Evaluation of Machine Learning and Traditional Statistical Models to Assess the Value of Stroke Genetic Liability for Prediction of Risk of Stroke Within the UK Biobank
Abstract
1. Introduction
2. Material and Method
2.1. Ethical Approval
2.2. Study Population

2.3. Genotyping and Imputation
2.4. Definition of the Outcome
2.5. Demographics and Clinical and Lifestyle Features
2.6. Computation of Genetic Liabilities
Selection of Genetic Variants
2.7. Data Preprocessing
3. Statistical Analysis
3.1. The Relationship Between Genetic Liability and Stroke
3.2. Prediction Models Development
4. Model Performance Assessment
Assessment of the Predictive Value of Genetic Liability
5. Results
5.1. Study Characteristics
| Characteristic | Overall (N = 243,399) | Non-Event (N = 241,426) | Stroke Event (N = 1973) | HR (95% CI) | p-Value |
|---|---|---|---|---|---|
| DM, yes; n (%) | 6939 (2.9%) | 6826 (2.8%) | 113 (5.7%) | 2.08(1.72, 2.51) | <0.001 |
| Hypertension, yes; n (%) | 116,216 (47.7%) | 114,840 (47.6%) | 1376 (69.7%) | 2.52(1.29, 2.78) | <0.001 |
| Sex, male; n (%) | 102,187 (42.0%) | 101,107 (41.9%) | 1080 (54.7%) | 1.67 (1.53, 1.83) | <0.001 |
| Age (years), mean (SD) | 55.4 (7.98) | 55.4 (7.98) | 60.0 (7.14) | 1.93 (1.83, 2.03) | <0.0001 |
| Body mass index (kg/m2), mean (SD) | 26.8 (4.57) | 26.8 (4.57) | 27.4 (4.83) | 1.12 (1.08, 1.17) | <0.001 |
| Total cholesterol (mmol/L), mean (SD) | 5.91 (1.06) | 5.91 (1.06) | 5.94 (1.09) | 1.03 (0.98, 1.07) | 0.30 * |
| LDL (mmol/L), mean (SD) | 4.68 (2.37) | 4.67 (2.36) | 5.03 (2.51) | 1.03 (1.03, 1.12) | 0.002 |
| Smoking | |||||
| Current; n (%) | 76,397 (31.4%) | 75,647 (31.3%) | 750 (38.0%) | REF | REF |
| Previous; n (%) | 2900 (1.2%) | 2855 (1.2%) | 45 (2.3%) | 1.58 (1.17, 2.13) | 0.003 |
| Never; n (%) | 164,102 (67.4%) | 162,924 67.5%) | 1178 (59.7%) | 0.73 (0.67, 0.80) | <0.001 |
| Alcohol | |||||
| Current; n (%) | 228,349 (93.8%) | 226,556(93.8%) | 1793 (90.9%) | REF | REF |
| Previous; n (%) | 7082 (2.9%) | 6996 (2.9%) | 86 (4.4%) | 1.55 (1.25, 1.93) | <0.001 |
| Never; n (%) | 7968 (3.3%) | 7874 (3.3%) | 94 (4.8%) | 1.50 (1.22, 1.85) | <0.001 |

5.2. The Association of Genetic Liability with Incident Stroke
| Genetic liability Level | HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|---|
| Model 1 | Model 2 | Model 3 | Model 4 | |||||
| Moderate risk | 1.06 (0.95, 1.18) | 0.31 | 1.06 (0.95, 1.18) | 0.31 | 1.05 (0.94, 1.17) | 0.04 | 1.05 (0.94, 1.17) | 0.40 |
| High risk | 1.15 (1.03, 1.28) | 0.01 | 1.16 (1.04, 1.30) | 0.01 | 1.14 (1.02, 1.27) | 0.02 | 1.14 (1.02, 1.27) | 0.02 |
| Genetic liability (continuous) | 1.08 (1.03, 1.13) | <0.001 | 1.08 (1.03, 1.13) | <0.001 | 1.07 (1.03, 1.12) | 0.002 | 1.07 (1.02, 1.12) | 0.003 |
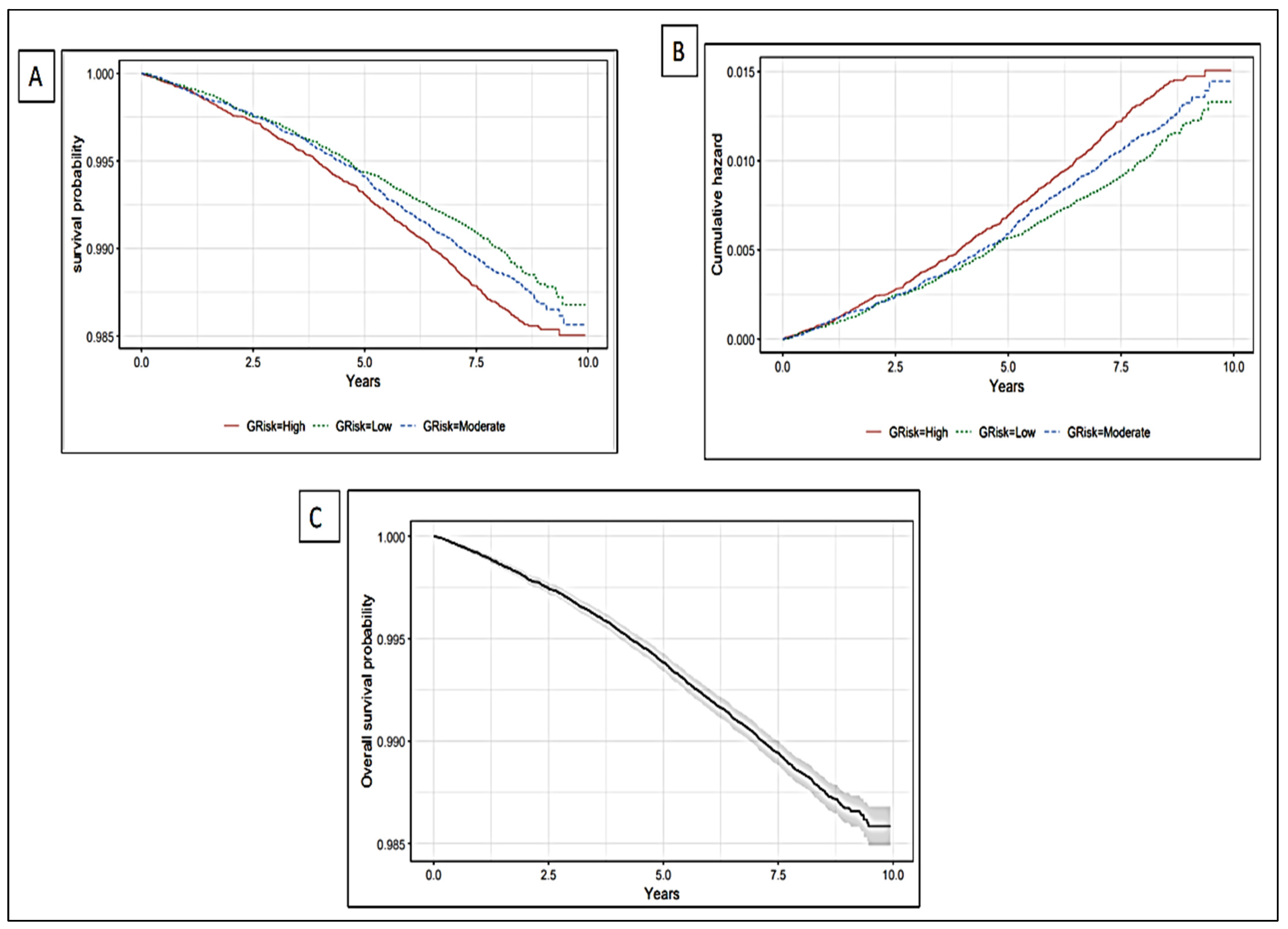

5.3. Prediction Value of the Conventional Factors
| Characteristics | Chi-Square | df | p-Value |
|---|---|---|---|
| Sex | 0.42 | 1 | 0.52 |
| Age | 0.82 | 1 | 0.37 |
| BMI | 7.49 | 1 | 0.01 |
| DM | 0.08 | 1 | 0.78 |
| HTN | 0.14 | 1 | 0.71 |
| LDL | 0.36 | 1 | 0.55 |
| Smoking | 1.40 | 1 | 0.24 |
| Alcohol | 0.12 | 1 | 0.73 |
| SGL | 2.48 | 1 | 0.12 |
| GLOBAL | 13.43 | 9 | 0.14 |
| Models | AUC 95%CI | NRI (95% CI) | p-Value for NRI | IDI (95% CI) | p-Value for IDI | Brier Score | ICI | |
|---|---|---|---|---|---|---|---|---|
| Coxph | Model 1 | 69.43 (67.30, 71.56) | REF | REF | REF | REF | 0.01 | 0.002 |
| Model 2 | 69.54 (67.40, 71.68) | 0.20 (0.119, 0.285) | 0.00 | 1.0 × 10−4 (0.000, 3.0 × 10−4) | 0.14 | 0.01 | 0.002 | |
| GBM | Model 1 | 69.34 (67.23, 71.50) | REF | REF | REF | REF | 0.01 | 0.001 |
| Model 2 | 69.38 (67.26, 71.50) | −0.11 (−0.193, −0.027) | 0.01 | 0.00 (−1.0 × 10−4, 1.0 × 10−4) | 0.61 | 0.01 | 0.001 | |
| DT ** | Model 1 | 61.40 (59.30, 63.40) | REF | REF | REF | REF | 0.01 | 0.001 |
| Model 2 | 61.40 (59.30, 63.40) | 0.00 (0.00, 0.00) | NaN | 0.00 (0.000, 0.000) | NaN | 0.01 | 0.001 | |
| DT | Model 1 | 67.58 (65.46, 69.70) | REF | REF | REF | REF | 0.01 | 0.001 |
| Model 2 | 67.58 (65.46, 69.70) | REF | REF | REF | REF | 0.01 | 0.001 | |
| RF | Model 1 | 65.62 (63.48, 67.75) | REF | REF | REF | REF | 0.01 | 0.003 |
| Model 2 | 65.35 (63.18, 67.52) | 0.17 (0.087, 0.249) | 5.0 × 10−5 | 0.00 (−7.0 × 10−4, 8.0 × 10−4) | 0.98 | 0.01 | 0.003 |
5.4. Prediction Value of Genetic Liability
6. Discussion
6.1. Main Findings
6.2. Implication
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthi, R.; Ikeda, T.; Feigin, V. Global, Regional and Country-Specific Burden of Ischaemic Stroke, Intracerebral Haemorrhage and Subarachnoid Haemorrhage: A Systematic Analysis of the Global Burden of Disease Study 2017. Neuroepidemiology 2020, 54, 171–179. [Google Scholar] [CrossRef]
- Dhamoon, M.S.; Tai, W.; Boden-Albala, B.; Rundek, T.; Paik, M.C.; Sacco, R.L.; Elkind, M.S.V. Risk of Myocardial Infarction or Vascular Death After First Ischemic Stroke: The Northern Manhattan Study. Stroke 2007, 38, 1752–1758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dhamoon, M.S.; Sciacca, R.R.; Rundek, T.; Sacco, R.L.; Elkind, M.S.V. Recurrent stroke and cardiac risks after first ischemic stroke: The Northern Manhattan study. Neurology 2006, 66, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Kyme, C. After ischemic stroke, patients are at higher risk of recurrent stroke than of cardiac events. Nat. Clin. Pract. Cardiovasc. Med. 2005, 2, 436. [Google Scholar] [CrossRef]
- Engstad, T.; Viitanen, M.; Arnesen, E. Predictors of Death Among Long-Term Stroke Survivors. Stroke 2003, 34, 2876–2880. [Google Scholar] [CrossRef]
- King, D.; Wittenberg, R.; Patel, A.; Quayyum, Z.; Berdunov, V.; Knapp, M. The future incidence, prevalence and costs of stroke in the UK. Age Ageing 2020, 49, 277–282. [Google Scholar] [CrossRef]
- Boehme, A.K.; Esenwa, C.; Elkind, M.S.V. Stroke Risk Factors, Genetics, and Prevention. Circ. Res. 2017, 120, 472–495. [Google Scholar] [CrossRef]
- Benjamin, E.J.; Blaha, M.J.; Chiuve, S.E.; Cushman, M.; Das, S.R.; Deo, R.; de Ferranti, S.D.; Floyd, J.; Fornage, M.; Gillespie, C.; et al. Heart Disease and Stroke Statistics—2017 Update: A Report From the American Heart Association. Circulation 2017, 135, e146–e603. [Google Scholar] [CrossRef]
- Bak, S.; Gaist, D.; Sindrup, S.H.; Skytthe, A.; Christensen, K. Genetic Liability in Stroke: A Long-Term Follow-Up Study of Danish Twins. Stroke 2002, 33, 769–774. [Google Scholar] [CrossRef]
- Malik, R.; Chauhan, G.; Traylor, M.; Okada, Y.; Giese, A.K.; Laan, S.; Chong, M.; Adams, H.; Ago, T.; Almgren, P.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef]
- Malik, R.; Rannikmäe, K.; Traylor, M.; Georgakis, M.K.; Sargurupremraj, M.; Markus, H.S.; Hopewell, J.C.; Debette, S.; Sudlow, C.L.M.; Dichgans, M. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Ann. Neurol. 2018, 84, 934–939. [Google Scholar] [CrossRef]
- American Heart Association News. New tool brings big changes to cardiovascular disease predictions. In Premium Official News; American Heart Association News: Dallas, TX, USA, 2023. [Google Scholar]
- Elias, M.F.; Sullivan, L.M.; D’Agostino, R.B.; Elias, P.K.; Beiser, A.; Au, R.; Seshadri, S.; DeCarli, C.; Wolf, P.A. Framingham Stroke Risk Profile and Lowered Cognitive Performance. Stroke 2004, 35, 404–409. [Google Scholar] [CrossRef]
- Bohr, A.; Memarzadeh, K. The rise of artificial intelligence in healthcare applications. Artificial Intelligence in Healthcare; Academic Press: Cambridge, MA, USA, 2020; pp. 25–60. [Google Scholar]
- Knowles, J.W.; Ashley, E.A. Cardiovascular disease: The rise of the genetic risk score. PLoS Med. 2018, 15, e1002546. [Google Scholar] [CrossRef] [PubMed]
- Traylor, M.; Farrall, M.; Holliday, E.G.; Sudlow, C.; Hopewell, J.C.; Cheng, Y.C.; Fornage, M.; Ikram, M.A.; Malik, R.; Bevan, S.; et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE Collaboration): A meta-analysis of genome-wide association studies. Lancet Neurol. 2012, 11, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Rutten-Jacobs, L.; Inouye, M. Risk Prediction Using Polygenic Risk Scores for Prevention of Stroke and Other Cardiovascular Diseases. Stroke 2021, 52, 2983–2991. [Google Scholar] [CrossRef] [PubMed]
- Gschwendtner, A.; Dichgans, M. Genetics of ischemic stroke. Nervenarzt 2013, 84, 166. [Google Scholar] [CrossRef]
- Della-Morte, D.; Guadagni, F.; Palmirotta, R.; Testa, G.; Caso, V.; Paciaroni, M.; Abete, P.; Rengo, F.; Ferroni, P.; Sacco, R.L.; et al. Genetics of ischemic stroke, stroke-related risk factors, stroke precursors and treatments. Pharmacogenomics 2012, 13, 595–613. [Google Scholar] [CrossRef]
- Mishra, A.; Malik, R.; Hachiya, T.; Jürgenson, T.; Namba, S.; Posner, D.C.; Kamanu, F.K.; Koido, M.; Le Grand, Q.; Shi, M.; et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 2022, 611, 115–123. [Google Scholar] [CrossRef]
- Myserlis, E.P.; Georgakis, M.K.; Demel, S.L.; Sekar, P.; Chung, J.; Malik, R.; Hyacinth, H.I.; Comeau, M.E.; Falcone, G.J.; Langefeld, C.D.; et al. A Genomic Risk Score Identifies Individuals at High Risk for Intracerebral Hemorrhage. Stroke 2023, 54, 973–982. [Google Scholar] [CrossRef]
- Rutten-Jacobs, L.C.; Larsson, S.C.; Malik, R.; Rannikmäe, K.; Sudlow, C.L.; Dichgans, M.; Markus, H.S.; Traylor, M. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: Cohort study of 306 473 UK Biobank participants. BMJ 2018, 363, k4168. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Sun, Z.; Sun, D.; Yu, C.; Guo, Y.; Sun, D.; Pang, Y.; Pei, P.; Yang, L.; Millwood, I.Y.; et al. Associations of polygenic risk scores with risks of stroke and its subtypes in Chinese. Stroke Vasc. Neurol. 2024, 9, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Abraham, G.; Malik, R.; Yonova-Doing, E.; Salim, A.; Wang, T.; Danesh, J.; Butterworth, A.S.; Howson, J.M.M.; Inouye, M.; Dichgans, M. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat. Commun. 2019, 10, 5819. [Google Scholar] [CrossRef]
- Verbaas, C.; Fornage, M.; Bis, J.C.; Choi, S.H.; Psaty, B.M.; Meigs, J.B.; Rao, M.; Nalls, M.; Fontes, J.D.; O’Donnell, C.J.; et al. Predicting Stroke Through Genetic Risk Functions the CHARGE Risk Score Project. Stroke 2014, 45, 403–412. [Google Scholar] [CrossRef]
- Bakker, M.K.; Kanning, J.P.; Abraham, G.; Martinsen, A.E.; Winsvold, B.S.; Zwart, J.A.; Bourcier, R.; Sawada, T.; Koido, M.; Kamatani, Y.; et al. Genetic Risk Score for Intracranial Aneurysms: Prediction of Subarachnoid Hemorrhage and Role in Clinical Heterogeneity. Stroke 2023, 54, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Bevan, S.; Nalls, M.A.; Holliday, E.G.; Devan, W.J.; Cheng, Y.C.; Ibrahim-Verbaas, C.A.; Verhaaren, B.F.; Bis, J.C.; Joon, A.Y.; et al. Multilocus Genetic Risk Score Associates with Ischemic Stroke in Case–Control and Prospective Cohort Studies. Stroke 2014, 45, 394–402. [Google Scholar] [CrossRef]
- Hachiya, T.; Hata, J.; Hirakawa, Y.; Yoshida, D.; Furuta, Y.; Kitazono, T.; Shimizu, A.; Ninomiya, T. Genome-Wide Polygenic Score and the Risk of Ischemic Stroke in a Prospective Cohort: The Hisayama Study. Stroke 2020, 51, 759–765. [Google Scholar] [CrossRef]
- Hachiya, T.; Kamatani, Y.; Takahashi, A.; Hata, J.; Furukawa, R.; Shiwa, Y.; Yamaji, T.; Hara, M.; Tanno, K.; Ohmomo, H.; et al. Genetic Predisposition to Ischemic Stroke: A Polygenic Risk Score. Stroke 2017, 48, 253–258. [Google Scholar] [CrossRef]
- Lynch, C.M.; Abdollahi, B.; Fuqua, J.D.; de Carlo, A.R.; Bartholomai, J.A.; Balgemann, R.N.; van Berkel, V.H.; Frieboes, H.B. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int. J. Med. Inform. 2017, 108, 1–8. [Google Scholar] [CrossRef]
- Chun, M.; Clarke, R.; Cairns, B.J.; Clifton, D.; Bennett, D.; Chen, Y.; Guo, Y.; Pei, P.; Lv, J.; Yu, C.; et al. Stroke risk prediction using machine learning: A prospective cohort study of 0.5 million Chinese adults. J. Am. Med. Inform. Assoc. 2021, 28, 1719–1727. [Google Scholar] [CrossRef]
- MacCarthy, G.; Pazoki, R. Using Machine Learning to Evaluate the Value of Genetic Liabilities in the Classification of Hypertension within the UK Biobank. J. Clin. Med. 2024, 13, 2955. [Google Scholar] [CrossRef] [PubMed]
- Schjerven, F.E.; Ingeström, E.M.L.; Steinsland, I.; Lindseth, F. Development of risk models of incident hypertension using machine learning on the HUNT study data. Sci. Rep. 2024, 14, 5609. [Google Scholar] [CrossRef] [PubMed]
- Wongvibulsin, S.; Wu, K.C.; Zeger, S.L. Clinical risk prediction with random forests for survival, longitudinal, and multivariate (RF-SLAM) data analysis. BMC Med. Res. Methodol. 2019, 20, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Niu, M.; Li, R.; Tu, R.; Liu, X.; Hou, J.; Mao, Z.; Wang, Z.; Wang, C. Genetic Risk Score Increased Discriminant Efficiency of Predictive Models for Type 2 Diabetes Mellitus Using Machine Learning: Cohort Study. Front. Public Health 2021, 9, 606711. [Google Scholar] [CrossRef]
- Datema, F.R.; Moya, A.; Krause, P.; Bäck, T.; Willmes, L.; Langeveld, T.; Baatenburg de Jong, R.J.; Blom, H.M. Novel head and neck cancer survival analysis approach: Random survival forests versus cox proportional hazards regression. Head Neck 2012, 34, 50–58. [Google Scholar] [CrossRef]
- Qiu, X.; Gao, J.; Yang, J.; Hu, J.; Hu, W.; Kong, L.; Lu, J.J. A Comparison Study of Machine Learning (Random Survival Forest) and Classic Statistic (Cox Proportional Hazards) for Predicting Progression in High-Grade Glioma after Proton and Carbon Ion Radiotherapy. Front. Oncol. 2020, 10, 551420. [Google Scholar] [CrossRef]
- Xu, L.; Cai, L.; Zhu, Z.; Chen, G. Comparison of the cox regression to machine learning in predicting the survival of anaplastic thyroid carcinoma. BMC Endocr. Disord. 2023, 23, 129. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Harding, D.; Slabaugh, G.; Marouli, E.; Deloukas, P. Prediction of atrial fibrillation and stroke using machine learning models in UK Biobank. Heliyon 2024, 10, e28034. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, Y.; Tan, Y.; Zhou, M.; Jiang, Y.; Liu, B. A comparison of random survival forest and Cox regression for prediction of mortality in patients with hemorrhagic stroke. BMC Med. Inform. Decis. Mak. 2023, 23, 215. [Google Scholar] [CrossRef]
- Chen, Y.; Chung, J.; Yeh, Y.; Lou, S.; Lin, H.; Lin, C.; Hsien, H.; Hung, K.; Yeh, S.J.; Shi, H. Predicting 30-Day Readmission for Stroke Using Machine Learning Algorithms: A Prospective Cohort Study. Front. Neurol. 2022, 13, 875491. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’connell, J.; et al. Genome-wide genetic data on ~500,000 UK biobank participants. bioRxiv 2017. [Google Scholar] [CrossRef]
- Welsh, S.; Peakman, T.; Sheard, S.; Almond, R. Comparison of DNA quantification methodology used in the DNA extraction protocol for the UK Biobank cohort. BMC Genom. 2017, 18, 26. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The Uk biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Marchini, J.; O’Connell, J.; Delaneau, O.; Sharp, K.; Kretzschmar, W.; Band, G.; McCarthy, S.; Petkova, D.; Bycroft, C.; Freeman, C.; et al. UK Biobank Phasing and Imputation Documentation Contributors to UK Biobank Phasing and Imputation. 2015. Available online: https://biobank.ctsu.ox.ac.uk/crystal/ukb/docs/impute_ukb_v1.pdf (accessed on 1 December 2023).
- Sacks, D.B.; Arnold, M.; Bakris, G.L.; Bruns, D.E.; Horvath, A.R.; Kirkman, M.S.; Lernmark, A.; Metzger, B.E.; Nathan, D.M. Guidelines and Recommendations for Laboratory Analysis in the Diagnosis and Management of Diabetes Mellitus. Clin. Chem. 2011, 57, e1–e47. [Google Scholar] [CrossRef]
- Flack, J.M.; Adekola, B. Blood pressure and the new ACC/AHA hypertension guidelines. Trends Cardiovasc. Med. 2020, 30, 160–164. [Google Scholar] [CrossRef]
- Chobanian, A.V.; Bakris, G.L.; Black, H.R.; Cushman, W.C.; Green, L.A.; Izzo, J.; Joseph, L.; Jones, D.W.; Materson, B.J.; Oparil, S.; et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: The JNC 7 Report. J. Am. Med. Assoc. 2003, 289, 2560–2571. [Google Scholar] [CrossRef]
- Pazoki, R.; Dehghan, A.; Evangelou, E.; Warren, H.; Gao, H.; Caulfield, M.; Elliott, P.; Tzoulaki, I. Genetic Predisposition to High Blood Pressure and Lifestyle Factors: Associations with Midlife Blood Pressure Levels and Cardiovascular Events. Circulation 2018, 137, 653–661. [Google Scholar] [CrossRef]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Chang, C.C. Data management and summary statistics with PLINK. Methods Mol. Biol. 2020, 2090, 49–65. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- Tabachnick, B.G.; Fidell, L.S. Using Multivariate Statistics, 6th ed.; Pearson: Boston, MA, USA, 2013. [Google Scholar]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Deo, S.V.; Deo, V.; Sundaram, V. Survival analysis—Part 2: Cox proportional hazards model. Indian J. Thorac. Cardiovasc. Surg. 2021, 37, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Abd ElHafeez, S.; D’Arrigo, G.; Leonardis, D.; Fusaro, M.; Tripepi, G.; Roumeliotis, S. Methods to Analyze Time-to-Event Data: The Cox Regression Analysis. Oxidative Med. Cell. Longev. 2021, 2021, 1302811. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning, 2nd ed.; corrected at 5 print ed.; Springer: New York, NY, USA, 2011. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Platt, J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Adv. Largin Margin Classif. 1999, 10, 61–74. [Google Scholar]
- Huang, Y.; Li, W.; Macheret, F.; Gabriel, R.A.; Ohno-Machado, L. A tutorial on calibration measurements and calibration models for clinical prediction models. J. Am. Med. Inform. Assoc. 2020, 27, 621–633. [Google Scholar] [CrossRef]
- Van Calster, B.; Nieboer, D.; Vergouwe, Y.; De Cock, B.; Pencina, M.J.; Steyerberg, E.W. A calibration hierarchy for risk models was defined: From utopia to empirical data. J. Clin. Epidemiol. 2016, 74, 167–176. [Google Scholar] [CrossRef]
- Miller, T.D.; Askew, J.W. Net reclassification improvement and integrated discrimination improvement: New standards for evaluating the incremental value of stress imaging for risk assessment. Circulation. Cardiovasc. Imaging 2013, 6, 496–498. [Google Scholar] [CrossRef]
- McKearnan, S.B.; Wolfson, J.; Vock, D.M.; Vazquez-Benitez, G.; O’Connor, P.J. Performance of the Net Reclassification Improvement for Nonnested Models and a Novel Percentile-Based Alternative. Am. J. Epidemiol. 2018, 187, 1327–1335. [Google Scholar] [CrossRef]
- Pencina, M.J.; D’Agostino, R.B., Sr.; Steyerberg, E.W. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat. Med. 2011, 30, 11–21. [Google Scholar] [CrossRef]
- Steyerberg, E.; Vickers, A.; Cook, N.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.; Kattan, M. Assessing the performance of prediction models: A framework for traditional and novel measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef]
- Clark, K.; Fu, W.; Liu, C.; Ho, P.; Wang, H.; Lee, W.; Chou, S.; Wang, L.; Tzeng, J. The prediction of Alzheimer’s disease through multi-trait genetic modeling. Front. Aging Neurosci. 2023, 15, 1168638. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Goddard, M.E. Multi-locus models of genetic risk of disease. Genome Med. 2010, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Namba, S.; Lopera, E.; Kerminen, S.; Tsuo, K.; Läll, K.; Kanai, M.; Zhou, W.; Favé, M.-J.; Bhatta, L.; et al. Global Biobank analyses provide lessons for developing polygenic risk scores across diverse cohorts. Cell Genom. 2023, 3, 100241. [Google Scholar] [CrossRef] [PubMed]
- Cárcel-Márquez, J.; Muiño, E.; Gallego-Fabrega, C.; Cullell, N.; Lledós, M.; Llucià-Carol, L.; Sobrino, T.; Campos, F.; Castillo, J.; Freijo, M.; et al. A Polygenic Risk Score Based on a Cardioembolic Stroke Multitrait Analysis Improves a Clinical Prediction Model for This Stroke Subtype. Front. Cardiovasc. Med. 2022, 9, 940696. [Google Scholar] [CrossRef]
- Jung, K.J.; Hwang, S.; Lee, S.; Kim, H.C.; Jee, S.H. Traditional and Genetic Risk Score and Stroke Risk Prediction in Korea. Korean Circ. J. 2018, 48, 731–740. [Google Scholar] [CrossRef]
- Du, M.; Haag, D.G.; Lynch, J.W.; Mittinty, M.N. Comparison of the Tree-Based Machine Learning Algorithms to Cox Regression in Predicting the Survival of Oral and Pharyngeal Cancers: Analyses Based on SEER Database. Cancers 2020, 12, 2802. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
MacCarthy, G.; Pazoki, R. Evaluation of Machine Learning and Traditional Statistical Models to Assess the Value of Stroke Genetic Liability for Prediction of Risk of Stroke Within the UK Biobank. Healthcare 2025, 13, 1003. https://doi.org/10.3390/healthcare13091003
MacCarthy G, Pazoki R. Evaluation of Machine Learning and Traditional Statistical Models to Assess the Value of Stroke Genetic Liability for Prediction of Risk of Stroke Within the UK Biobank. Healthcare. 2025; 13(9):1003. https://doi.org/10.3390/healthcare13091003
Chicago/Turabian StyleMacCarthy, Gideon, and Raha Pazoki. 2025. "Evaluation of Machine Learning and Traditional Statistical Models to Assess the Value of Stroke Genetic Liability for Prediction of Risk of Stroke Within the UK Biobank" Healthcare 13, no. 9: 1003. https://doi.org/10.3390/healthcare13091003
APA StyleMacCarthy, G., & Pazoki, R. (2025). Evaluation of Machine Learning and Traditional Statistical Models to Assess the Value of Stroke Genetic Liability for Prediction of Risk of Stroke Within the UK Biobank. Healthcare, 13(9), 1003. https://doi.org/10.3390/healthcare13091003






