Identifying Robust Risk Factors for Knee Osteoarthritis Progression: An Evolutionary Machine Learning Approach
Abstract
1. Introduction
2. Methods
2.1. Dataset Description
2.2. Problem Definition
2.3. Data Pre-Processing
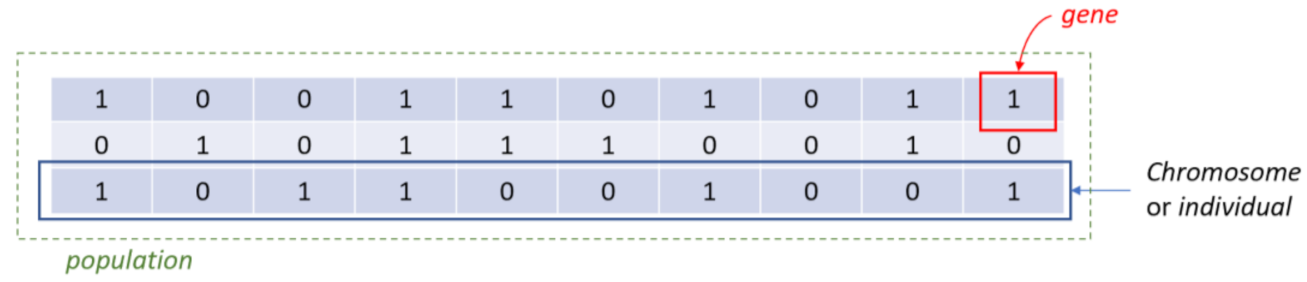
2.4. Feature Selection
- Step 1. InitializationA group of k chromosomes are randomly generated, forming the initial population of individuals.
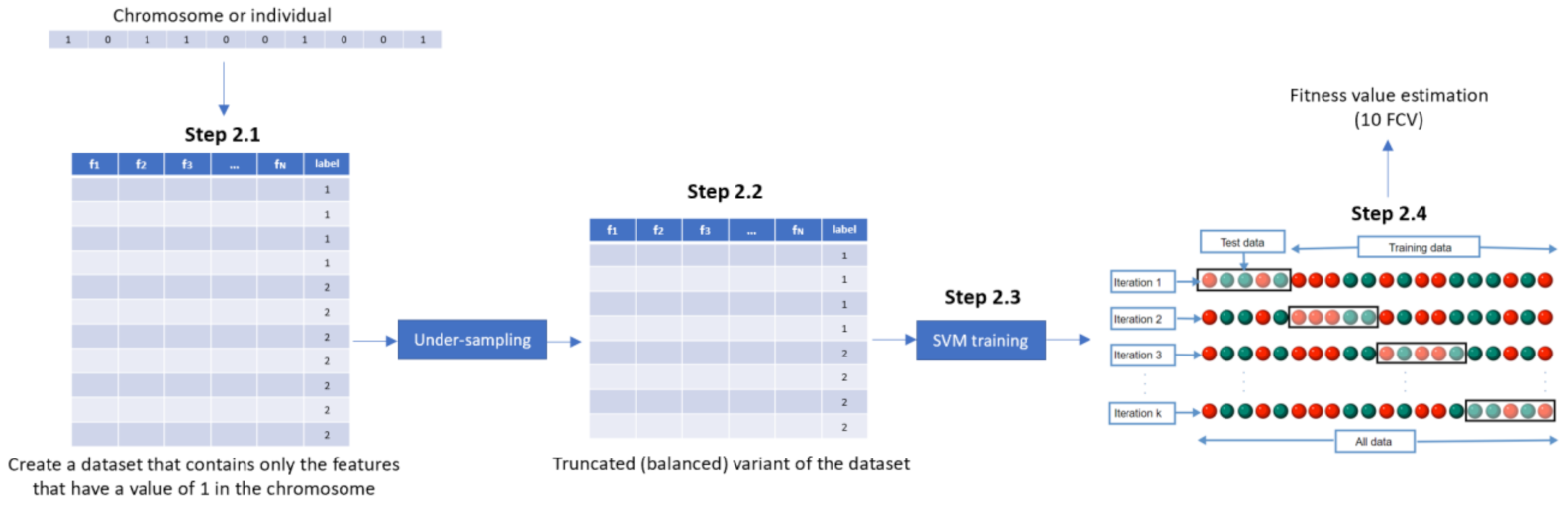
- Step 2. Fitness assignmentA fitness value is assigned to each chromosome in the population. Specifically, the process of measuring fitness in GenWrapper can be summarized as follows. The following 3-step process (Figure 3) is repeated for each of the chromosomes of the population:
- Step 2.1. From the training dataset, we keep only the features that have a value of 1 in the current chromosome. This creates a truncated training set.
- Step 2.2. Random undersampling on the majority class is performed on the truncated training set. This action leads to a balanced variant of the truncated training set.
- Step 2.3. A classifier is trained on the newly produced balanced dataset. Linear support vector machines (SVMs) have been chosen as the main classification criterion due to their generalization capability.
- Step 2.4. A k-fold cross-validation scheme is employed to validate the classifier performance that is finally assigned as a fitness value to the specific individual.
- Step 3. Termination conditionThe algorithm stops if the average relative change in the best fitness function value over Κ generations is less than or equal to a pre-determined threshold.
- Step 4. Generation of a new populationIn case the termination criterion is not satisfied, a new population of individuals is generated by applying the following three GA operators:Selection operator: The best individuals are selected according to their fitness value.Crossover operator: This operator recombines the selected individuals to generate a new population.Mutation operator: Mutated versions of the new individuals are created by randomly changing genes in the chromosomes (e.g., by flipping a 0 to 1 and vice versa).
- Step 5. The algorithm returns to step 2.
- Step 6: Final feature ranking determinationUpon termination of the GA algorithm, the features are ranked with respect to the number of times that they have been selected in all the individuals (chromosomes) of the final population.
- Step 6.1. A feature gets a vote when it has a value of 1 in a chromosome of the final generation.
- Step 6.2. Step 6.1 is repeated for all the chromosomes of the final generation and the features’ votes are summed up.
- Step 6.3. Features are ranked in descending order with respect to the total number of votes received.
2.5. Learning
2.6. Validation
- Step 1. Random undersampling is applied on the majority class, and the retained samples along with those from the minority class form a balanced binary dataset.
- Step 2. A classifier is built on the balanced binary dataset and its accuracy is calculated using 10-fold cross-validation (10FCV).
- Step 3. Steps 1 and 2 are repeated 10 times, each one using a different randomly generated balanced dataset.
- Step 4. The final performance is calculated by averaging the obtained 10FCV classification accuracies. The resulting final performance will be referred to here as mean 10FCV.
2.7. Explainability
3. Results
3.1. Selection Criterion
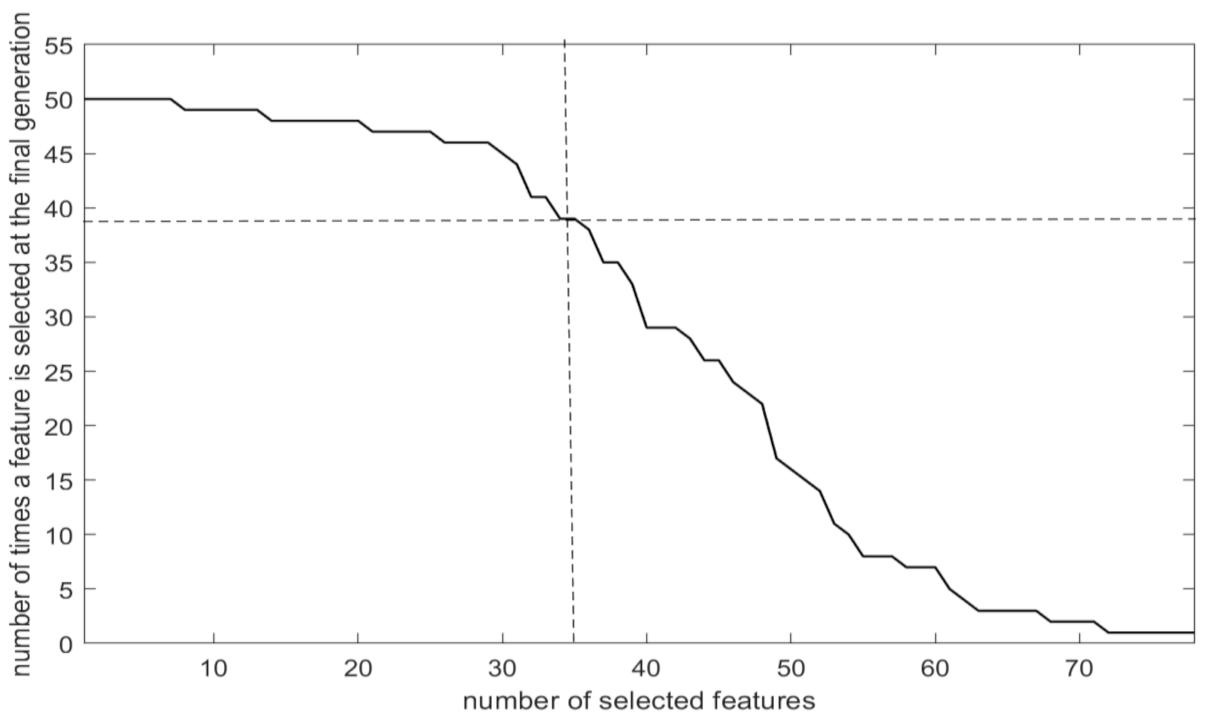
3.2. Features Selected
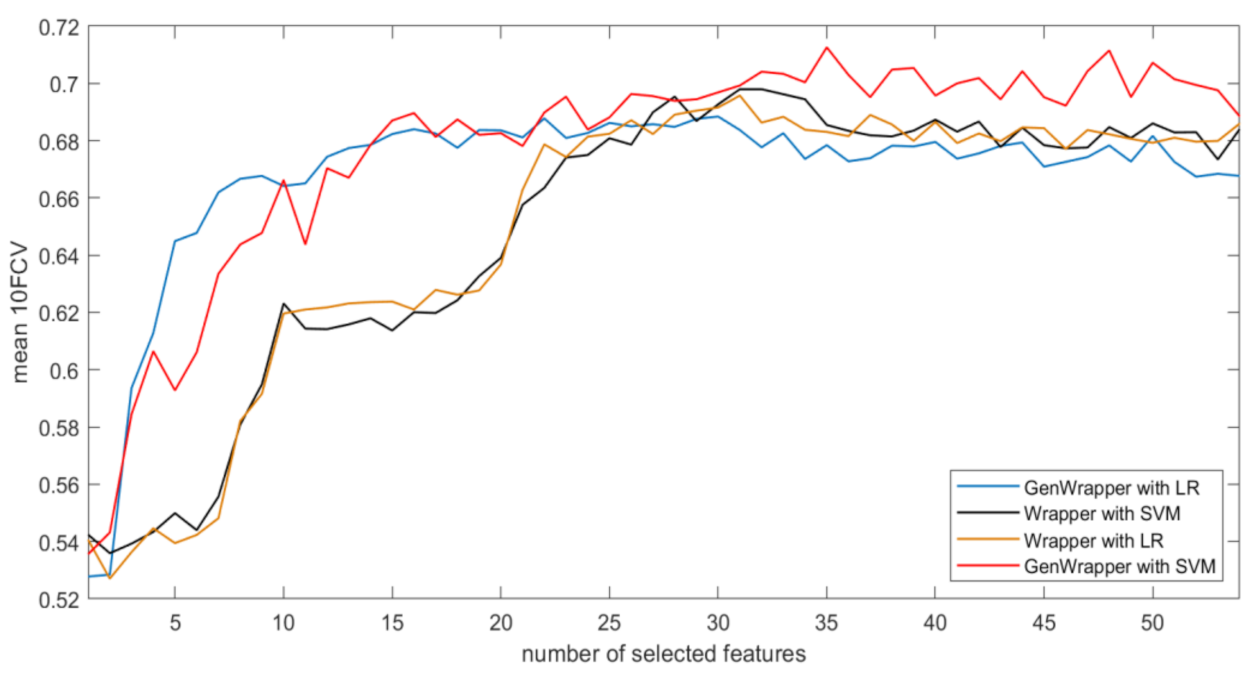
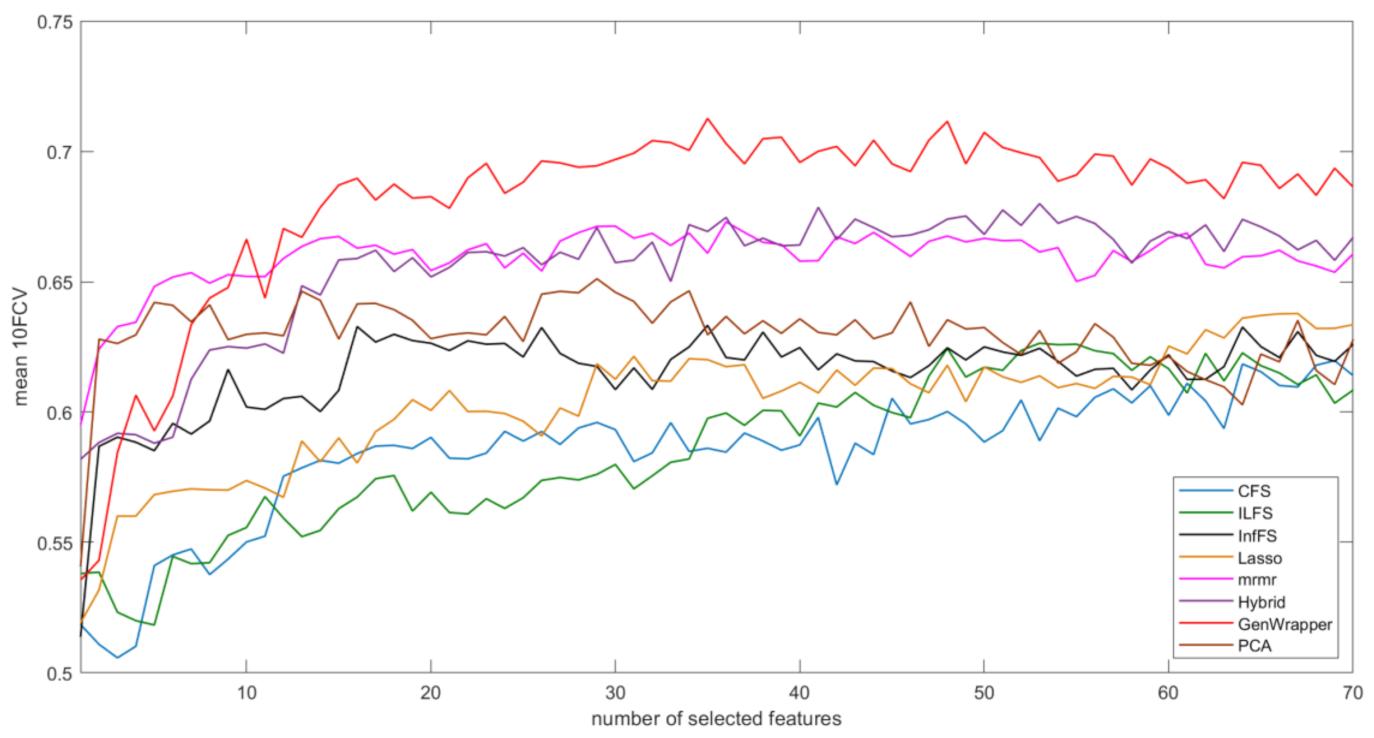
3.3. Comparative Analysis
- GenWrapper significantly outperforms the classical wrapper FS, especially for a small number of selected features (up to 20). This superiority is proven for both SVM and LR;
- GenWrapper employing SVM gives the best overall performance (71.25% at 35 selected features).
3.4. Explainability Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Selected Features | Description | Feature Category |
|---|---|---|
| P01SVLKOST | Left knee baseline X-ray: evidence of knee osteophytes | Medical imaging outcome |
| P01BMI | Body mass index | Subject characteristics |
| V00SUPCA | Block Brief 2000: average daily nutrients from vitamin supplements, calcium (mg) | Nutrition |
| V00EDCV | Highest grade or year of school completed | Behavioral |
| V00FFQ59 | Block Brief 2000: ice cream/frozen yogurt/ice cream bars, eat how often, past 12 months | Nutrition |
| V00KQOL2 | Quality of life: modified lifestyle to avoid potentially damaging activities to knee(s) | Behavioral |
| V00CHNFQCV | Chondroitin sulfate frequency of use, past 6 months | Medical history |
| V00WOMSTFR | Right knee: WOMAC Stiffness Score | Symptoms |
| V00FFQSZ13 | Block Brief 2000: french fries/fried potatoes/hash browns, how much each time | Nutrition |
| V00KQOL4 | Quality of life: in general, how much difficulty have with knee(s) | Behavioral |
| P01HEIGHT | Average height (mm) | Subject characteristics |
| V00lfTHPL | Left Flexion MAX Force High Production Limit | Physical exam |
| V00rkdefcv | Right knee exam: alignment varus or valgus | Physical exam |
| V00FFQ19 | Block Brief 2000: green beans/green peas, eat how often, past 12 months | Nutrition |
| V00FFQ33 | Block Brief 2000: beef steaks/roasts/pot roast (including in frozen dinners/sandwiches), eat how often, past 12 months | Nutrition |
| KPLKN1 | Left knee pain: twisting/pivoting on knee, last 7 days | Symptoms |
| PASE2 | Leisure activities: walking, past 7 days | Physical activity |
| V00INCOME | Yearly income | Behavioral |
| V00PA130CV | How often climb up total of 10 or more flights of stairs during typical week, past 30 days | Physical activity |
| V00CESD9 | How often thought my life had been a failure, past week | Behavioral |
| PASE6 | Leisure activities: muscle strength/endurance, past 7 days | Physical activity |
| DIRKN16 | Right knee difficulty: heavy chores, last 7 days | Symptoms |
| V00SUPB2 | Block Brief 2000: average daily nutrients from vitamin supplements, B2 (mg) | Nutrition |
| STEPST1 | 20-meter walk: trial 1 number of steps | Physical exam |
| V00FFQ12 | Block Brief 2000: any other fruit (e.g., grapes/melon/strawberries/peaches), eat how often, past 12 months | Nutrition |
| KSXRKN1 | Right knee symptoms: swelling, last 7 days | Symptoms |
| V00lfmaxf | Left Flexion MAX Force | Physical exam |
| V00rfTHPL | Right Flexion MAX Force High Production Limit | Physical exam |
| RKALNMT | Right knee exam: alignment, degrees (valgus negative) | Physical exam |
| CEMPLOY | Current employment | Behavioral |
| V00KOOSYML | Left knee: KOOS Symptoms Score | Symptoms |
| V00WPLKN2 | Left knee pain: stairs, last 7 days | Symptoms |
| V00RA | Charlson Comorbidity: have rheumatoid arthritis | Medical history |
| V00SUPFOL | Block Brief 2000: average daily nutrients from vitamin supplements, folate (mcg) | Nutrition |
| V00RXCHOND | Rx Chondroitin sulfate use indicator | Medical history |
References
- Silverwood, V.; Blagojevic-Bucknall, M.; Jinks, C.; Jordan, J.; Protheroe, J.; Jordan, K. Current evidence on risk factors for knee osteoarthritis in older adults: A systematic review and meta-analysis. Osteoarthr. Cartil. 2015, 23, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, I.N.; Kemp, J.L.; Crossley, K.M.; Culvenor, A.G.; Hinman, R.S. Hip and Knee Osteoarthritis Affects Younger People, Too. J. Orthop. Sport. Phys. Ther. 2017, 47, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Antony, B.; Jones, G.; Jin, X.; Ding, C. Do early life factors affect the development of knee osteoarthritis in later life: A narrative review. Arthritis Res. Ther. 2016, 18, 202. [Google Scholar] [CrossRef] [PubMed]
- Toivanen, A.T.; Heliövaara, M.; Impivaara, O.; Arokoski, J.P.; Knekt, P.; Lauren, H.; Kröger, H. Obesity, physically demanding work and traumatic knee injury are major risk factors for knee osteoarthritis—A population-based study with a follow-up of 22 years. Rheumatology 2010, 49, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Lespasio, M.J.; Piuzzi, N.S.; Husni, M.E.; Muschler, G.F.; Guarino, A.; Mont, M.A. Knee osteoarthritis: A primer. Perm. J. 2017, 21, 16–183. [Google Scholar] [CrossRef] [PubMed]
- Jack Farr, I.; Miller, L.E.; Block, J.E. Quality of life in patients with knee osteoarthritis: A commentary on nonsurgical and surgical treatments. Open Orthop. J. 2013, 7, 619. [Google Scholar] [CrossRef]
- Ntakolia, C.; Kokkotis, C.; Moustakidis, S.; Tsaopoulos, D. A machine learning pipeline for predicting joint space narrowing in knee osteoarthritis patients. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE), Cincinnati, OH, USA, 26–28 October 2020; pp. 934–941. [Google Scholar]
- Moustakidis, S.; Papandrianos, N.I.; Christodolou, E.; Papageorgiou, E.; Tsaopoulos, D. Dense neural networks in knee osteoarthritis classification: A study on accuracy and fairness. Neural Comput. Appl. 2020, 1–13. [Google Scholar] [CrossRef]
- Moustakidis, S.; Christodoulou, E.; Papageorgiou, E.; Kokkotis, C.; Papandrianos, N.; Tsaopoulos, D. Application of machine intelligence for osteoarthritis classification: A classical implementation and a quantum perspective. Quantum Mach. Intell. 2019. [Google Scholar] [CrossRef]
- Kokkotis, C.; Moustakidis, S.; Giakas, G.; Tsaopoulos, D. Identification of Risk Factors and Machine Learning-Based Prediction Models for Knee Osteoarthritis Patients. Appl. Sci. 2020, 10, 6797. [Google Scholar] [CrossRef]
- Alexos, A.; Kokkotis, C.; Moustakidis, S.; Papageorgiou, E.; Tsaopoulos, D. Prediction of pain in knee osteoarthritis patients using machine learning: Data from Osteoarthritis Initiative. In Proceedings of the 2020 11th International Conference on Information, Intelligence, Systems and Applications (IISA), Piraeus, Greece, 15–17 July 2020; pp. 1–7. [Google Scholar]
- Alexos, A.; Moustakidis, S.; Kokkotis, C.; Tsaopoulos, D. Physical Activity as a Risk Factor in the Progression of Osteoarthritis: A Machine Learning Perspective. In Proceedings of the 14th International Conference—LION 14, Athens, Greece, 24–28 May 2020; pp. 16–26. [Google Scholar]
- Kokkotis, C.; Moustakidis, S.; Papageorgiou, E.; Giakas, G.; Tsaopoulos, D. Machine Learning in Knee Osteoarthritis: A Review. Osteoarthr. Cartil. Open 2020, 2, 100069. [Google Scholar] [CrossRef]
- Jamshidi, A.; Pelletier, J.-P.; Martel-Pelletier, J. Machine-learning-based patient-specific prediction models for knee osteoarthritis. Nat. Rev. Rheumatol. 2019, 15, 49–60. [Google Scholar] [CrossRef]
- Lazzarini, N.; Runhaar, J.; Bay-Jensen, A.C.; Thudium, C.S.; Bierma-Zeinstra, S.M.A.; Henrotin, Y.; Bacardit, J. A machine learning approach for the identification of new biomarkers for knee osteoarthritis development in overweight and obese women. Osteoarthr. Cartil. 2017, 25, 2014–2021. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Almajalid, R.; Shan, J.; Zhang, M. A Novel Method to Predict Knee Osteoarthritis Progression on MRI Using Machine Learning Methods. IEEE Trans. Nanobiosci. 2018. [Google Scholar] [CrossRef]
- Halilaj, E.; Le, Y.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Modeling and predicting osteoarthritis progression: Data from the osteoarthritis initiative. Osteoarthr. Cartil. 2018, 26, 1643–1650. [Google Scholar] [CrossRef]
- Pedoia, V.; Haefeli, J.; Morioka, K.; Teng, H.L.; Nardo, L.; Souza, R.B.; Ferguson, A.R.; Majumdar, S. MRI and biomechanics multidimensional data analysis reveals R2 -R1rho as an early predictor of cartilage lesion progression in knee osteoarthritis. J. Magn. Reson. Imaging JMRI 2018, 47, 78–90. [Google Scholar] [CrossRef]
- Abedin, J.; Antony, J.; McGuinness, K.; Moran, K.; O’Connor, N.E.; Rebholz-Schuhmann, D.; Newell, J. Predicting knee osteoarthritis severity: Comparative modeling based on patient’s data and plain X-ray images. Sci. Rep. 2019, 9, 5761. [Google Scholar] [CrossRef]
- Nelson, A.; Fang, F.; Arbeeva, L.; Cleveland, R.; Schwartz, T.; Callahan, L.; Marron, J.; Loeser, R. A machine learning approach to knee osteoarthritis phenotyping: Data from the FNIH Biomarkers Consortium. Osteoarthr. Cartil. 2019, 27, 994–1001. [Google Scholar] [CrossRef] [PubMed]
- Widera, P.; Welsing, P.M.; Ladel, C.; Loughlin, J.; Lafeber, F.P.; Dop, F.P.; Larkin, J.; Weinans, H.; Mobasheri, A.; Bacardit, J. Multi-classifier prediction of knee osteoarthritis progression from incomplete imbalanced longitudinal data. arXiv 2019, arXiv:1909.13408. [Google Scholar] [CrossRef] [PubMed]
- Tiulpin, A.; Klein, S.; Bierma-Zeinstra, S.M.; Thevenot, J.; Rahtu, E.; van Meurs, J.; Oei, E.H.; Saarakkala, S. Multimodal machine learning-based knee osteoarthritis progression prediction from plain radiographs and clinical data. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Jamshidi, A.; Leclercq, M.; Labbe, A.; Pelletier, J.-P.; Abram, F.; Droit, A.; Martel-Pelletier, J. Identification of the most important features of knee osteoarthritis structural progressors using machine learning methods. Ther. Adv. Musculoskelet. Dis. 2020, 12. [Google Scholar] [CrossRef]
- Wang, Y.; You, L.; Chyr, J.; Lan, L.; Zhao, W.; Zhou, Y.; Xu, H.; Noble, P.; Zhou, X. Causal Discovery in Radiographic Markers of Knee Osteoarthritis and Prediction for Knee Osteoarthritis Severity With Attention–Long Short-Term Memory. Front. Public Health 2020, 8, 845. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-Z.; Meng, H.-H.; Lu, W.-C.; Yang, J.Y.; Yang, M.Q. Asymmetric bagging and feature selection for activities prediction of drug molecules. BMC Bioinform. 2008, 9, S7. [Google Scholar] [CrossRef]
- Fu, G.-H.; Wu, Y.-J.; Zong, M.-J.; Pan, J. Hellinger distance-based stable sparse feature selection for high-dimensional class-imbalanced data. BMC Bioinform. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Nimankar, S.S.; Vora, D. Designing a Model to Handle Imbalance Data Classification Using SMOTE and Optimized Classifier. In Data Management, Analytics and Innovation; Springer: New York, NY, USA, 2020; pp. 323–334. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Han, H.; Wang, W.-Y.; Mao, B.-H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In Proceedings of the International Conference on Intelligent Computing—ICIC 2005, Hefei, China, 23–26 August 2005; pp. 878–887. [Google Scholar]
- Yen, S.-J.; Lee, Y.-S. Cluster-based under-sampling approaches for imbalanced data distributions. Expert Syst. Appl. 2009, 36, 5718–5727. [Google Scholar] [CrossRef]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 1322–1328. [Google Scholar]
- Chawla, N.V.; Lazarevic, A.; Hall, L.O.; Bowyer, K.W. SMOTEBoost: Improving prediction of the minority class in boosting. In Proceedings of the 7th European Conference on Principles and Practice of Knowledge Discovery in Databases, Cavtat-Dubrovnik, Croatia, 22–26 September 2003; pp. 107–119. [Google Scholar]
- Hanifah, F.S.; Wijayanto, H.; Kurnia, A. SMOTEBagging algorithm for imbalanced dataset in logistic regression analysis (case: Credit of bank X). Appl. Math. Sci. 2015, 9, 6857–6865. [Google Scholar]
- Elkan, C. The foundations of cost-sensitive learning. In Proceedings of the Seventeenth International Joint Conference on Artificial Intelligence, Seattle, WA, USA, 4–10 August 2001; pp. 973–978. [Google Scholar]
- Ling, C.X.; Sheng, V.S. Cost-Sensitive Learning and the Class Imbalance Problem. Encycl. Mach. Learn. 2008, 2011, 231–235. [Google Scholar]
- Shin, H.J.; Eom, D.-H.; Kim, S.-S. One-class support vector machines—An application in machine fault detection and classification. Comput. Ind. Eng. 2005, 48, 395–408. [Google Scholar] [CrossRef]
- Seo, K.-K. An application of one-class support vector machines in content-based image retrieval. Expert Syst. Appl. 2007, 33, 491–498. [Google Scholar] [CrossRef]
- Ertekin, S.; Huang, J.; Giles, C.L. Active learning for class imbalance problem. In Proceedings of the 30th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Amsterdam, The Netherlands, 23–27 July 2007; pp. 823–824. [Google Scholar]
- Attenberg, J.; Ertekin, S. Class imbalance and active learning. Imbalanced Learn. Found. Algorithms Appl. 2013, 101–149. [Google Scholar] [CrossRef]
- Dodge, Y.; Commenges, D. The Oxford Dictionary of Statistical Terms; Oxford University Press on Demand: Oxford, UK, 2006. [Google Scholar]
- Shanker, M.; Hu, M.Y.; Hung, M.S. Effect of data standardization on neural network training. Omega 1996, 24, 385–397. [Google Scholar] [CrossRef]
- Rockel, J.S.; Zhang, W.; Shestopaloff, K.; Likhodii, S.; Sun, G.; Furey, A.; Randell, E.; Sundararajan, K.; Gandhi, R.; Zhai, G.; et al. A classification modeling approach for determining metabolite signatures in osteoarthritis. PLoS ONE 2018, 13, e0199618. [Google Scholar] [CrossRef]
- Gornale, S.S.; Patravali, P.U.; Marathe, K.S.; Hiremath, P.S. Determination of Osteoarthritis Using Histogram of Oriented Gradients and Multiclass SVM. Int. J. Image Graph. Signal Process. 2017, 9, 41–49. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4765–4774. [Google Scholar]
- Roffo, G.; Melzi, S.; Castellani, U.; Vinciarelli, A. Infinite latent feature selection: A probabilistic latent graph-based ranking approach. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 1398–1406. [Google Scholar]
- Roffo, G.; Melzi, S.; Cristani, M. Infinite feature selection. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 4202–4210. [Google Scholar]
- Shahbaz, M.B.; Wang, X.; Behnad, A.; Samarabandu, J. On efficiency enhancement of the correlation-based feature selection for intrusion detection systems. In Proceedings of the 2016 IEEE 7th Annual Information Technology, Electronics and Mobile Communication Conference (IEMCON), Vancouver, BC, Canada, 13–15 October 2016; pp. 1–7. [Google Scholar]
- Hagos, D.H.; Yazidi, A.; Kure, Ø.; Engelstad, P.E. Enhancing security attacks analysis using regularized machine learning techniques. In Proceedings of the 2017 IEEE 31st International Conference on Advanced Information Networking and Applications (AINA), Taipei, Taiwan, 27–29 March 2017; pp. 909–918. [Google Scholar]
- Nguyen, H.T.; Franke, K.; Petrovic, S. Towards a generic feature-selection measure for intrusion detection. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 1529–1532. [Google Scholar]
- Cooper, C.; Snow, S.; McAlindon, T.E.; Kellingray, S.; Stuart, B.; Coggon, D.; Dieppe, P.A. Risk factors for the incidence and progression of radiographic knee osteoarthritis. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 2000, 43, 995–1000. [Google Scholar] [CrossRef]
- Hartley, A.; Hardcastle, S.A.; Paternoster, L.; McCloskey, E.; Poole, K.E.; Javaid, M.K.; Aye, M.; Moss, K.; Granell, R.; Gregory, J. Individuals with High Bone Mass have increased progression of radiographic and clinical features of knee osteoarthritis. Osteoarthr. Cartil. 2020, 28, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Blagojevic, M.; Jinks, C.; Jeffery, A.; Jordan, K. Risk factors for onset of osteoarthritis of the knee in older adults: A systematic review and meta-analysis. Osteoarthr. Cartil. 2010, 18, 24–33. [Google Scholar] [CrossRef]
- Heidari, B. Knee osteoarthritis prevalence, risk factors, pathogenesis and features: Part I. Casp. J. Intern. Med. 2011, 2, 205. [Google Scholar]
- Santos, M.S.; Soares, J.P.; Abreu, P.H.; Araujo, H.; Santos, J. Cross-validation for imbalanced datasets: Avoiding overoptimistic and overfitting approaches [research frontier]. IEEE Comput. Intell. Mag. 2018, 13, 59–76. [Google Scholar] [CrossRef]





| Category | Description | Number of Features from Baseline | Number of Features from Visit 1 |
|---|---|---|---|
| Subject characteristics | Includes anthropometric parameters (Body mass index (BMI), height, etc.) | 36 | 9 |
| Symptoms | Questionnaire data regarding arthritis symptoms and general arthritis or health-related function and disability | 120 | 80 |
| Behavioral | Includes variables of participants’ quality level of daily routine and social behavior | 61 | 43 |
| Medical history | Questionnaire results regarding a participant’s arthritis-related and general health histories and medications | 123 | 51 (only medications) |
| Medical imaging outcome | Medical imaging outcomes (e.g., joint space narrowing and osteophytes) | 21 | - |
| Nutrition | Block Food Frequency questionnaire | 224 | - |
| Physical activity | Questionnaire data regarding leisure activities, etc. | 24 | 24 |
| Physical exam | Participants’ measurements, including knee and hand exams, walking tests and other performance measures | 115 | 26 |
| Number of features (subtotal): | 724 | 233 | |
| Total number of features: | 957 | ||
| Parameter | Description | Selected Value |
|---|---|---|
| Population size | Number of individual solutions in the population | 50 |
| Number of generations | Maximum number of generations before the algorithm halts | 100 |
| Mutation rate | Probability rate of being mutated | 0.1 |
| Crossover Fraction | The fraction of the population at the next generation, not including elite children, that the crossover function creates. | 0.8 |
| Elite Count | Positive integer specifying how many individuals in the current generation are guaranteed to survive into the next generation | 5 |
| StallGenLimit | The algorithm stops if the weighted average change in the fitness function value over StallGenLimit generations is less than Function tolerance | 50 |
| Tolerance | 1 × 10−3 |
| FS Criterion | 10FCV Accuracy Performed 10 Times | ||||
|---|---|---|---|---|---|
| Average | Min | Max | Std | No. of Features | |
| Feature subset extracted from the “best” individual solution of the final generation | 70.10% | 67.59% | 72.04% | 1.13% | 42 |
| Proposed feature ranking | 71.25% | 69.22% | 73.33% | 1.57% | 35 |
| Selected Features | Feature Category | Description |
|---|---|---|
| P01BMI, P01HEIGHT | Subject characteristics | Anthropometric parameters including height and BMI |
| KSXRKN1, V00WOMSTFR, KPLKN1, V00WPLKN2, DIRKN16, V00KOOSYML, V00INCOME | Symptoms | Symptoms related to pain, swelling, stiffness and knee difficulty |
| V00EDCV, V00KQOL4, V00KQOL2, V00CESD9, CEMPLOY | Behavioral | Participants’ quality level of daily routine and social behavior and social status |
| V00RXCHOND, V00RA, V00CHNFQCV | Medical history | Questionnaire data regarding a participant’s general health histories and medications |
| P01SVLKOST | Medical imaging outcome | Medical imaging outcomes (e.g., osteophytes) |
| V00SUPCA, V00FFQ59, V00FFQSZ13, V00FFQ33, V00SUPB2, V00FFQ12, V00SUPFOL, V00FFQ19 | Nutrition | Block Food Frequency questionnaire for daily average, how much each time or for past 12 months |
| PASE2, PASE6, V00PA130CV | Physical activity | Questionnaire results regarding activities during typical week or past 7 days |
| RKALNMT, V00lfmaxf, V00rfTHPL, V00lfTHPL, STEPST1, V00rkdefcv | Physical exam | Physical measurements of participants, including tests and other performance measures |
| Approach | Best Accuracy (Mean 10FCV) | Number of Features | Statistical Comparison * | Execution Time (sec) ** |
|---|---|---|---|---|
| GenWrapper | 71.25 | 35 | - | 311.6 |
| Wrapper | 69.79 | 31 | p < 0.001 | 10.2 |
| CFS | 61.97 | 69 | p < 0.001 | 0.1 |
| ILFS | 63.63 | 82 | p < 0.001 | 0.5 |
| Inf-FS | 63.32 | 35 | p < 0.001 | 0.1 |
| Lasso | 64.41 | 94 | p < 0.001 | 21.2 |
| Mrmr | 67.29 | 36 | p < 0.001 | 2.3 |
| Hybrid | 67.85 | 41 | p < 0.001 | 15.5 |
| PCA | 65.11 | 29 | p < 0.001 | <0.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkotis, C.; Moustakidis, S.; Baltzopoulos, V.; Giakas, G.; Tsaopoulos, D. Identifying Robust Risk Factors for Knee Osteoarthritis Progression: An Evolutionary Machine Learning Approach. Healthcare 2021, 9, 260. https://doi.org/10.3390/healthcare9030260
Kokkotis C, Moustakidis S, Baltzopoulos V, Giakas G, Tsaopoulos D. Identifying Robust Risk Factors for Knee Osteoarthritis Progression: An Evolutionary Machine Learning Approach. Healthcare. 2021; 9(3):260. https://doi.org/10.3390/healthcare9030260
Chicago/Turabian StyleKokkotis, Christos, Serafeim Moustakidis, Vasilios Baltzopoulos, Giannis Giakas, and Dimitrios Tsaopoulos. 2021. "Identifying Robust Risk Factors for Knee Osteoarthritis Progression: An Evolutionary Machine Learning Approach" Healthcare 9, no. 3: 260. https://doi.org/10.3390/healthcare9030260
APA StyleKokkotis, C., Moustakidis, S., Baltzopoulos, V., Giakas, G., & Tsaopoulos, D. (2021). Identifying Robust Risk Factors for Knee Osteoarthritis Progression: An Evolutionary Machine Learning Approach. Healthcare, 9(3), 260. https://doi.org/10.3390/healthcare9030260








