Abstract
(1) Background: Chemerin, or the RARRES2 (Retinoic Acid Receptor Responder 2) gene, is found to be associated with an increased incidence of insulin resistance, endothelial dysfunction, type 2 diabetes (T2D), and coronary artery disease (CAD). This study investigates associations of RARRES2 rs17173608 with insulin resistance and the severity of CAD in non-obese T2D patients in relation to the clinical and genetic factors. (2) Methods: A total of 300 patients with T2D and CAD were recruited in this study. The associations of insulin resistance and the severity of CAD with RARRES2 rs17173608 and clinical factors were assessed. The genotyping procedures were performed using the TaqMan method. The significant associations (p ≤ 0.05) from preliminary tests were employed to carry out the secondary analysis. (3) Results: RARRES2 rs17173608 (TT, TG, and GG polymorphisms in the preliminary analysis; TG and GG polymorphisms in a secondary analysis) was associated with insulin resistance and the severity of CAD in both the preliminary and secondary analysis (all p-values were < 0.05). Additionally, in the secondary analysis, FPG and ACEI were also associated with insulin resistance and the severity of CAD (all p-values were < 0.05). (4) Conclusion: From the preliminary findings, rs17173608 is a significant predictor of insulin resistance and the severity of CAD.
1. Introduction
Chemerin, also known as retinoic acid receptor responder protein 2 (RARRES2) and tazarotene50 induced gene 2 [1], is a chemoattractant protein involved in the pathogenesis of metabolic syndrome [2]. It is secreted in the liver, acts as a chemotactic agent, and is highly stimulated by elements of the innate immune system, such as plasmacytoid dendritic cells and macrophages [2]. Chemerin induces angiogenesis of endothelial cells and results in endothelial dysfunction [3]. Besides which, it can activate inflammatory response and oxidative stress in adipose tissue, results in insulin resistance and further enhance endothelial dysfunction [4]. Endothelial dysfunction and insulin resistance cause the progression of type 2 diabetes (T2D) and atherosclerotic coronary artery disease (CAD).
The CAD is classified as severe when the atherosclerotic plaques narrow down the vessels with stenosis by more than 50% [5]. In a study by Dahpy et al., serum chemerin levels were higher in patients with T2D and metabolic syndromes than nondiabetics and patients without metabolic syndromes [4]. There is growing evidence showing a relationship between coronary atherosclerosis and chemerin. Several cross-sectional studies showed an independent correlation between chemerin concentrations and CAD. Chemerin is moderately heritable, with 16–25% of variations ascribed to genetic factors [5].
The encoding gene of chemerin, RARRES2, is also associated with an increased incidence of metabolic syndrome and inflammatory diseases [6]. Metabolic syndrome independently predicts the development of T2D and CAD [7]. T2D patients have a higher cardiovascular morbidity and mortality rate. The diabetic vascular disease causes a two–fourfold increase in CAD occurrence [8]. In Malaysia, 3.6 million people have diabetes, and most of them have T2D [9]. Meanwhile, CAD is the leading cause of death in T2D patients in Malaysia [10]. However, the relationships between RARRES2, inflammation, metabolic syndromes, insulin resistance, and CAD have not been completely elucidated.
Genetic polymorphisms can play an essential role in phenotypic variation disease susceptibility [11]. Studies show that the single nucleotide polymorphisms (SNPs) of the RARRES2 gene is associated with an increased incidence of T2D and CAD [12,13]. The RARRES2 rs17173608 SNP had been associated with metabolic syndromes and obesity. It is located in the potential site of mutation, intron 2 of RARRES2 [14,15]. However, the literature on the association of rs17173608 with insulin resistance and the severity of CAD in T2D patients is lacking. In a previous study conducted by Mehanna et al., the minor G allele of rs17173608 SNP showed its significant association with metabolic syndromes in Egyptian women [16]. In another study, the TT genotype of rs17173608 was more widely distributed in T2D patients than non-diabetics [4]. In this study, we evaluated the association rs17173608 with insulin resistance and the severity of CAD in Malaysian T2D patients for the first time.
2. Methods and Materials
2.1. Participants
We conducted a cross-sectional study with 300 patients from the diabetic clinic and cardiology clinics of University Malaya Medical Centre (UMMC) after the approval of the UMMC Medical Ethics Committee (ref number: 20158-1552). Written, informed consent was obtained from the participants before the blood samples were taken. For analysis, the patients were divided into three groups; T2D patients with CAD (n = 150), patients with T2D only (n = 90), and patients with CAD only (n = 60). The T2D patients with CAD group were the case group of this study, whilst the other two were the control groups. We excluded patients with type 1 diabetes, critically ill patients, asthma patients, and obese patients (more than 30.0 kg/m2) [15].
2.2. Sample Size Calculation
Quanto software version 1.2.4 was used to calculate the sample size (USC, Los Angeles, CA, USA). The sample size for detecting the association between disease and SNP marker was positively affected by disease prevalence, disease allele frequency, and inheritance model. According to the National Cardiovascular Disease Database, only about 20% of CAD patients have T2D [17]. At the same time, we had very stringent inclusion and exclusion criteria. We only included patients that had stable CAD with more than 50% stenosis. We excluded obese patients as obesity may influence the outcome of the study, as rs17173608 had been associated with obesity in previous studies [4,15]. Thus, the sample size is smaller. From the calculation, the sample size was 300 (case (T2D patients with CAD patients) = 150 + controls (T2D only patients + CAD only patients) = 150, Total = 300) (Table 1).

Table 1.
Sample size calculation.
2.3. Demographic and Clinical Information
The demographic factors, such as gender, race, and family history of T2D and CAD, were obtained from the electronic medical record system (EMR) of University Malaya Medical Centre (UMMC) and the National Cardiovascular Disease Database (NCVD). Age was calculated based on the patients’ birth years. The EMR also provided information on the types and number of comorbidities, pharmacological treatments and concomitant drugs, and the duration of T2D and CAD.
2.4. Anthropometric Measurement
The anthropometric parameters of height and weight were obtained for each participant. BMI (kg/m2) was calculated by dividing weight by the square of the height [18].
2.5. Biochemical Parameters
After at least eight hours of fasting, blood samples were collected. A YSI 7100 multiparameter bioanalytical system [19] was used to analyse the plasma glucose concentrations. In the meantime, plasma insulin concentrations were determined using the Santo Advia Centaur XP immunoassay system [20]. The Beckman Coulter Immage 800 immunochemistry system was used to measure high-sensitivity C-reactive protein (hs–CRP) levels, and the G8 HPLC analyzer was used to measure A1C concentrations [21]. Triglyceride (TG), total cholesterol (TC), and high-density lipoprotein cholesterol (HDL-c) levels were measured using an automated analyser, the Dimension® RxL Max® integrated chemistry system [22]. The homeostasis model assessment—insulin resistance (HOMA-IR) was calculated by multiplying fasting plasma insulin (FPI) and fasting plasma glucose (FPG) and dividing by 22.5 [23]. Serum chemerin concentrations were measured using an ELISA kit and read at 450 nm using a microplate reader [24].
2.6. Selection of SNP
The candidate gene approach was used to discover the SNP. Initially, risk variants and candidate SNPs associated with T2D and CAD were identified. The putative SNPs were then identified by investigating their relevance to the disease mechanism. The selected SNPs were then evaluated by identifying existing gene variants and determining which variants result in proteins with altered functions that may influence the trait of interest [25], with rs17173608 being chosen because it had an intronic functional consequence on gene regulation. The rs17173608 variant is located on the second intron of the chemerin gene and has previously been linked to T2D and CAD [26].
2.7. Genetic Analysis
The Qiagen DNA blood mini kits (QIAGEN N.V., Venlo, The Netherlands) were used for DNA extraction. The DNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Polymorphism rs17173608 was analysed using real-time polymerase chain reaction using the TaqMan fluorogenic 5′ nuclease assay. The StepOnePlusTM real-time PCR system assay (Applied Biosystems, Foster City, CA, USA) was used to amplify the polymorph. Applied Biosystems StepOnePlusTM real-time software was used to compute the results (Thermo Fisher Scientific, Waltham, MA, USA).
2.8. Statistical Analysis
The IBM Statistical Package for the Social Sciences (SPSS) (Armok, NY, USA) software version 26.0 was used to analyse the data. Descriptive statistics, expressed as frequency, were used to summarise patients’ demographic and clinical characteristics (N, %). Continuous variables were summarised as mean ± SD (normally distributed) or median ± IQR (not normally distributed). Chi-square and Fisher exact test were used to study the differences in categorical variables. The significance of all continuous demographics and biochemical parameters among the groups was assessed using one-way ANOVA and the Kruskal–Wallis test. HWE analyses were applied to compare the observed and expected genotype frequencies of subjects by a goodness-of-fit chi-square using the genepop software (http://genepop.curtin.edu.au) (accessed on 5 January 2021) [27]. For the HWE test, P-values greater than 0.05 were deemed significant. Binary and multinomial logistic regression tests (univariate and multivariate) were employed in the preliminary and secondary analysis, after which the age, race, BMI (Body Mass Index), and gender were adjusted as covariates.
Binary logistic regression was used to analyse the association model of insulin resistance with rs17173608 and clinical factors. Meanwhile, multinomial logistic regression analysis was used to assess the association model of the severity of CAD with rs17173608 and clinical factors. The preliminary analysis’s significant associations (p ≤ 0.05) were used to carry out the secondary tests. The cut-off point of insulin resistance was determined using a plotted receiver-operating characteristic (ROC) curve. The point on the ROC curve with maximum Youden index (sensitivity-(1-specificity)) and the point with the shortest distance from the point (0, 1) ((1-sensitivity)2 + (1-specificity)2) were calculated to find the optimal threshold of HOMA-IR and a cut-off point of 7.17 obtained [28].
2.9. Operational Definitions
- Insulin resistance: HOMA-IR value of more than 7.17 from the ROC curve is categorised as insulin resistance (IR) whilst less than 7.17 is categorised as insulin-sensitive (IS).
- The severity of CAD: one, two, or three arteries narrowed by 50% of stenosis. One vessel = single-vessel disease (SVD), two vessels = double-vessel disease (DVD), three vessels = triple-vessel disease (TVD). TVD is the most severe type of vessel disease (3 major vessels involved) [5].
- Obesity: BMI more than or equal to 30 kg/m2 [15].
- Preliminary analysis: one-way ANOVA, Chi-square, Fisher exact test, Kruskal–Wallis test, one-sample T-test, binary logistic regression (univariate tests), multinomial logistic regression (univariate tests).
- Secondary analysis: binary logistic regression (multivariate tests), multinomial logistic regression (multivariate tests).
- Univariate tests: p values were obtained by running binary/multinomial analysis for each variable separately.
- Multivariate tests: involved a set of multiple variables that were significant from the univariate tests. p values were obtained for each of the variables involved by running the binary/multinomial analysis as a group.
3. Results
3.1. Demographic and Clinical Data
Table 2 shows the demographic and clinical data of the study population. The number of males was higher in T2D patients with CAD, and CAD-only patients. The average age of patients was higher in the CAD-only group compared to the other two groups. Indian patients were more prone to T2D with or without CAD, in comparison with Malay and Chinese patients. Patients with only CAD had a lower BMI than T2D patients with CAD and T2D-only patients. Most of the patients had a family history of T2D and CAD.

Table 2.
Demographic and clinical factors of the study population (n = 300).
3.2. Hardy–Weinberg Equilibrium Test for SNP rs17173608
The SNP rs17173608 did not deviate from the Hardy–Weinberg Equilibrium (p > 0.05) in T2D patients with CAD (p = 0.98), T2D-only (p = 0.92), and CAD-only (p = 0.22) groups. Thus, the SNP was included in the subsequent analyses.
3.3. Associations of Clinical Factors with rs17173608
From the preliminary statistical analysis, among the OHA- and insulin- combination treatments, biguanides and insulin were associated with rs17173608. Among the mono-therapy treatments, biguanides were associated with rs17173608 in T2D patients with CAD. Sulphonylureas and CCBs were found to be associated with rs17173608 in the T2D-only group. Meanwhile, FPG, FPI, and A1C levels were significantly associated with rs17173608 in CAD-only groups. Among all these significant associations, the association of FPG with rs17173608 was the strongest, with an OR value of 6.053. Table 3 demonstrates the associations of clinical factors with rs17173608 in the study population.

Table 3.
Associations of clinical factors with rs17173608 in subjects (n = 300).
3.4. Associations of rs17173608 with Insulin Resistance and Severity of CAD
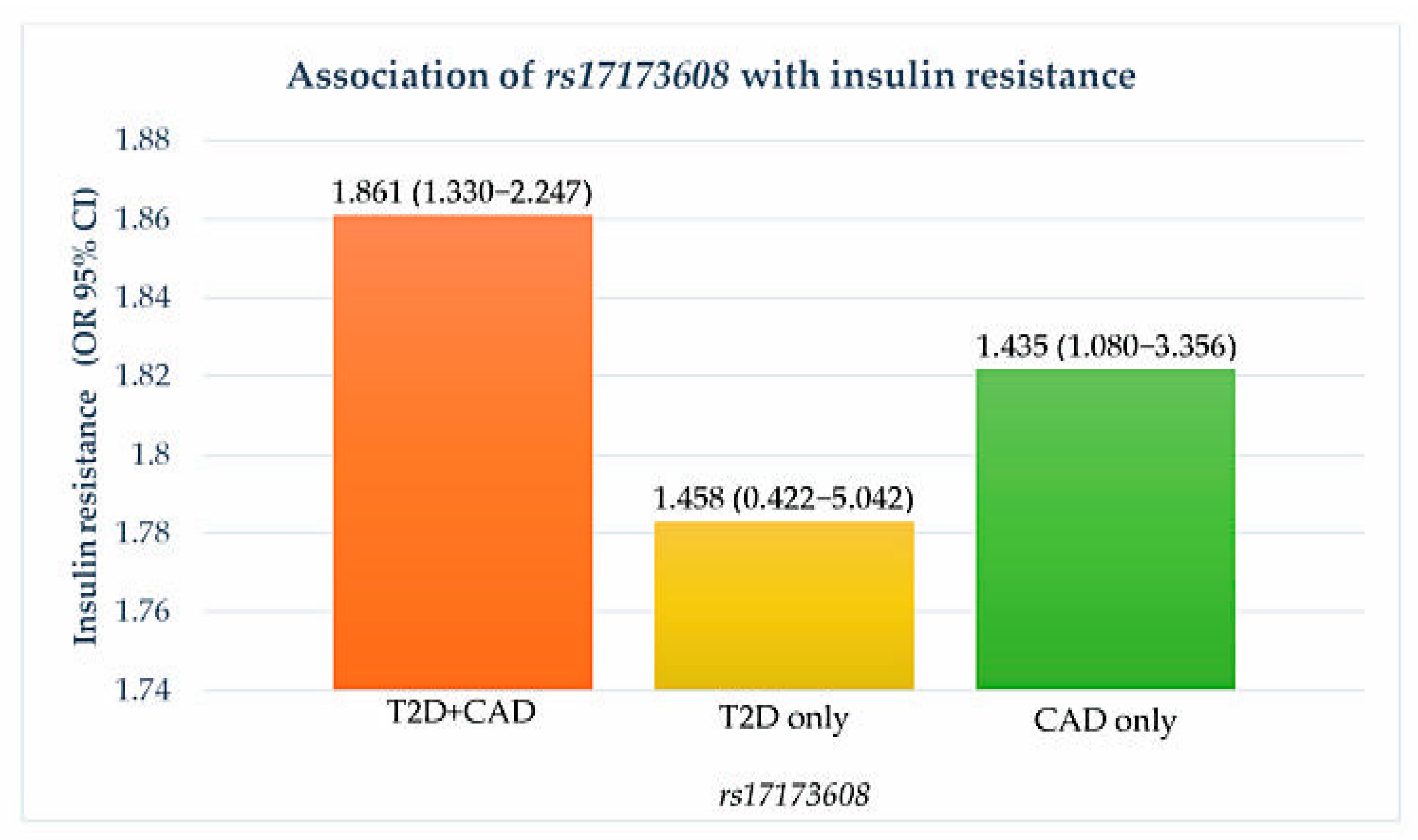
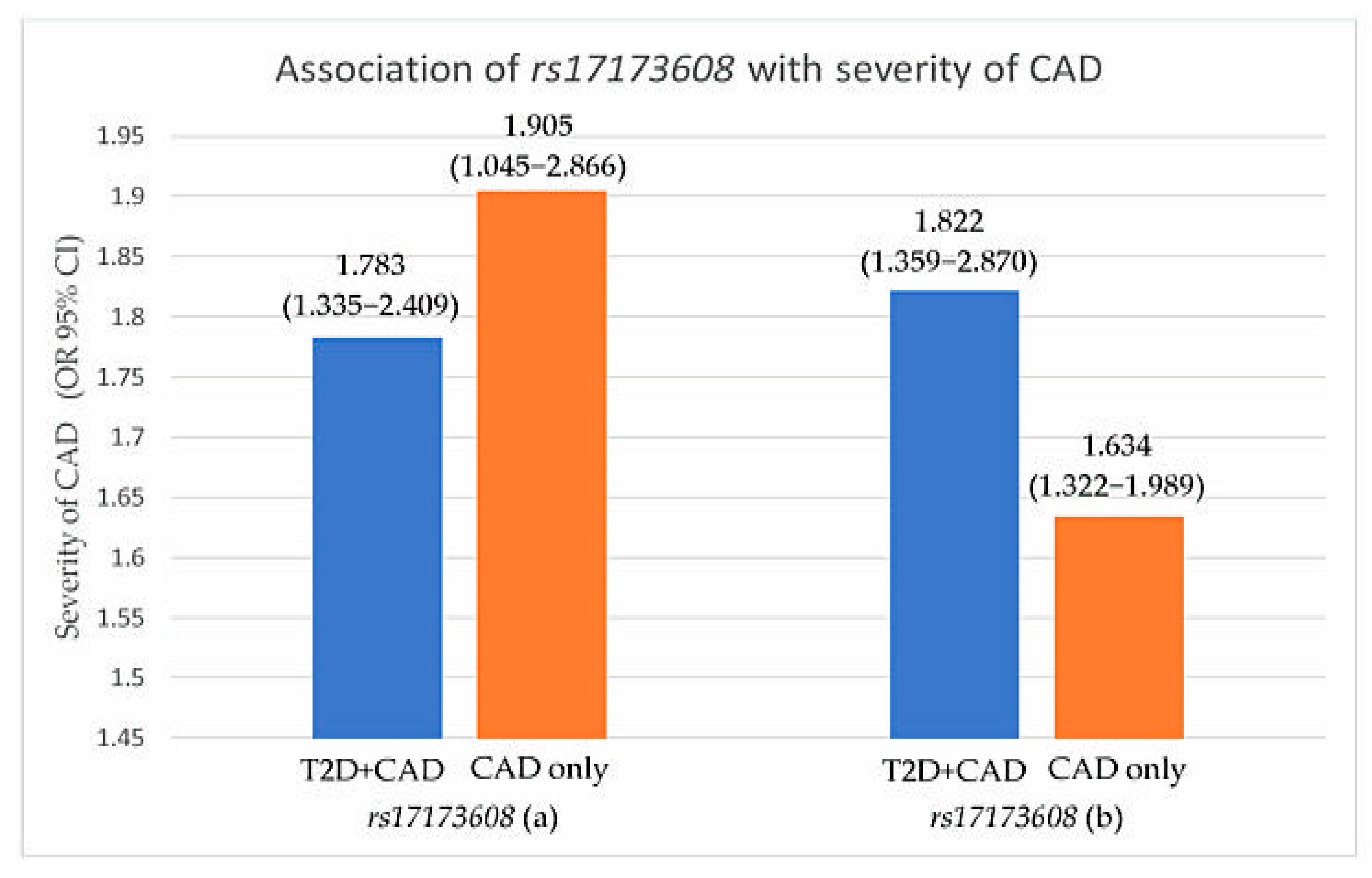
Preliminary associations of rs17173608 with insulin resistance and the severity of CAD are presented in Table 4, Figure 1 and Figure 2. The preliminary results showed significant correlations between rs17173608 and insulin resistance and the severity of CAD in T2D patients with CAD and CAD-only groups. Secondary analysis using binary and multinomial logistic regression was carried out to find out the associations.

Table 4.
Associations of rs17173608 with insulin resistance and severity of CAD.
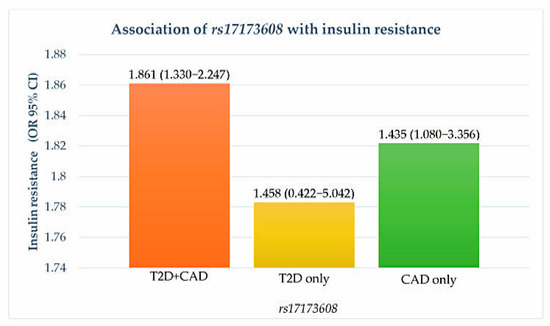
Figure 1.
Association of rs17173608 with insulin resistance. OR: odds ratio; CI: confidence interval.
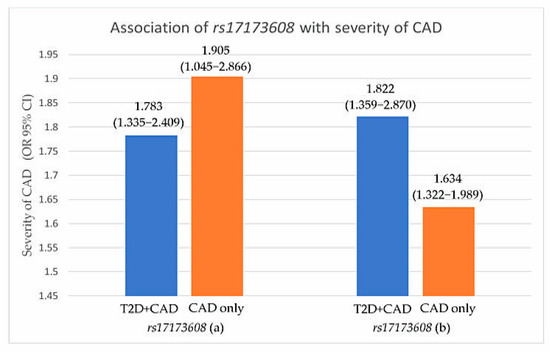
Figure 2.
Association of rs17173608 with severity of CAD. (a) SVD vs. DVD, (b) SVD vs. TVD. OR: odds ratio; CI: confidence interval.
3.5. Association Models of Insulin Resistance and Severity of CAD in Correlation with rs17173608 and Clinical Factors (Secondary Analysis)
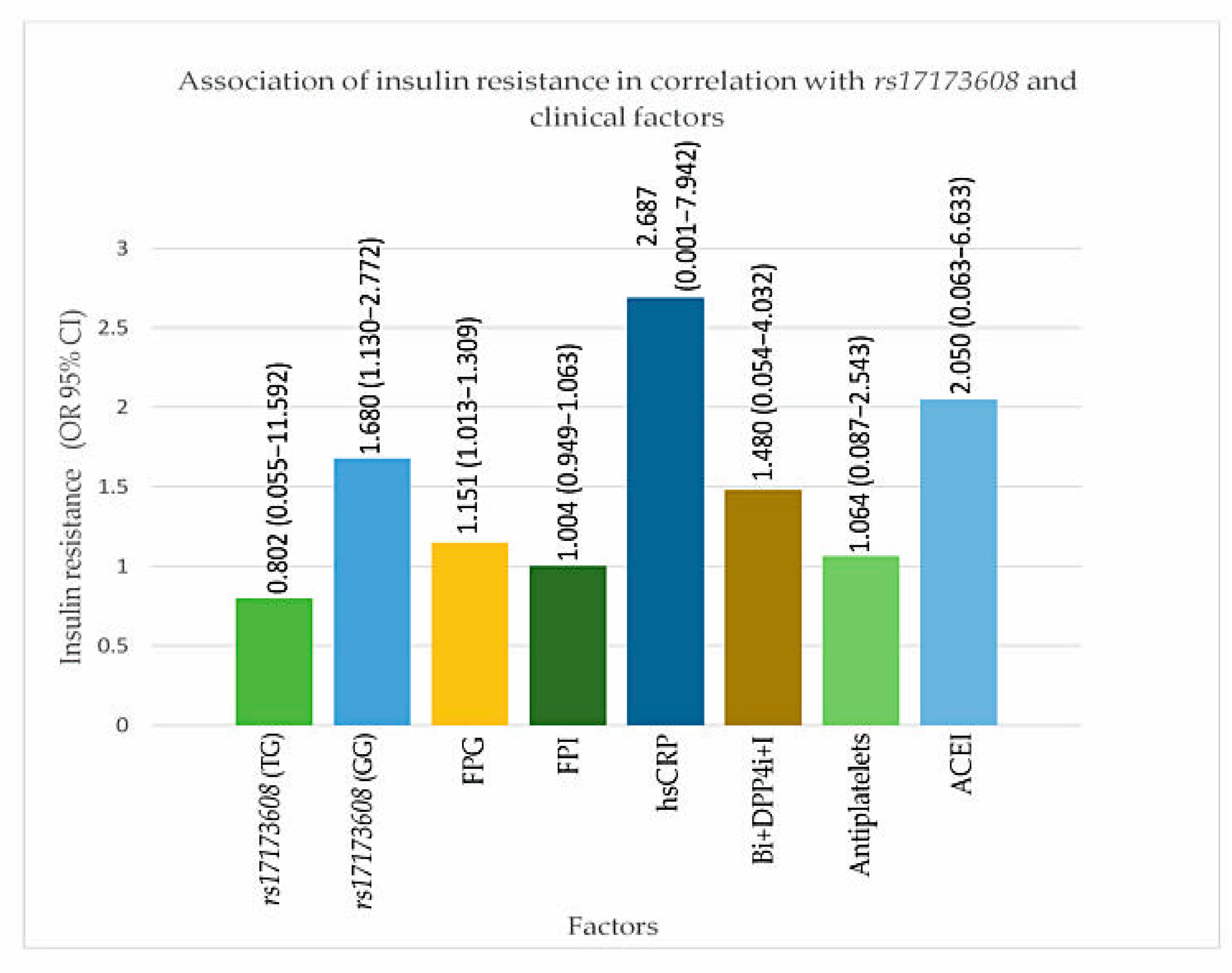
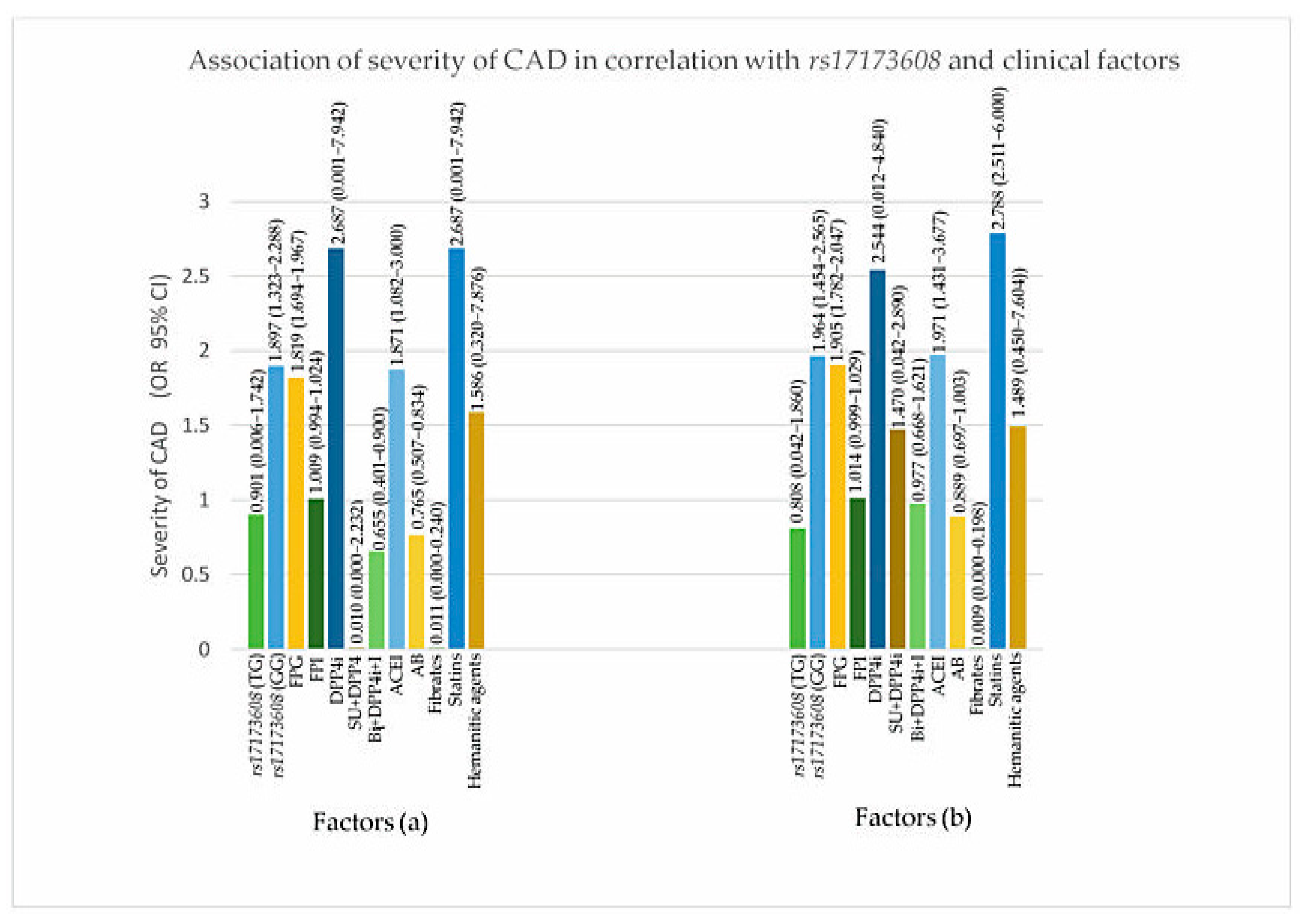
From the secondary analysis, FPG (p value for IR = 0.047, DVD = 0.017, TVD = 0.032), ACEI (p value for IR = 0.024, DVD = 0.017, TVD = 0.025), rs17173608 TG polymorphism (p value for IR = 0.015, DVD = 0.028, TVD = 0.036), and rs17173608 GG polymorphism (p value for IR = 0.012, DVD = 0.042, TVD = 0.048) were associated with insulin resistance and the severity of CAD. Table 5 and Table 6, Figure 3 and Figure 4 show the secondary analysis of insulin resistance and the severity of CAD in correlation with rs17173608 and clinical factors. Appendix A contains a summary of the findings of this study.

Table 5.
Associations of insulin resistance in correlation with rs17173608 and clinical factors.

Table 6.
Associations of severity of CAD in correlation with rs17173608 and clinical factors.
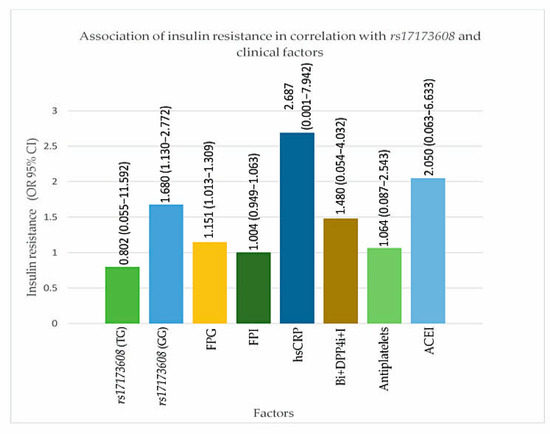
Figure 3.
Association of insulin resistance in correlation with rs17173608 and clinical factors. FPG: Figure 4. i: dipeptidyl peptidase-4 inhibitor; I: insulin. OR: odds ratio; CI: confidence interval.
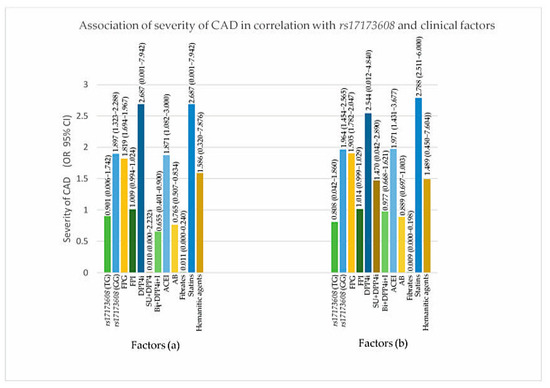
Figure 4.
Association of severity of CAD in correlation with rs17173608 and clinical factors. (a) SVD vs. DVD, (b) SVD vs. TVD. FPG: fasting plasma glucose; FPI: fasting plasma insulin; AB: alpha blockers; ACEI: angiotensin-converting-enzyme inhibitor; DPP4i: dipeptidyl peptidase-4 inhibitor; I: insulin; SU: sulphonylureas.
4. Discussion
Insulin resistance and the severity of CAD were postulated to come independently from “common soil”. The demographical and clinical factors associated with insulin resistance may be associated with CAD patients. The study by Arambewela et al. found that the prevalence of T2D with CAD was higher in males than females [29], and the study by Krishnan et al. found that the prevalence of CAD in non-diabetic CAD patients was higher in males than females [30]. Gender differences exist because, regardless of T2D, males develop CAD 7 to 10 years earlier than females [31]. Females are more likely to develop CAD as a result of massive hormonal changes following menopause [32].
Similarly, more males than females were found in T2D patients with CAD and CAD-only patients in our study. Nonetheless, females were diagnosed with T2D at a higher rate than males in T2D-only patients. The National Health and Morbidity Survey (NHMS) 2019 found that the prevalence of T2D was higher in females than in males [33]. Several studies have found that the prevalence of T2D and CAD varies across ethnic groups living in the same country. It has been postulated to be associated with environmental and genetic factors. Indian patients had the highest prevalence in our study, followed by Malay and Chinese patients in the T2D with CAD and T2D-only groups. Tee and Yap [34] discovered similar results in the Malaysian population [34]. This could be because Indians have a higher inherent risk of T2D and CAD in Malaysia [34].
FPG and A1C levels were higher in T2D patients with CAD than in T2D patients without CAD in a previous study [35]. As a result, the findings of the studies contradicted our findings. Meanwhile, LDL-c levels in T2D patients without CAD were higher than in T2D patients with CAD and CAD patients without T2D [35]. The results for FPI and HOMA-IR in the same study contradicted our findings, as the values were higher in T2D patients without CAD [35]. BMI is said to have an effect on glycaemic control in people with diabetes and other metabolic disorders. The BMIs of T2D-only patients were higher than the other two groups in this study, which may have resulted in an increase in FPG, A1C, LDL-c, FPI, and HOMA-IR levels in T2D patients only when compared to the other groups of patients.
Furthermore, previous studies were conducted on obese individuals, yielding contradictory results [35]. In a previous study, CAD patients had higher hs–CRP, HDL-c, and total cholesterol levels, which were comparable to the findings of our study. [36]. rs17173608 is a RARRES2 gene variant, and to the best of our knowledge, this is the first study to associate rs17173608 with demographic and clinical factors of insulin resistance and severity of CAD. In the preliminary associations, some elements were discovered to be associated with rs17173608. Gender was associated with rs17173608. rs17173608 was associated with increased visceral fat mass in non-obese subjects in one study [37]. Men tend to store more fat in their visceral depots. As a result, the association of rs17173608 with the male gender is most likely due to gender-related body fat distribution [38]. Furthermore, according to Table 2, the majority of T2D patients with CAD were not receiving biguanide treatment (Yes = 47, No = 103).
As a result of the significant association of rs17173608 with biguanides in preliminary analysis, it was hypothesised that the administration of biguanides to these patients would reduce the expression of RARRES2 and thus chemerin. In the T2D-only group, however, many patients were on sulphonylureas. Sulphonylureas are the second-line drugs in T2D management, and the use is associated with an adverse vascular function, which results in endothelial dysfunction and cardiovascular events [39]. SNP rs17173608 was associated with insulin resistance in our study and previous studies [36]. Besides this, RARRES2 SNP rs17173608 was associated with the severity of CAD in the study by Lachine et al. [40]. Our findings were consistent with previous studies, despite differences in the study population and pharmacological treatments. Secondary analyses were performed to determine the additional association of rs17173608 polymorphisms (TT, TG, and GG) with insulin resistance and the severity of CAD in T2D patients with CAD. The secondary analysis revealed a significant association of rs17173608 polymorphisms (TG and GG) with insulin resistance and the severity of CAD.
Strength and Limitations
An essential strength of this study was the homogenous sample of individuals. All the samples were non-obese individuals. This enabled researchers to draw conclusions about the impact of rs1717308 in a non-obese T2D population with and without CAD. Furthermore, this was the first study to associate rs17173608 with both insulin resistance and the severity of CAD. The main constraint was the small sample size. Nonetheless, the samples were chosen using very strict inclusion and exclusion criteria in order to reduce the impact of confounding variables that could interfere with the outcome of this association study.
5. Conclusions
Among Malaysia’s three major ethnic groups, Indians had the highest prevalence of insulin resistance and the severity of CAD in the study population. The preliminary and secondary analyses revealed that the TG and GG polymorphisms of rs17173608 were significantly associated with insulin resistance and the severity of CAD. Thus, the findings indicate that rs17173608 is a significant predictor of insulin resistance and the severity of CAD.
Author Contributions
S.P. was responsible for clinical sample collection, laboratory work, interpretation of data and writing the whole manuscript. H.Z.H. and W.A.W.A. involved in the conceptual design of the study and revised the manuscript critically for important intellectual content. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by University Malaya (Grant number: RP024B-14HTM and PG173-2016A).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of UMMC (protocol code: 20158-1552, date of approval: 22/10/2015).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available as they contain the information that could compromise the privacy of research participants.
Acknowledgments
The authors would like to thank University Malaya for the financial and technical support in completing the study (Grant number: RP024B-14HTM and PG173-2016A). In addition, the authors would like to thank all the participants of this study and the nurses of UMMC who helped with blood sample collection.
Conflicts of Interest
The authors have no other relevant affiliations or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter of materials discussed in the manuscript apart from those disclosed. No writing assistance was utilised in the production of this manuscript.
Appendix A

Table A1.
Summary points.
Table A1.
Summary points.
Chemerin and RARRES2
|
Insulin resistance and severity of CAD
|
Demographic and clinical factors
|
rs17173608
|
Conclusion
|
References
- Landgraf, K.; Friebe, D.; Ullrich, T.; Kratzsch, J.; Dittrich, K.; Herberth, G.; Adams, V.; Kiess, W.; Erbs, S.; Körner, A. Chemerin as a Mediator between Obesity and Vascular Inflammation in Children. J. Clin. Endocrinol. Metab. 2012, 97, E556–E564. [Google Scholar] [CrossRef] [PubMed]
- Helfer, G.; Wu, Q.F. Chemerin: A multifaceted adipokine involved in metabolic disorders. J. Endocrinol. 2018, 238, R79–R94. [Google Scholar] [CrossRef] [PubMed]
- Dimitriadis, G.K.; Kaur, J.; Adya, R.; Kaltsas, G.; Tan, B.K.; Randeva, H.S. Chemerin induces endothelial cell inflammation: Activation of nuclear factor-kappa beta and monocyte-endothelial adhesion. Oncotarget 2018, 9, 16678–16690. [Google Scholar] [CrossRef]
- Dahpy, M.; Khairallah, M.; Azoz, N.; Ezzat, G. The associations among RARRES2 rs17173608 gene polymorphism, serum chemerin, and non-traditional lipid profile in patients with metabolic syndrome. Egypt. J. Med. Hum. Genet. 2020, 21, 1. [Google Scholar] [CrossRef]
- Buechler, C.; Feder, S.; Haberl, E.M.; Aslanidis, C. Chemerin Isoforms and Activity in Obesity. Int. J. Mol. Sci. 2019, 20, 1128. [Google Scholar] [CrossRef]
- Er, L.K.; Wu, S.; Hsu, L.A.; Teng, M.S.; Sun, Y.C.; Ko, Y.L. Pleiotropic Associations of RARRES2 Gene Variants and Circulating Chemerin Levels: Potential Roles of Chemerin Involved in the Metabolic and Inflammation-Related Diseases. Mediat. Inflamm. 2018, 2018, 4670521. [Google Scholar] [CrossRef]
- Martín-Timón, I.; Sevillano-Collantes, C.; Segura-Galindo, A.; Del Cañizo-Gómez, F.J. Type 2 diabetes and cardiovascular disease: Have all risk factors the same strength? World J. Diabetes 2014, 5, 444–470. [Google Scholar] [CrossRef]
- Einarson, T.R.; Acs, A.; Ludwig, C.; Panton, U.H. Prevalence of cardiovascular disease in type 2 diabetes: A systematic literature review of scientific evidence from across the world in 2007–2017. Cardiovasc. Diabetol. 2018, 17, 83. [Google Scholar] [CrossRef]
- Hussein, Z.; Taher, S.W.; Gilcharan Singh, H.K.; Chee Siew Swee, W. Diabetes Care in Malaysia: Problems, New Models, and Solutions. Ann. Glob. Health 2015, 81, 851–862. [Google Scholar] [CrossRef]
- Chang, C.T.; Lee, P.Y.; Cheah, W.L. The prevalence of cardiovascular risk factors in the young and middle-aged rural population in Sarawak, Malaysia. Malays. J. Med. Sci. 2012, 19, 27–34. [Google Scholar]
- Knight, J.C. Regulatory polymorphisms underlying complex disease traits. J. Mol. Med. 2005, 83, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Er, L.K.; Hsu, L.A.; Juang, J.J.; Wu, S.; Lin, J.F.; Ko, Y.L. Circulating Chemerin Levels, but not the RARRES2 Polymorphisms, Predict the Long-Term Outcome of Angiographically Confirmed Coronary Artery Disease. Int. J. Mol. Sci. 2019, 20, 1174. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Jiang, J.; Huo, Y.; Chen, D. Genetic predisposition to type 2 diabetes is associated with severity of coronary artery disease in patients with acute coronary syndromes. Cardiovasc. Diabetol. 2019, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Rezaei, H.; Eskandari-Nasab, E.; Kaykhaei, M.; Zakeri, Z.; Taheri, M. Association between chemerin rs17173608 and vaspin rs2236242 gene polymorphisms and the metabolic syndrome, a preliminary report. Gene 2012, 510, 113–117. [Google Scholar] [CrossRef]
- Yamauchi, E.; Suzuki, Y.; So, K.H.; Suzuki, K.; Katoh, K.; Roh, S.G. Single Nucleotide Polymorphism in the Coding Region of Bovine Chemerin Gene and Their Associations with Carcass Traits in Japanese Black Cattle. Asian-Australas. J. Anim. Sci. 2015, 28, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Mehanna, E.; Mesbah, N.; Ghattas, M.; Saleh, S.; Abo-Elmatty, D. Association of chemerin Rs17173608 and vaspin Rs2236242 gene polymorphisms with metabolic syndrome in Egyptian women. Endocr. Res. 2015, 41, 43–48. [Google Scholar] [CrossRef] [PubMed]
- National Cardiovascular Disease Database. Acrm.org.my. 2021. Available online: http://www.acrm.org.my/ncvd/ (accessed on 1 March 2021).
- Dorian, J.; Coates, A.M.; Banks, S. The Impact of Altered Timing of Eating, Sleep and Work Patterns on Human Health; MDPI: Basel, Switzerland, 2018. [Google Scholar]
- Ysi.com. 2021. Available online: https://www.ysi.com/File%20Library/Documents/Guides/YSI-Biochemistry-Analyzer365Replacement-Guide_v1.pdf. (accessed on 8 March 2021).
- ADVIA Centaur CP Immunoassay System Siemens-healthineers.com. 2021. Available online: https://www.siemens-healthineers.com/en368my/immunoassay/systems/advia-centaur-cp-immunoassay-sys (accessed on 8 March 2021).
- Beckmancoulter.com. 2021. Available online: https://beckmancoulter.com/wsrportal/techdocs?docname=/cis/A38059/AC/EN_CCRP.pdf (accessed on 8 March 2020).
- Fliser, E.; Jerkovic, K.; Vidovic, T.; Gorenjak, M. Investigation of unusual high serum indices for lipemia in clear serum samples on Siemens analysers Dimension. Biochem. Med. 2012, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.Q.; Li, Q.; Rentfro, A.R.; Fisher-Hoch, S.P.; McCormick, J.B. The definition of insulin resistance using HOMA-IR for Americans of Mexican descent using machine learning. PLoS ONE 2011, 6, e21041. [Google Scholar] [CrossRef]
- Human Chemerin Quantikine ELISA www.rndsystems.com. 2021. Available online: https://www.rndsystems.com/products/human-chemerin380quantikine-elisa_dchm00#assay-procedure (accessed on 8 March 2021).
- Kwon, J.M.; Goate, A.M. The candidate gene approach. Alcohol. Res. Health. 2000, 24, 164–168. [Google Scholar]
- Movahed, Z.; Kohan, L.; Fallahi, S.; Tabiee, O. Influence of chemerin rs17173608 polymorphism on polycystic ovary syndrome susceptibility. Taiwan. J. Obstet Gynecol. 2015, 54, 280–283. [Google Scholar] [CrossRef]
- Genepop on the Web.Genepop.curtin.edu.au. 2021. Available online: https://genepop.curtin.edu.au/ (accessed on 28 January 2021).
- Hoo, Z.H.; Candlish, J.; Teare, D. What is an ROC curve? Emerg. Med. J. 2016, 34, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Arambewela, M.H.; Somasundaram, N.P.; Jayasekara, H.B.P.R.; Fernando, K.R.A.S.; Kusumsiri, D.P. Prevalence of Chronic Complications, Their Risk Factors, and the Cardiovascular Risk Factors among Patients with Type 2 Diabetes Attending the Diabetic Clinic at a Tertiary Care Hospital in Sri Lanka. J. Diab. Res. 2018, 2018, 4504287. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.N.; Zachariah, G.; Venugopal, K.; Jeyaseelan, L.; Thankappan, K.R. Prevalence of coronary artery disease and its risk factors in Kerala, South India: A community-based cross-sectional study. BMC Cardiovasc. Disord. 2016, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.G.; Chen, X.Y.; Wang, L.; Lau, A.; Wong, A.; Thomas, G.N.; Tomlinson, B.; Liu, R.; Chan, J.C.; Leung, T.W.; et al. Sex differences in epidemiology and risk factors of acute coronary syndrome in Chinese patients with type 2 diabetes: A long-term prospective cohort study. PLoS ONE 2015, 10, e0122031. [Google Scholar] [CrossRef]
- Barrett-Connor, E. Menopause, atherosclerosis, and coronary artery disease. Curr. Opin. Pharmacol. 2013, 13, 186–191. [Google Scholar] [CrossRef]
- National Health and Morbidity Survey 2015-VOLUME, II. Available online: https://www.researchgate.net/publication/305213149_National_Health_and_Morbidity_Survey_2015-VOLUME_II_Non419Communicable_Diseases_Risk_Factors_Other_Health_Problems. (accessed on 28 March 2020).
- Tee, E.S.; Yap, R.W.K. Type 2 diabetes mellitus in Malaysia: Current trends and risk factors. Eur. J. Clin. Nutr. 2017, 71, 844–849. [Google Scholar] [CrossRef]
- Goldfine, A.B.; Phua, E.J.; Abrahamson, M.J. Glycemic management in patients with coronary artery disease and prediabetes or type 2 diabetes mellitus. Circulation 2014, 129, 2567–2573. [Google Scholar] [CrossRef]
- Dai, W.; Zhang, Z.; Zhao, S. Baseline levels of serum high sensitivity C reactive protein and lipids in predicting the residual risk of cardiovascular events in Chinese population with stable coronary artery disease: A prospective cohort study. Lipids Health Dis. 2018, 17, 273. [Google Scholar] [CrossRef]
- Müssig, K.; Staiger, H.; Machicao, F.; Claussen, C.D.; Stefan, N.; Fritsche, A.; Häring, H.U. RARRES2, encoding the novel adipokine chemerin, is a genetic determinant of disproportionate regional body fat distribution: A comparative magnetic resonance imaging study. Metabolism 2009, 58, 519–524. [Google Scholar] [CrossRef]
- Karastergiou, K.; Smith, S.R.; Greenberg, A.S.; Fried, S.K. Sex differences in human adipose tissues-the biology of pear shape. Biol. Sex Differ. 2012, 3, 13. [Google Scholar] [CrossRef]
- Sola, D.; Rossi, L.; Schianca, G.P.; Corlianò, F.; Fra, G.P.; Bartoli, E.; Derosa, G. Sulfonylureas and their use in clinical practice. Arch. Med. Sci. 2015, 11, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Lachine, N.A.; Elnekiedy, A.A.; Megallaa, M.H.; Khalil, G.I.; Sadaka, M.A.; Rohoma, K.H.; Kassab, H.S. Serum chemerin and high-sensitivity C reactive protein as markers of subclinical atherosclerosis in Egyptian patients with type 2 diabetes. Ther. Adv. Endocrinol. Metab. 2016, 7, 47–56. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).