for signs of redness, blanching, and tissue loss, and take note of any other signs of infection or tissue damage. The healthcare provider may also use a probe or another instrument to assess the depth of the wound and evaluate any underlying tissue damage [
19]. Accurate pressure ulcer classification is crucial, but healthcare providers can misclassify them due to various reasons. These include a lack of training and experience, limited information, observer bias, and variability in wound appearance. Factors such as the patient’s skin color, age, and overall health can also influence the visual appearance of a pressure ulcer, making it difficult to classify the wound without additional information [
20]. Pressure ulcer classification requires a comprehensive assessment, considering various factors [
21]. Deep learning, a subtype of machine learning inspired by the human brain, has shown potential in image classification by automatically learning complex patterns [
22]. Computer vision and deep learning techniques have been widely adopted in various domains, including medical imaging, security, and image classification [
23,
24,
25,
26]. These techniques can detect and classify pressure ulcers by identifying visual features, such as color intensity, texture, depth, border, undermining, and tunneling, enabling earlier treatment and prevention of complications.
Table 2 outlines the detectable features of pressure ulcers at each stage, which can be utilized by computer vision techniques, such as convolutional neural networks (CNNs) for accurate and efficient detection and classification.
Hence, we conclude that pressure ulcers are serious medical conditions that require accurate and timely detection and classification. While current methods rely on manual inspection, there has been increasing interest in developing automated approaches that can provide more accurate and efficient results. Various computer vision techniques, such as CNNs and manual image processing, have been explored for this task. However, previous studies have limitations, such as small dataset sizes, reliance on limited feature extraction methods, and lack of generalizability. Therefore, a more comprehensive and accurate solution to pressure ulcer detection and classification is needed [
27,
28,
29,
30,
31,
32,
33].
To address this need, we propose a YOLO-based model for the detection and classification of pressure ulcers in medical images. Our method leverages the benefits of deep learning, such as its ability to automatically learn features, and employs data augmentation techniques to improve the model’s performance. We also use a dataset compiled from multiple sources with manually labeled images, which provides a more comprehensive and diverse set of training examples. Our study aims to accurately and efficiently detect and classify pressure ulcers, potentially leading to better patient outcomes and reduced healthcare costs.
Therefore, our work highlights the importance of image processing, computer vision, machine learning, and deep learning in the context of pressure ulcer detection and classification. By using a more comprehensive dataset and a more advanced deep learning model, we aim to address some of the limitations of previous studies and provide a more accurate and efficient solution to this critical healthcare problem. Our main contributions are as follows. We:
Developed a YOLO-based deep learning model for the detection and classification of pressure ulcers in medical images, which can accurately classify pressure ulcers into four stages based on severity.
Created a new dataset with the stage-wise classification of pressure ulcers, as no such dataset was previously available, by manually labeling images with bounding boxes and polygonal regions of interest using YOLO.
Added a large number of images of non-pressure ulcers, including surgical wounds and burns, to the dataset to make it more representative of real-world scenarios.
Utilized data augmentation techniques, such as rotation and flipping, to increase the dataset size and improve the model’s performance.
Compared the performance of the proposed model to other cutting-edge deep learning models as well as to conventional pressure ulcer detection and classification methods, indicating the model’s advantage in terms of accuracy and effectiveness.
Demonstrated the potential of the proposed model to aid in the timely diagnosis, treatment, and prevention of pressure ulcers, potentially improving patient outcomes and reducing healthcare costs.
Contributed to the field by providing a new dataset and an accurate and efficient solution for the detection and classification of pressure ulcers, which has the potential to advance the state-of-the-art in this important area of medical image analysis.
In
Section 2, we will first provide a comprehensive literature review of existing methods for pressure ulcer detection and classification, highlighting their strengths and limitations. In
Section 3, we will present the materials and methods used in our study, including details of the dataset and image labeling process, as well as a description of the YOLO-based deep learning model and data augmentation techniques employed.
Section 4 will report and discuss the results of our training and evaluation, including a comparison of our model’s performance with that of previous studies. In
Section 5, we will provide an in-depth analysis of the model’s strengths and limitations, and discuss potential directions for future research. Finally, in
Section 6, we will conclude our work with a summary of our key findings and contributions, as well as a discussion of the potential impact of our model on clinical practice and patient outcomes.
2. Literature Review
In recent years, a significant amount of research has been devoted to the use of deep learning in medical imaging. The literature review for this study primarily focuses on deep learning techniques applied to various medical imaging tasks, including object detection, image segmentation, and image classification.
Figure 3 illustrates a hierarchy of tasks and algorithms for analyzing medical images using deep learning.
As seen in
Figure 3, one area of research has been the development of convolutional neural networks (CNNs) for object detection in medical images. Researchers have applied CNNs to detect various structures in medical images, such as tumors, organs, and blood vessels. For example, Salama et al. and Agnes et al. [
34,
35] used a CNN to detect breast tumors in mammography images, Gao et al. and Monkam et al. [
36,
37] used a CNN to detect lung nodules in CT scans, and Li et al. and Ting et al. [
38,
39] used a CNN to detect retinal abnormalities in eye images. These studies have demonstrated the potential of CNNs for accurate and efficient object detection in medical images. Another method for identifying and segmenting liver tumors in multi-phase CT images is the phase attention mask R-CNN proposed by [
40]. This method selectively extracts features from each phase using an attention network for each scale and outperforms other methods for segmenting liver tumors in terms of segmentation accuracy. Several other notable studies have explored the application of RCNN, Faster RCNN, and Fast RCNN in the automatic detection and segmentation of medical images, including works by [
41,
42,
43,
44,
45,
46].
Deep learning is being increasingly used for image segmentation in medical imaging. Researchers have developed deep learning algorithms for segmenting different parts of the body in medical photographs, such as the brain in MRI images [
47], the lungs in CT scans [
48], and the retina in eye images [
49].
In addition to object detection and image segmentation, researchers have employed deep learning to classify medical images. The main focus of this study has been on creating algorithms for classifying medical images into specified categories, such as normal and abnormal, benign and malignant. For instance, Esteva et al. [
50] used an intelligent algorithm to categorize the images of skin lesions, Shu et al. [
51] used such a method to categorize mammography images, and Dansana et al. [
52] used a deep learning system to categorize images of chest X-rays into normal and abnormal. Hence, we found extensive literature studies that have demonstrated that deep learning can successfully and properly categorize diseases in medical images.
Since our research focus is also mainly on object detection and classification in pressure ulcer images, we outline several noteworthy publications that have used deep learning algorithms for detection and classification tasks in medical images in
Table 3.
YOLO (you only look once) is a recent deep learning approach used for object detection in medical images [
60]. Compared to traditional CNN, YOLO is designed to be fast and efficient, making it well-suited for real-time object detection in medical images. YOLO divides the image into a grid and predicts bounding boxes and class probabilities for each cell. For example, in the case of lung cancer screening, the bounding boxes would represent the location of the lung nodules, and the class probabilities would indicate whether the nodule is cancerous or not. This can aid radiologists in the early detection and diagnosis of lung cancer, resulting in better patient outcomes. In medical imaging, YOLO has been applied to the detection of various structures, including tumors, organs, and blood vessels. For example, researchers have used YOLO to detect lung nodules in CT scans and retinal abnormalities in eye images; Boonrod et al. [
61] used YOLO to detect abnormal cervical vertebrae in X-ray images, achieving higher accuracy and a faster processing time compared to traditional object detection methods. Similarly, Wojaczek et al. [
62] used YOLO to detect the location and shape of prostate cancer in magnetic resonance images. Compared to traditional CNNs, YOLO has been shown to be faster and more efficient, while still achieving similar or even better performance in object detection tasks. Furthermore, YOLO is designed to be easy to train and implement, making it accessible to researchers and practitioners who may have limited experience with deep learning [
63,
64,
65].
Various studies have explored the use of automated image analysis, deep learning, vision, and machine learning techniques for pressure ulcer detection, classification, and segmentation. These studies have demonstrated the feasibility of recognizing complicated structures in biomedical images with high accuracy, including the use of convolutional neural networks for tissue classification and segmentation, as well as the simultaneous segmentation and classification of stage 1–4 pressure injuries. However, these studies have their limitations, including the need for labeled datasets and the requirement for the manual selection of parameters. The authors of [
66] proposed a system that utilized a LiDAR sensor and deep learning models for automatically assessing pressure injuries, achieving satisfactory accuracy with U-Net outperforming Mask R-CNN; Zahia et al. [
67] used CNNs for automatic tissue classification in pressure injuries, achieving an overall classification accuracy of 92.01%; Liu et al. [
68] developed a system using deep learning algorithms to identify pressure ulcers and achieved high accuracy with the Inception-ResNet-v2 model; Fergus et al. [
69] used a faster region-based convolutional neural network and a mobile platform to classify pressure ulcers; Swerdlow et al. [
70] applied the Mask-R-CNN algorithm for simultaneous segmentation and classification of stage 1–4 pressure injuries; Elmogy et al. [
71] proposed a tissue classification system for pressure ulcers using a 3D-CNN.
Table 4 summarizes these studies.
In this study, YOLO is employed for the detection and classification of pressure ulcers into four stages, and non-pressure ulcer images are also considered. Augmentation techniques are used to enhance the quality and quantity of the available dataset. By collecting images from various sources, the study aims to overcome the limitations of previous studies and provide a more accurate and robust system for pressure ulcer detection and classification. This study’s results will be important for medical professionals and caregivers to make informed decisions on the prevention and treatment of pressure ulcers.
4. Results
We conducted our experiments on a Colab GPU with YOLOv5 version 7.0-114-g3c0a6e6, Python version 3.8.10, and Torch version 1.13.1+cu116. We trained the model using the following hyperparameters: a learning rate (lr0) of 0.01, momentum of 0.937, weight decay of 0.0005, and batch size of 18. We used the stochastic gradient descent (SGD) optimizer for 500 epochs with patience of 100, and saved the best model weights.
Based on the YOLOv5s model we trained, we achieved good results in terms of the overall mAP and individual class performance. The model achieved an overall mAP50 of 0.769 and mAP50-95 of 0.398 on the validation set. This means that the model was able to accurately detect and classify pressure ulcers with a high degree of confidence.
Figure 6 shows the loss values for the box loss, object loss, and class loss at each epoch during the training process. The box loss represents the difference between the predicted and ground-truth bounding box coordinates, the object loss represents the confidence score for each object detected in an image, and the class loss represents the probability of each detected object belonging to a specific class.
The goal of training an object detection model is to minimize the total loss, which is a combination of box loss, object loss, and class loss. The loss values should exhibit a decreasing trend as the training progresses, indicating an improvement in the model’s ability to detect different stages of pressure ulcers in the images.
Moreover, from
Figure 7, it appears that the precision, recall, and mean average precision (mAP) are all increasing with training epochs. This could indicate that the model improves over time and becomes more accurate at identifying the correct stages of pressure ulcers in the images.
From
Table 5, it can be observed that the model attained high precision and recall scores for the non-pressure ulcer (NonPU) and Stage 1 categories, suggesting that the model accurately detected and classified these classes. However, the recall score for Stage 2 was relatively low at 0.164, indicating that the model may have missed some instances of this category, possibly due to the small number of images available for this class.
Stage 3 and Stage 4 of our model have shown promising results in detecting and classifying pressure ulcers. Our proposed model achieved a mean average precision (mAP50) of 0.749 and 0.729, respectively, indicating a high level of accuracy in identifying and localizing pressure ulcers in the images. In Stage 3, the model accurately identified 26 instances of pressure ulcers with a precision (P) of 0.659 and recall (R) of 0.692, demonstrating its ability to detect and classify pressure ulcers even in challenging image conditions. In Stage 4, the model identified 22 instances of pressure ulcers with a precision (P) of 0.615 and recall (R) of 0.864, indicating its capability to correctly identify and classify a significant number of pressure ulcers with high accuracy.
Overall, the results of our YOLOv5s model suggest that it performed well in accurately detecting and classifying pressure ulcers in the images we used for training and validation. Hence, we can infer that these results demonstrate the potential of the YOLOv5 model for detecting and classifying pressure ulcers in medical images, which could have significant implications for improving patient care and outcomes.
In addition to the metrics presented in
Table 5, we also generated additional evaluation metrics to further analyze the performance of our YOLOv5 model. The precision confidence curve, recall confidence curve, precision–recall (PR) curves, F1 score curves, and confusion matrices can be found in
Figure 7,
Figure 8,
Figure 9,
Figure 10, and
Figure 11, respectively. These evaluation metrics provide a more detailed understanding of the model’s ability to accurately detect and classify pressure ulcers in the images. The PR curves and F1 score curves show the trade-off between precision and recall for different decision thresholds, while the confusion matrices provide information on the number of true positives, true negatives, false positives, and false negatives for each class.
6. Conclusions
Our study demonstrates the effectiveness of a deep learning-based approach for the automated detection and classification of pressure ulcers. By utilizing the YOLOv5 model, we achieved high accuracy in identifying pressure ulcers of different stages and non-pressure ulcers. Our proposed model outperformed existing CNN-based methods, showcasing the superiority of YOLOv5 for this task.
We also developed a diverse dataset of pressure ulcers and non-pressure ulcers, which we augmented to enhance the model’s robustness to variations in input data. Our model achieved high precision and recall scores for non-pressure ulcers and Stage 1 pressure ulcers, which are easier to identify due to their distinct features. However, the recall score for Stage 2 pressure ulcers was relatively low due to the limited number of images available for this class. To improve the model’s performance, we recommend collecting more images of Stage 2 pressure ulcers.
Our study’s findings demonstrate the potential of deep learning-based systems for automated pressure ulcer detection and classification, offering a promising solution for earlier intervention and improved patient outcomes. This technology could also reduce the workload of healthcare professionals, allowing them to focus on other essential tasks. To further advance this field, we plan to investigate a tailored YOLOv5 model trained on a larger and more diverse dataset, including images from different populations, races, and age groups. Additionally, we will explore the use of transfer learning by leveraging pre-trained models on other medical imaging datasets.