The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents
Abstract
:1. Introduction
2. Therapeutic Drug Monitoring of Antimicrobial Agents
2.1. Antimicrobial Quantification Methods
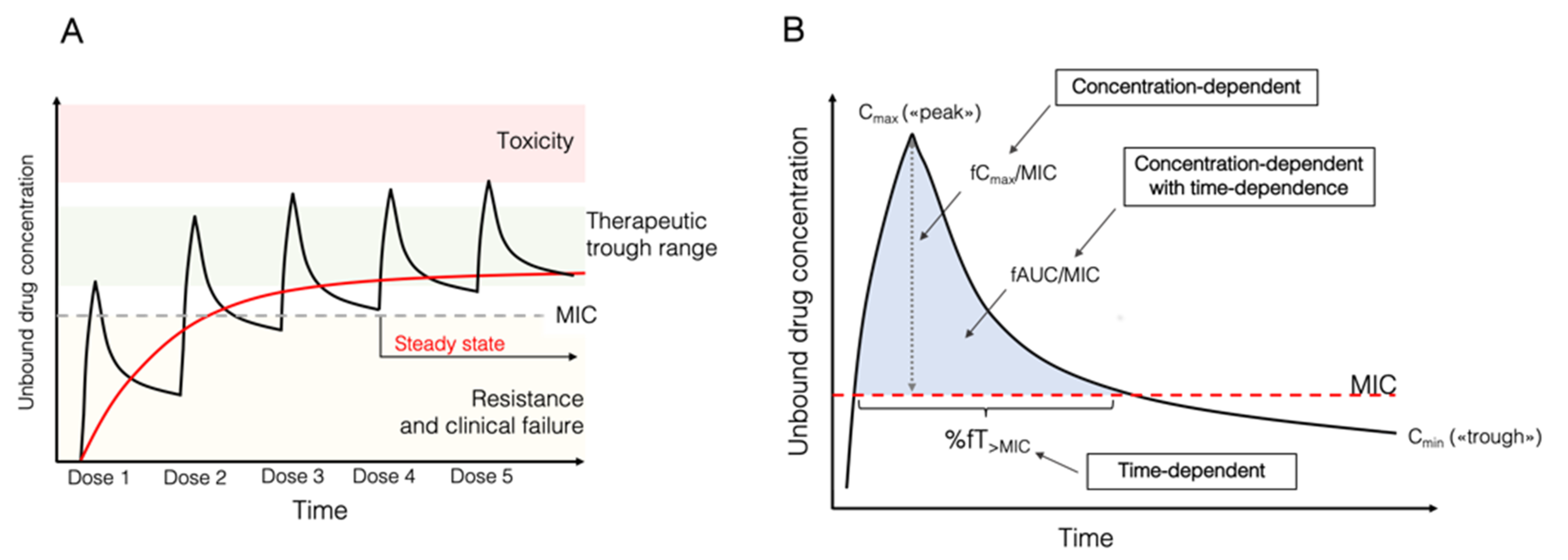
2.2. Pharmacokinetic and Pharmacodynamic Indices of Antimicrobial Agents
- Time-dependent agents. The antimicrobial effect is dependent on the cumulative percentage of time over a 24 h period that the free (unbound) antimicrobial concentration surpasses the MIC (fT%>MIC). For these antimicrobials, the longer the concentration remains at serum levels above the MIC, the greater the effectiveness. Increasing their concentration any more than three or four times that of the MIC does not ultimately make a difference. Typical examples are beta-lactams.
- Concentration-dependent agents. The antimicrobial effect is dependent on the peak concentration in a dosing interval divided by the MIC (Cmax/MIC), with different target levels, usually Cmax/MIC >8–10. These antimicrobials would achieve optimal effect with higher immediate serum concentrations, and extending the time after which the concentration remains above the MIC would have little effect. Typical examples are aminoglycosides and daptomycin.
- Concentration-dependent with time-dependent agents. The effect is defined by the AUC0–24 h over a 24 h period divided by the MIC (AUC0–24 h/MIC). This ratio indicates the importance of both time and concentration for optimal effect. Examples are fluoroquinolones, tigecycline, linezolid, and glycopeptides. Specific targets vary according to the antimicrobial.
3. Surface Enhanced Raman Scattering and Therapeutic Drug Monitoring
Clinical Applications of SERS, including TDM, Quantitative SERS
4. SERS Applications for TDM of Antimicrobial Agents
4.1. Time-Dependent Agents
4.2. Concentration-Dependent Agents
4.3. Concentration-Dependent Agents with Time Dependence
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fleischmann-Struzek, C.; Mellhammar, L.; Rose, N.; Cassini, A.; Rudd, K.E.; Schlattmann, P.; Allegranzi, B.; Reinhart, K. Incidence and Mortality of Hospital- and ICU-Treated Sepsis: Results from an Updated and Expanded Systematic Review and Meta-Analysis. Intensive Care Med. 2020, 46, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, Regional, and National Sepsis Incidence and Mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.A.; Abdul-Aziz, M.H.; Lipman, J.; Mouton, J.W.; Vinks, A.A.; Felton, T.W.; Hope, W.W.; Farkas, A.; Neely, M.N.; Schentag, J.J.; et al. Individualised Antibiotic Dosing for Patients Who Are Critically Ill: Challenges and Potential Solutions. Lancet Infect. Dis. 2014, 14, 498–509. [Google Scholar] [CrossRef] [Green Version]
- Rea, R.S.; Capitano, B.; Bies, R.; Bigos, K.L.; Smith, R.; Lee, H. Suboptimal Aminoglycoside Dosing in Critically Ill Patients. Ther. Drug Monit. 2008, 30, 674–681. [Google Scholar] [CrossRef]
- Muller, A.E.; Huttner, B.; Huttner, A. Therapeutic Drug Monitoring of β-Lactams and Other Antibiotics in the Intensive Care Unit: Which Agents, Which Patients and Which Infections? Drugs 2018, 78, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, A.; Ewoldt, T.M.J.; Hunfeld, N.G.M.; Muller, A.E.; Rietdijk, W.J.R.; Polinder, S.; van Gelder, T.; Endeman, H.; Koch, B.C.P. The Effect of Therapeutic Drug Monitoring of β-Lactam and Fluoroquinolones on Clinical Outcome in Critically Ill Patients: The DOLPHIN Trial Protocol of a Multi-Centre Randomised Controlled Trial. BMC Infect. Dis. 2020, 20, 57. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.A.; Taccone, F.S.; Udy, A.A.; Vincent, J.-L.; Jacobs, F.; Lipman, J. Vancomycin Dosing in Critically Ill Patients: Robust Methods for Improved Continuous-Infusion Regimens. Antimicrob. Agents Chemother. 2011, 55, 2704–2709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashbee, H.R.; Barnes, R.A.; Johnson, E.M.; Richardson, M.D.; Gorton, R.; Hope, W.W. Therapeutic Drug Monitoring (TDM) of Antifungal Agents: Guidelines from the British Society for Medical Mycology. J. Antimicrob. Chemother. 2014, 69, 1162–1176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blot, S.I.; Pea, F.; Lipman, J. The Effect of Pathophysiology on Pharmacokinetics in the Critically Ill Patient—Concepts Appraised by the Example of Antimicrobial Agents. Adv. Drug Deliv. Rev. 2014, 77, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, K.; Daniels, R.; Kissoon, N.; Machado, F.R.; Schachter, R.D.; Finfer, S. Recognizing Sepsis as a Global Health Priority—A WHO Resolution. N. Engl. J. Med. 2017, 377, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Wicha, S.G.; Märtson, A.-G.; Nielsen, E.I.; Koch, B.C.P.; Friberg, L.E.; Alffenaar, J.-W.; Minichmayr, I.K.; International Society of Anti-Infective Pharmacology (ISAP), the PK/PD Study Group of the European Society of Clinical Microbiology, Infectious Diseases (EPASG). From Therapeutic Drug Monitoring to Model-Informed Precision Dosing for Antibiotics. Clin. Pharmacol. Ther. 2021, 109, 928–941. [Google Scholar] [CrossRef] [PubMed]
- The Infection Section of European Society of Intensive Care Medicine (ESICM); Pharmacokinetic/Pharmacodynamic and Critically Ill Patient Study Groups of European Society of Clinical Microbiology and Infectious Diseases (ESCMID); Infectious Diseases Group of International Association of Therapeutic Drug Monitoring and Clinical Toxicology (IATDMCT); Infections in the ICU and Sepsis Working Group of International Society of Antimicrobial Chemotherapy (ISAC); Abdul-Aziz, M.H.; Alffenaar, J.-W.C.; Bassetti, M.; Bracht, H.; Dimopoulos, G.; Marriott, D.; et al. Antimicrobial Therapeutic Drug Monitoring in Critically Ill Adult Patients: A Position Paper. Intensive Care Med. 2020, 46, 1127–1153. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Cotta, M.O.; Roberts, J.A. Pharmacokinetics/Pharmacodynamics of β-Lactams and Therapeutic Drug Monitoring: From Theory to Practical Issues in the Intensive Care Unit. Semin. Respir. Crit. Care Med. 2019, 40, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Märtson, A.-G.; Burch, G.; Ghimire, S.; Alffenaar, J.-W.C.; Peloquin, C.A. Therapeutic Drug Monitoring in Patients with Tuberculosis and Concurrent Medical Problems. Expert Opin. Drug Metab. Toxicol. 2021, 17, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, Y.; Li, X.; Zhang, Y.; Chen, Q.; Ye, Z.; Alqarni, Z.; Bell, S.E.J.; Xu, Y. Towards Practical and Sustainable SERS: A Review of Recent Developments in the Construction of Multifunctional Enhancing Substrates. J. Mater. Chem. C 2021, 9, 11517–11552. [Google Scholar] [CrossRef]
- Jaworska, A.; Fornasaro, S.; Sergo, V.; Bonifacio, A. Potential of Surface Enhanced Raman Spectroscopy (SERS) in Therapeutic Drug Monitoring (TDM). A Critical Review. Biosensors 2016, 6, 47. [Google Scholar] [CrossRef] [Green Version]
- Frosch, T.; Knebl, A.; Frosch, T. Recent Advances in Nano-Photonic Techniques for Pharmaceutical Drug Monitoring with Emphasis on Raman Spectroscopy. Nanophotonics 2019, 9, 19–37. [Google Scholar] [CrossRef]
- Tommasini, M.; Zanchi, C.; Lucotti, A.; Bombelli, A.; Villa, N.S.; Casazza, M.; Ciusani, E.; de Grazia, U.; Santoro, M.; Fazio, E.; et al. Laser-Synthesized SERS Substrates as Sensors toward Therapeutic Drug Monitoring. Nanomaterials 2019, 9, 677. [Google Scholar] [CrossRef] [Green Version]
- Sloan-Dennison, S.; O’Connor, E.; Dear, J.W.; Graham, D.; Faulds, K. Towards Quantitative Point of Care Detection Using SERS Lateral Flow Immunoassays. Anal. Bioanal. Chem. 2022, 1–9. [Google Scholar] [CrossRef]
- Kaza, M.; Karaźniewicz-Łada, M.; Kosicka, K.; Siemiątkowska, A.; Rudzki, P.J. Bioanalytical Method Validation: New FDA Guidance vs. EMA Guideline. Better or Worse? J. Pharm. Biomed. Anal. 2019, 165, 381–385. [Google Scholar] [CrossRef]
- Mabilat, C.; Gros, M.F.; Nicolau, D.; Mouton, J.W.; Textoris, J.; Roberts, J.A.; Cotta, M.O.; van Belkum, A.; Caniaux, I. Diagnostic and Medical Needs for Therapeutic Drug Monitoring of Antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 791–797. [Google Scholar] [CrossRef] [Green Version]
- Roberts, J.A.; Lipman, J. Pharmacokinetic Issues for Antibiotics in the Critically Ill Patient. Crit. Care Med. 2009, 37, 840–851, quiz 859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlier, M.; Stove, V.; Wallis, S.C.; De Waele, J.J.; Verstraete, A.G.; Lipman, J.; Roberts, J.A. Assays for Therapeutic Drug Monitoring of β-Lactam Antibiotics: A Structured Review. Int. J. Antimicrob. Agents 2015, 46, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A. Immunoassays and Issues with Interference in Therapeutic Drug Monitoring. In Clinical Challenges in Therapeutic Drug Monitoring; Elsevier: Amsterdam, The Netherlands, 2016; pp. 17–44. ISBN 978-0-12-802025-8. [Google Scholar]
- Manohar, M.; Marzinke, M.A. Application of Chromatography Combined with Mass Spectrometry in Therapeutic Drug Monitoring. In Clinical Challenges in Therapeutic Drug Monitoring; Elsevier: Amsterdam, The Netherlands, 2016; pp. 45–70. ISBN 978-0-12-802025-8. [Google Scholar]
- Committee for Human Medicinal Products. Draft ICH Guideline M10 on Bioanalytical Method Validation; EMA/CHMP/ICH/172948/2019; European Medicines Agency: London, UK, 2019.
- Decosterd, L.A.; Widmer, N.; André, P.; Aouri, M.; Buclin, T. The Emerging Role of Multiplex Tandem Mass Spectrometry Analysis for Therapeutic Drug Monitoring and Personalized Medicine. TrAC Trends Anal. Chem. 2016, 84, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Belanger, C.R.; Hancock, R.E.W. Testing Physiologically Relevant Conditions in Minimal Inhibitory Concentration Assays. Nat. Protoc. 2021, 16, 3761–3774. [Google Scholar] [CrossRef]
- Mouton, J.W.; Muller, A.E.; Canton, R.; Giske, C.G.; Kahlmeter, G.; Turnidge, J. MIC-Based Dose Adjustment: Facts and Fables. J. Antimicrob. Chemother. 2018, 73, 564–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Huang, S.-C.; Hu, S.; Yan, S.; Ren, B. Fundamental Understanding and Applications of Plasmon-Enhanced Raman Spectroscopy. Nat. Rev. Phys. 2020, 2, 253–271. [Google Scholar] [CrossRef]
- Han, X.X.; Rodriguez, R.S.; Haynes, C.L.; Ozaki, Y.; Zhao, B. Surface-Enhanced Raman Spectroscopy. Nat. Rev. Methods Primer 2021, 1, 87. [Google Scholar] [CrossRef]
- Le Ru, E.C.; Etchegoin, P.G. A Quick Overview of Surface-Enhanced Raman Spectroscopy. In Principles of Surface-Enhanced Raman Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2009; pp. 1–27. ISBN 978-0-444-52779-0. [Google Scholar]
- Ding, S.-Y.; You, E.-M.; Tian, Z.-Q.; Moskovits, M. Electromagnetic Theories of Surface-Enhanced Raman Spectroscopy. Chem. Soc. Rev. 2017, 46, 4042–4076. [Google Scholar] [CrossRef] [PubMed]
- Pilot, R.; Signorini, R.; Durante, C.; Orian, L.; Bhamidipati, M.; Fabris, L. A Review on Surface-Enhanced Raman Scattering. Biosensors 2019, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Mosier-Boss, P. Review of SERS Substrates for Chemical Sensing. Nanomaterials 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhao, X.; Yi, Z.; Glushenkov, A.M.; Kong, L. Plasmonic Substrates for Surface Enhanced Raman Scattering. Anal. Chim. Acta 2017, 984, 19–41. [Google Scholar] [CrossRef] [PubMed]
- Khlebtsov, B.; Khlebtsov, N. Surface-Enhanced Raman Scattering-Based Lateral-Flow Immunoassay. Nanomaterials 2020, 10, 2228. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-Activated Platforms for Immunoassay: Probes, Encoding Methods, and Applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-Enhanced Raman Spectroscopy for Bioanalysis: Reliability and Challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Panikar, S.S.; Cialla-May, D.; De la Rosa, E.; Salas, P.; Popp, J. Towards Translation of Surface-Enhanced Raman Spectroscopy (SERS) to Clinical Practice: Progress and Trends. TrAC Trends Anal. Chem. 2021, 134, 116122. [Google Scholar] [CrossRef]
- Cailletaud, J.; De Bleye, C.; Dumont, E.; Sacré, P.-Y.; Netchacovitch, L.; Gut, Y.; Boiret, M.; Ginot, Y.-M.; Hubert, P.; Ziemons, E. Critical Review of Surface-Enhanced Raman Spectroscopy Applications in the Pharmaceutical Field. J. Pharm. Biomed. Anal. 2018, 147, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.E.J.; Charron, G.; Cortés, E.; Kneipp, J.; Chapelle, M.L.; Langer, J.; Procházka, M.; Tran, V.; Schlücker, S. Towards Reliable and Quantitative Surface—Enhanced Raman Scattering (SERS): From Key Parameters to Good Analytical Practice. Angew. Chem. Int. Ed. 2020, 59, 5454–5462. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.E.J.; Sirimuthu, N.M.S. Quantitative Surface-Enhanced Raman Spectroscopy. Chem. Soc. Rev. 2008, 37, 1012–1024. [Google Scholar] [CrossRef]
- Goodacre, R.; Graham, D.; Faulds, K. Recent Developments in Quantitative SERS: Moving towards Absolute Quantification. TrAC Trends Anal. Chem. 2018, 102, 359–368. [Google Scholar] [CrossRef] [Green Version]
- Masson, J.-F. The Need for Benchmarking Surface-Enhanced Raman Scattering (SERS) Sensors. ACS Sens. 2021, 6, 3822–3823. [Google Scholar] [CrossRef]
- Girmatsion, M.; Mahmud, A.; Abraha, B.; Xie, Y.; Cheng, Y.; Yu, H.; Yao, W.; Guo, Y.; Qian, H. Rapid Detection of Antibiotic Residues in Animal Products Using Surface-Enhanced Raman Spectroscopy: A Review. Food Control 2021, 126, 108019. [Google Scholar] [CrossRef]
- Guo, Y.; Girmatsion, M.; Li, H.-W.; Xie, Y.; Yao, W.; Qian, H.; Abraha, B.; Mahmud, A. Rapid and Ultrasensitive Detection of Food Contaminants Using Surface-Enhanced Raman Spectroscopy-Based Methods. Crit. Rev. Food Sci. Nutr. 2020, 61, 3555–3568. [Google Scholar] [CrossRef] [PubMed]
- Balan, C.; Pop, L.-C.; Baia, M. IR, Raman and SERS Analysis of Amikacin Combined with DFT-Based Calculations. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 214, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Huang, J.; Sun, Y.; Deng, R.; Teng, M.; Li, Q.; Yang, Y.; Hu, X.; Zhang, Z.; Zhang, G. A SERS-Based Multiple Immuno-Nanoprobe for Ultrasensitive Detection of Neomycin and Quinolone Antibiotics via a Lateral Flow Assay. Microchim. Acta 2018, 185, 84. [Google Scholar] [CrossRef]
- McKeating, K.S.; Couture, M.; Dinel, M.-P.; Garneau-Tsodikova, S.; Masson, J.-F. High Throughput LSPR and SERS Analysis of Aminoglycoside Antibiotics. Analyst 2016, 141, 5120–5126. [Google Scholar] [CrossRef] [Green Version]
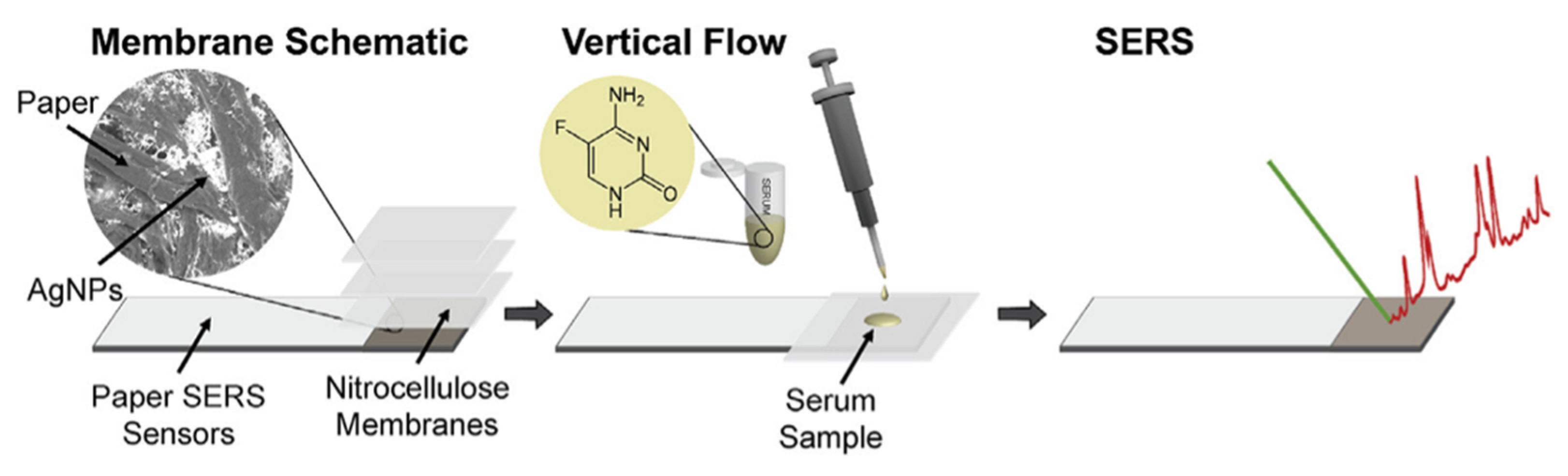
- Berger, A.G.; Restaino, S.M.; White, I.M. Vertical-Flow Paper SERS System for Therapeutic Drug Monitoring of Flucytosine in Serum. Anal. Chim. Acta 2017, 949, 59–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
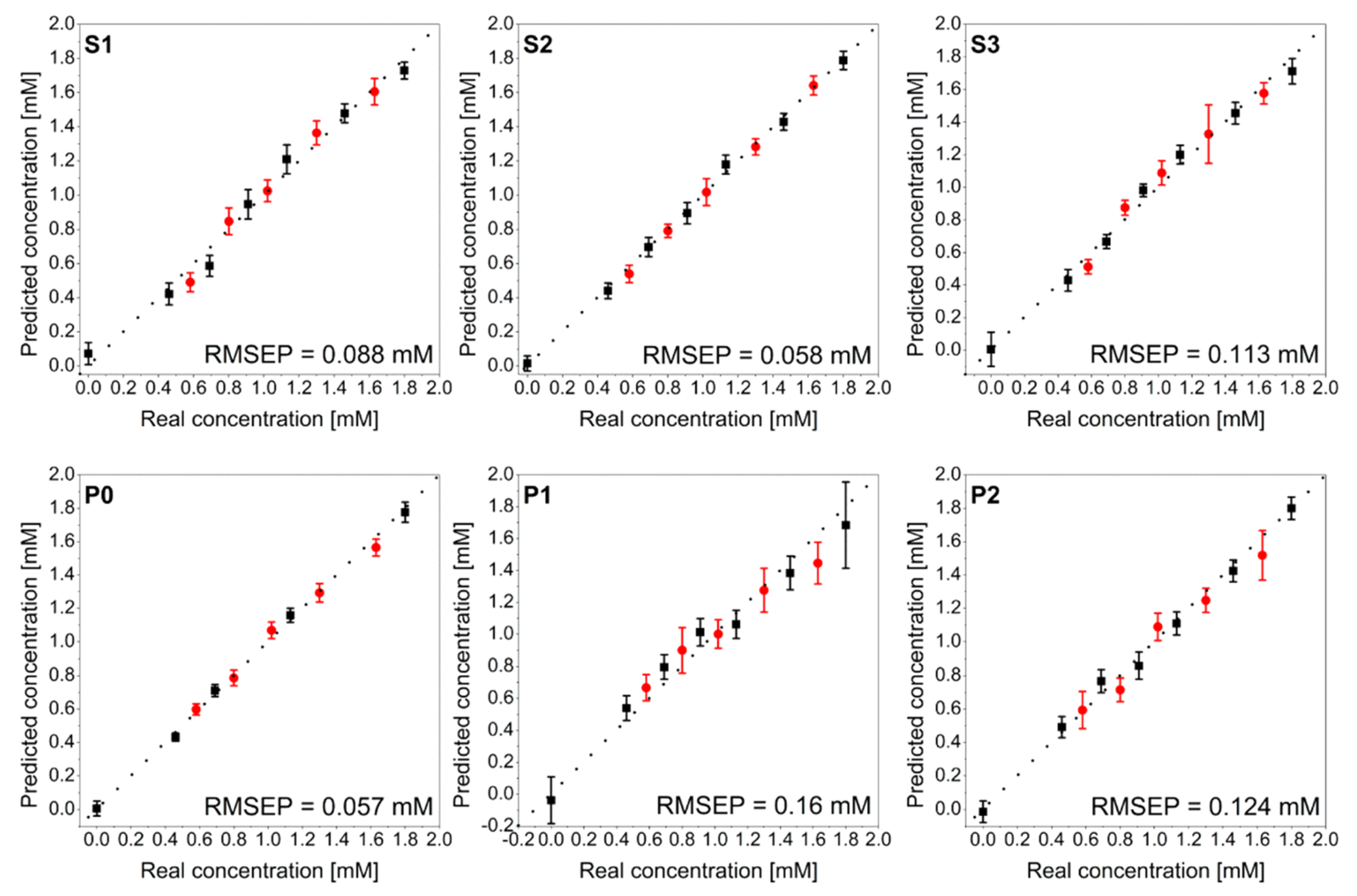
- Liu, H.; Wang, Y.; Wang, N.; Liu, M.; Liu, S. The Determination of Plasma Voriconazole Concentration by Surface-Enhanced Raman Spectroscopy Combining Chemometrics. Chemom. Intell. Lab. Syst. 2019, 193, 103833. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.-S.; Wu, J.; Zhang, Z.; An, D. Surface-Enhanced Raman Spectra of Some Anti-Tubercle Bacillus Drugs. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2000, 56, 2637–2644. [Google Scholar] [CrossRef]
- Muneer, S.; Sarfo, D.K.; Ayoko, G.A.; Islam, N.; Izake, E.L. Gold-Deposited Nickel Foam as Recyclable Plasmonic Sensor for Therapeutic Drug Monitoring in Blood by Surface-Enhanced Raman Spectroscopy. Nanomaterials 2020, 10, 1756. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Markin, A.V. Application of Aluminum Hydroxide for Improvement of Label-Free SERS Detection of Some Cephalosporin Antibiotics in Urine. Biosensors 2019, 9, 91. [Google Scholar] [CrossRef] [Green Version]
- Markina, N.E.; Ustinov, S.N.; Zakharevich, A.M.; Markin, A.V. Copper Nanoparticles for SERS-Based Determination of Some Cephalosporin Antibiotics in Spiked Human Urine. Anal. Chim. Acta 2020, 1138, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Chen, H.; Zhao, S.; Huang, J. Quantitative Analysis of Ceftazidime Using SERS Based on Silver Nanoparticles Substrate. Nanophoton. MicroNano Opt. IV 2018, 10823, 1082313. [Google Scholar] [CrossRef]
- Markina, N.E.; Volkova, E.K.; Zakharevich, A.M.; Goryacheva, I.Y.; Markin, A.V. SERS Detection of Ceftriaxone and Sulfadimethoxine Using Copper Nanoparticles Temporally Protected by Porous Calcium Carbonate. Microchim. Acta 2018, 185, 481. [Google Scholar] [CrossRef] [PubMed]
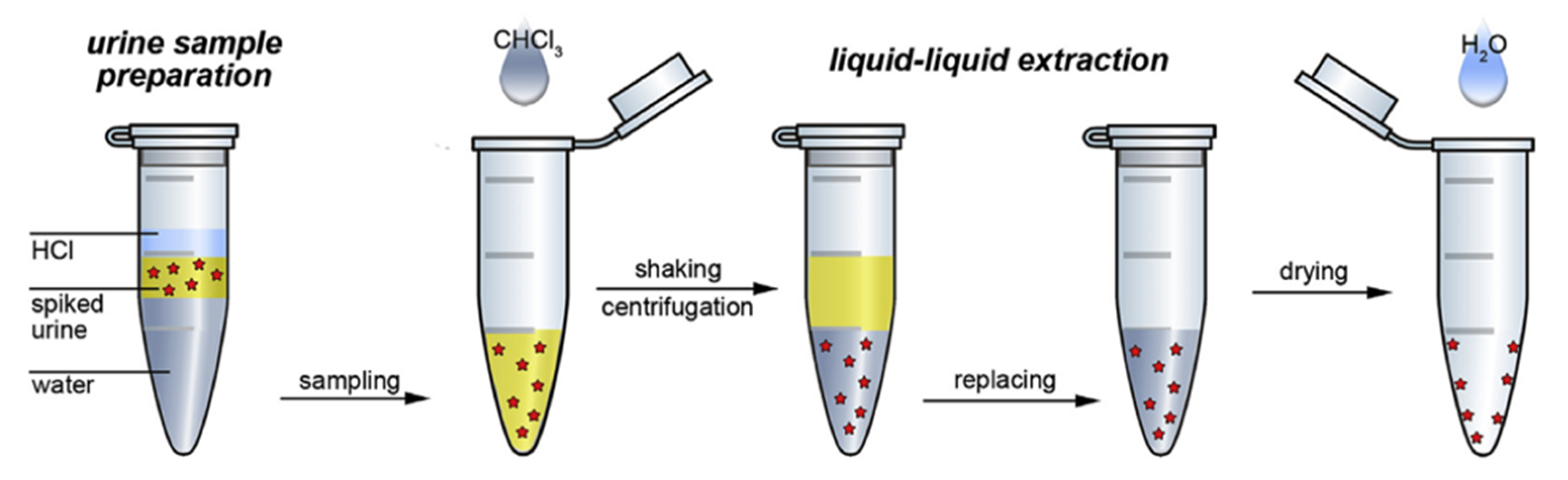
- Markina, N.E.; Goryacheva, I.Y.; Markin, A.V. Sample Pretreatment and SERS-Based Detection of Ceftriaxone in Urine. Anal. Bioanal. Chem. 2018, 410, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Sang, Q.; Yang, M.; Du, J.; Yang, L.; Jiang, X.; Han, X.; Zhao, B. Detection of Several Quinolone Antibiotic Residues in Water Based on Ag-TiO2 SERS Strategy. Sci. Total Environ. 2020, 702, 134956. [Google Scholar] [CrossRef] [PubMed]
- Hidi, I.J.; Heidler, J.; Weber, K.; Cialla-May, D.; Popp, J. Ciprofloxacin: PH-Dependent SERS Signal and Its Detection in Spiked River Water Using LoC-SERS. Anal. Bioanal. Chem. 2016, 408, 8393–8401. [Google Scholar] [CrossRef]
- Hong, K.Y.; de Albuquerque, C.D.L.; Poppi, R.J.; Brolo, A.G. Determination of Aqueous Antibiotic Solutions Using SERS Nanogratings. Anal. Chim. Acta 2017, 982, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Guo, X.; Wu, Q.; Barman, J.; Su, S.; Huang, B.; Biao, T.; Zhang, Z.; Zhan, Q. Facile Silicone Oil-Coated Hydrophobic Surface for Surface Enhanced Raman Spectroscopy of Antibiotics. RSC Adv. 2019, 9, 14109–14115. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Meng, Y.; Jiao, X.; Huang, W.; Fan, D.; Liu, T.C. Facile Synthesis of an Economic 3D Surface-Enhanced Raman Scattering Platform for Ultrasensitive Detection of Antibiotics. Food Anal. Methods 2020, 13, 1947–1955. [Google Scholar] [CrossRef]
- Hidi, I.J.; Jahn, M.; Pletz, M.W.; Weber, K.; Cialla-May, D.; Popp, J. Toward Levofloxacin Monitoring in Human Urine Samples by Employing the LoC-SERS Technique. J. Phys. Chem. C 2016, 120, 20613–20623. [Google Scholar] [CrossRef]
- Bindesri, S.D.; Alhatab, D.S.; Brosseau, C.L. Development of an Electrochemical Surface-Enhanced Raman Spectroscopy (EC-SERS) Fabric-Based Plasmonic Sensor for Point-of-Care Diagnostics. Analyst 2018, 143, 4128–4135. [Google Scholar] [CrossRef]
- Tian, Y.; Li, G.; Zhang, H.; Xu, L.; Jiao, A.; Chen, F.; Chen, M. Construction of Optimized Au@Ag Core-Shell Nanorods for Ultralow SERS Detection of Antibiotic Levofloxacin Molecules. Opt. Express 2018, 26, 23347. [Google Scholar] [CrossRef]
- Liu, S.; Huang, J.; Chen, Z.; Chen, N.; Pang, F.; Wang, T.; Hu, L. Raman Spectroscopy Measurement of Levofloxacin Lactate in Blood Using an Optical Fiber Nano—Probe. J. Raman Spectrosc. 2015, 46, 197–201. [Google Scholar] [CrossRef]
- Liu, S.; Rong, M.; Zhang, H.; Chen, N.; Pang, F.; Chen, Z.; Wang, T.; Yan, J. In Vivo Raman Measurement of Levofloxacin Lactate in Blood Using a Nanoparticle-Coated Optical Fiber Probe. Biomed. Opt. Express 2016, 7, 810. [Google Scholar] [CrossRef] [Green Version]
- Mohaghegh, F.; Tehrani, A.M.; Rana, D.; Winterhalter, M.; Materny, A. Detection and Quantification of Small Concentrations of Moxifloxacin Using Surface—Enhanced Raman Spectroscopy in a Kretschmann Configuration. J. Raman Spectrosc. 2021, 52, 1617–1629. [Google Scholar] [CrossRef]
- Mamián-López, M.B.; Poppi, R.J. Quantification of Moxifloxacin in Urine Using Surface-Enhanced Raman Spectroscopy (SERS) and Multivariate Curve Resolution on a Nanostructured Gold Surface. Anal. Bioanal. Chem. 2013, 405, 7671–7677. [Google Scholar] [CrossRef]
- El-Zahry, M.R.; Lendl, B. Structure Elucidation and Degradation Kinetic Study of Ofloxacin Using Surface Enhanced Raman Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 193, 63–70. [Google Scholar] [CrossRef] [PubMed]
- März, A.; Trupp, S.; Rösch, P.; Mohr, G.J.; Popp, J. Fluorescence Dye as Novel Label Molecule for Quantitative SERS Investigations of an Antibiotic. Anal. Bioanal. Chem. 2012, 402, 2625–2631. [Google Scholar] [CrossRef] [PubMed]
- Filgueiras, A.L.; Paschoal, D.; Santos, H.F.D.; Sant’Ana, A.C. Adsorption Study of Antibiotics on Silver Nanoparticle Surfaces by Surface-Enhanced Raman Scattering Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 136, 979–985. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, G.; Guan, X.; Zhao, L. Rapid Preparation of Surface-Enhanced Raman Substrate in Microfluidic Channel for Trace Detection of Amoxicillin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 235, 63–70. [Google Scholar] [CrossRef]
- Anh, N.T.; Dinh, N.X.; Tuan, H.V.; Doan, M.Q.; Anh, N.H.; Khi, N.T.; Trang, V.T.; Tri, D.Q.; Le, A.-T. Eco-Friendly Copper Nanomaterials-Based Dual-Mode Optical Nanosensors for Ultrasensitive Trace Determination of Amoxicillin Antibiotics Residue in Tap Water Samples. Mater. Res. Bull. 2022, 147, 111649. [Google Scholar] [CrossRef]
- EL-Zahry, M.R.; Refaat, I.H.; Mohamed, H.A.; Rosenberg, E.; Lendl, B. Utility of Surface Enhanced Raman Spectroscopy (SERS) for Elucidation and Simultaneous Determination of Some Penicillins and Penicilloic Acid Using Hydroxylamine Silver Nanoparticles. Talanta 2015, 144, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Ashley, J.; Wu, K.; Hansen, M.F.; Schmidt, M.S.; Boisen, A.; Sun, Y. Quantitative Detection of Trace Level Cloxacillin in Food Samples Using Magnetic Molecularly Imprinted Polymer Extraction and Surface-Enhanced Raman Spectroscopy Nanopillars. Anal. Chem. 2017, 89, 11484–11490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, K.; Wu, C.; Deng, Z.; Guo, Y.; Peng, B. Preparation of Silver Decorated Silica Nanocomposite Rods for Catalytic and Surface-Enhanced Raman Scattering Applications. RSC Adv. 2015, 5, 52726–52736. [Google Scholar] [CrossRef]
- Li, Y.-T.; Qu, L.-L.; Li, D.-W.; Song, Q.-X.; Fathi, F.; Long, Y.-T. Rapid and Sensitive In-Situ Detection of Polar Antibiotics in Water Using a Disposable Ag–Graphene Sensor Based on Electrophoretic Preconcentration and Surface-Enhanced Raman Spectroscopy. Biosens. Bioelectron. 2013, 43, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Ji, W.; Yao, W. Rapid Surface Enhanced Raman Scattering Detection Method for Chloramphenicol Residues. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015, 144, 125–130. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, X.; Yin, H.; Meng, Q.; Zhao, Y.; Liu, L.; Wu, Z.; Xu, H. Quantitative and Sensitive Detection of Chloramphenicol by Surface-Enhanced Raman Scattering. Sensors 2017, 17, 2962. [Google Scholar] [CrossRef] [Green Version]
- Xiao, D.; Jie, Z.; Ma, Z.; Ying, Y.; Guo, X.; Wen, Y.; Yang, H. Fabrication of Homogeneous Waffle-like Silver Composite Substrate for Raman Determination of Trace Chloramphenicol. Microchim. Acta 2020, 187, 593. [Google Scholar] [CrossRef]
- Li, Y.; Tang, S.; Zhang, W.; Cui, X.; Zhang, Y.; Jin, Y.; Zhang, X.; Chen, Y. A Surface-Enhanced Raman Scattering-Based Lateral Flow Immunosensor for Colistin in Raw Milk. Sens. Actuators B Chem. 2019, 282, 703–711. [Google Scholar] [CrossRef]
- Sutherland, W.S.; Laserna, J.J.; Angebranndt, M.J.; Winefordner, J.D. Surface-Enhanced Raman Analysis of Sulfa Drugs on Colloidal Silver Dispersion. Anal. Chem. 1990, 62, 689–693. [Google Scholar] [CrossRef]
- Markina, N.E.; Markin, A.V.; Weber, K.; Popp, J.; Cialla-May, D. Liquid-Liquid Extraction-Assisted SERS-Based Determination of Sulfamethoxazole in Spiked Human Urine. Anal. Chim. Acta 2020, 1109, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Huang, J.; Sun, Y.; Yin, M.; Hu, M.; Hu, X.; Zhang, Z.; Zhang, G. Utilization of a Lateral Flow Colloidal Gold Immunoassay Strip Based on Surface-Enhanced Raman Spectroscopy for Ultrasensitive Detection of Antibiotics in Milk. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 197, 107–113. [Google Scholar] [CrossRef]
- Gatta, M.M.F.; Mendez, M.E.; Romano, S.; Calvo, M.V.; Dominguez-Gil, A.; Lanao, J.M. Pharmacokinetics of Amikacin in Intensive Care Unit Patients. J. Clin. Pharm. Ther. 1996, 21, 417–421. [Google Scholar] [CrossRef]
- Roberts, J.A.; Paul, S.K.; Akova, M.; Bassetti, M.; De Waele, J.J.; Dimopoulos, G.; Kaukonen, K.-M.; Koulenti, D.; Martin, C.; Montravers, P.; et al. DALI: Defining Antibiotic Levels in Intensive Care Unit Patients: Are Current Lactam Antibiotic Doses Sufficient for Critically Ill Patients? Clin. Infect. Dis. 2014, 58, 1072–1083. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Broll, M.; Kaever, V.; Burhenne, H.; Hafer, C.; Clajus, C.; Knitsch, W.; Burkhardt, O.; Kielstein, J.T. Pharmacokinetics of Ampicillin/Sulbactam in Critically Ill Patients with Acute Kidney Injury Undergoing Extended Dialysis. Clin. J. Am. Soc. Nephrol. CJASN 2012, 7, 385–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ismail, R.; Teh, L.K.; Choo, E.K. Chloramphenicol in Children: Dose, Plasma Levels and Clinical Effects. Ann. Trop. Paediatr. 1998, 18, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Nahid, P.; Mase, S.R.; Migliori, G.B.; Sotgiu, G.; Bothamley, G.H.; Brozek, J.L.; Cattamanchi, A.; Cegielski, J.P.; Chen, L.; Daley, C.L.; et al. Treatment of Drug-Resistant Tuberculosis. An Official ATS/CDC/ERS/IDSA Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2019, 200, e93–e142. [Google Scholar] [CrossRef]
- Tsuji, B.T.; Pogue, J.M.; Zavascki, A.P.; Paul, M.; Daikos, G.L.; Forrest, A.; Giacobbe, D.R.; Viscoli, C.; Giamarellou, H.; Karaiskos, I.; et al. International Consensus Guidelines for the Optimal Use of the Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-Infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacotherapy 2019, 39, 10–39. [Google Scholar] [CrossRef] [PubMed]
- Galar, A.; Muñoz, P.; Valerio, M.; Cercenado, E.; García-González, X.; Burillo, A.; Sánchez-Somolinos, M.; Juárez, M.; Verde, E.; Bouza, E. Current Use of Daptomycin and Systematic Therapeutic Drug Monitoring: Clinical Experience in a Tertiary Care Institution. Int. J. Antimicrob. Agents 2019, 53, 40–48. [Google Scholar] [CrossRef] [PubMed]


| Method | Multianalyte | Pros | Cons | Cost/Analysis |
|---|---|---|---|---|
| Immunoassay | No | Commercially available for most AM; simple to perform; direct from primary tube | Low accuracy; interferences from other drugs; may measure combined activity of parent and metabolites; time-consuming calibration. | High |
| LC-MS/MS | Yes | High sensitivity and accuracy | Initial costs; complexity of technology; not widely available; time consuming. | Medium |
| HPLC with UV/DAD detection | Yes | Widespread technology; commercially available assays; high accuracy | Manual sample preparation; subject to interference from various substances; run times may be slow. | Low |
| SERS | Yes | No need for sample preparation; fast measurement; ready for POC-T | Very often high RSD of the SERS substrates; method optimization needed for each drug. | Low |
| Family | Analytes | Substrate | Laser | Matrix | LOD | LOQ | Ref |
|---|---|---|---|---|---|---|---|
| Aminoglycosides | Amikacin | cAg | 532 | water | na | na | [49] |
| Neomycin | SERS-LFA | 633 | milk | 0.37 pg/mL | na | [50] | |
| Tobramycin | Au-DTNB NPs | 633 | human serum | na | na | [51] | |
| Antifungal | 5-flucytosine | sAg | 785 | serum | 12.1 μg/mL | na | [52] |
| Voriconazole | cAu | 785 | plasma | na | na | [53] | |
| AntiTB | Isoniazid | cAG | 488 | water | 5 ng | na | [54] |
| Carbapenems | Meropenem | sAu | 785 | plasma | na | na | [55] |
| Cephalosporins | Cefazolin | cAg | 638 | urine | na | na | [56] |
| Cefazolin | cCu | 638 | urine | 8.8 μg/mL | 90 ng/mL | [57] | |
| Cefoperazone | cAg | 638 | urine | na | na | [56] | |
| Cefoperazone | cCu | 638 | urine | 36 μg/mL | 1 pM | [57] | |
| Cefotaxime | cAg | 638 | urine | na | na | [56] | |
| Ceftazidime | sAg | 785 | water | na | na | [58] | |
| Ceftriaxone | CaCO3-Cu-NPs | 633 | water | 5 μM | 89 ng/mL | [59] | |
| Ceftriaxone | cCu | 638 | urine | 7.5 μg/mL | 19 μg/mL | [57] | |
| Ceftriaxone | cAg | 638 | urine | na | na | [56] | |
| Ceftriaxone | cAg | 473 | urine | 0.4 μg/mL | 45 μg/mL | [60] | |
| Cefuroxime | cAg | 638 | urine | na | na | [56] | |
| Fluoroquinolones | Ciprofloxacin | Ag-TiO2 NPs | 633 | water | 70.8 pM | 34 μg/mL | [61] |
| Ciprofloxacin | SERS-LFA | 633 | milk | 0.57 pg/mL | na | [50] | |
| Ciprofloxacin | cAg | 514 | water | 0.1 μM | 0.1 mM | [62] | |
| Ciprofloxacin | sAg | 785 | water | na | na | [63] | |
| Ciprofloxacin | Au NRs | 633 | water | 0.01 ppm | 92 μg/mL | [64] | |
| Ciprofloxacin | sAg | 785 | water | 0.351 ppb | 2 μg/mL | [65] | |
| Enoxacin | Ag-TiO2 NPs | 633 | water | 315 pM | 280 μg/mL | [61] | |
| Levofloxacin | cAg | 532 | urine | 0.07 mM | na | [66] | |
| Levofloxacin | EC-SERS | 785 | synthetic urine | na | na | [67] | |
| Levofloxacin | Au@Ag NRs | 633 | water | 0.37 ng/L | na | [68] | |
| Levofloxacin | cAu | 633 | mice blood | na | 1 ppm | [69] | |
| Levofloxacin | optical fiber nano-probe | 633 | mice blood | na | na | [70] | |
| Moxifloxacin | sAg | 514 | water | 100 nM | na | [71] | |
| Moxifloxacin | sAu | 785 | urine | 0.085 μg/mL | na | [72] | |
| Ofloxacin | cAg | 532 | methanol | 23.5 ng/mL | na | [73] | |
| Macrolide | Erythromycin | cAg | 532 | water/ethanol | na | na | [74] |
| Monobactam | Aztreonam | cAg | 633 | water | na | na | [75] |
| Penicillins | Amoxicillin | sAg | 785 | water | 2.4 nM | na | [76] |
| Amoxicillin | cCu, Cu-GO | 785 | water | 0.0157 μM | na | [77] | |
| Ampicillin | cAg | 533 | water | 27 ng/mL | na | [78] | |
| Cloxacillin | sAu | 780 | methanol/acetic acid (9:1) | 7.8 pmol | 0.15 μg/mL | [79] | |
| Penicillin G | SiO2@Ag-NRs | 532 | water | 0.001 nM | 72.6 ng/mL | [80] | |
| Penicillin G | Ag-GO | 785 | water | 0.3 nM | na | [81] | |
| Penicillin G | cAg | 533 | water | 29 ng/mL | na | [78] | |
| Phenicols | Chloramphenicol | cAu | 785 | water | 0.1 μg/mL | na | [82] |
| Chloramphenicol | cAu | 785 | eye drops | 0.01 μM | na | [83] | |
| Chloramphenicol | sAg | 633 | honey | 4 ng/mL | na | [84] | |
| Polymixins | Colistin | SERS-LFIA | 785 | milk | 0.10 ng/mL | na | [85] |
| Sulfanilamides | Sulfadiazine | cAg | 514 | water | 1 ng/mL | na | [86] |
| Sulfamethoxazole | cAg | 488 | urine | 1.7 μg/mL | na | [87] |
| DRUG | Effective Concentrations (μM) | Toxic Concentrations (Cmin, μM) | Protein Bound Fraction (%) | TDM | SERS Method | Ref |
|---|---|---|---|---|---|---|
| Amikacin | 7–14 (trough); 25–43 (peak) | >14 (trough); >55 (peak) | <10 | required | + | [89] |
| Amoxicillin | 86–172 | >988 | 18 | recommended for ICU patients | ++ | [90] |
| Ampicillin-sulbactam | 23–106 | >106 | 28 | recommended for ICU patients | ++ | [91] |
| Aztreonam | 207–376 | 56 (36–43) | recommended for ICU patients | + | [12] | |
| Cefazolin | 88–176 | 74–86 | recommended for ICU patients | +++ | [90] | |
| Cefepime | 10–73 | >46 | 20 | recommended for ICU patients | +++ | [90] |
| Cefotaxime | 65–132 | 35–45 | recommended for ICU patients | +++ | [90] | |
| Ceftazidime | 64–146 | <10 | recommended for ICU patients | ++ | [90] | |
| Ceftriaxone | 44–120 | 95 | recommended for ICU patients | ++ | [90] | |
| Chloramphenicol | 31–77 | >77 | 50–60 | required | ++ | [92] |
| Ciprofloxacin | 3–30 | unclear | 20–40 | recommended for ICU patients | ++ | [12] |
| Clofazimine | 1–4 | 99.99 | recommended (TB) | nr | [93] | |
| Cloxacillin | 46–115 | 95 | recommended for ICU patients | + | [12] | |
| Colistin | unclear | >2 | 69–74 | required with prolonged dosing | ++ | [94] |
| Daptomycin | <15 | >15 | 90–93 | neither recommended nor discouraged | nr | [95] |
| Ertapenem | 11–21 (trough) | 85–95 cd | recommended for ICU patients | nr | [90] | |
| Fluconazole | >33–49 (trough) | 11–12 | neither recommended nor discouraged | nr | [12] | |
| 5-Flucytosine | 194–387 (trough); 387–775 (peak) | >775 | 3–4 | neither recommended nor discouraged | +++ | [12] |
| Gentamycin | <4 (trough); 10–21 (peak) | >4 (trough); >25 (peak) | 0–30 | required | nr | [12] |
| Isoniazid | 22–51 (peak, 300 mg dose) *; 66–131 (peak, 900 mg dose) * | >146 | <10–15 | recommended | ++ | [93] |
| Itraconazole | 0.7–1.4 | <6 | 99.8 | required | nr | [12] |
| Levofloxacin | 0.83–36 | unclear | 24–38 | recommended for ICU/TB patients | +++ | [12,93] |
| Linezolid | 6–21 | >21 | 31 | recommended for ICU/TB patients | nr | [12,93] |
| Meropenem | 21–42 | >145 | ~2 | recommended for ICU/TB patients | +++ | [12,93] |
| Moxifloxacin | 3–12 | unclear | 50 | recommended for ICU/TB patients | +++ | [12,93] |
| Ofloxacin | 2–28 | unclear | 32 | recommended for ICU patients | + | [12] |
| Oxacillin | 1–5 | >411 | 94.2 +/− 2.1 | recommended for ICU patients | nr | [90] |
| Piperacillin | 155–310 (Css) | >1000 | 51.7 | recommended for ICU patients | nr | [90] |
| Rifampicin | 10–29 (Peak) * | >67 | 86.1–88.9 | recommended for ICU patients | nr | [12] |
| Sulfamethoxazole | unclear | 395–592 | 70 | neither recommended nor discouraged | +++ | [12] |
| Ticarcillin | ~843 | 45 | recommended for ICU patients | nr | [12] | |
| Tigecycline | 0.1–1 | 71–89 | recommended for ICU patients | nr | [12] | |
| Tobramycin | <4 (trough); 10–21 (peak) | >4 (through); >26 (peak); | <10 | required | +++ | [12] |
| Vancomycin | 3–7 (trough); 14–28 (peak) | >7 (trough); >55 (peak) | 55 | required | nr | [12] |
| Voriconazole | 1–6 (trough) | 14–17 | 58 | required | +++ | [12] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fornasaro, S.; Cialla-May, D.; Sergo, V.; Bonifacio, A. The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents. Chemosensors 2022, 10, 128. https://doi.org/10.3390/chemosensors10040128
Fornasaro S, Cialla-May D, Sergo V, Bonifacio A. The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents. Chemosensors. 2022; 10(4):128. https://doi.org/10.3390/chemosensors10040128
Chicago/Turabian StyleFornasaro, Stefano, Dana Cialla-May, Valter Sergo, and Alois Bonifacio. 2022. "The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents" Chemosensors 10, no. 4: 128. https://doi.org/10.3390/chemosensors10040128
APA StyleFornasaro, S., Cialla-May, D., Sergo, V., & Bonifacio, A. (2022). The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents. Chemosensors, 10(4), 128. https://doi.org/10.3390/chemosensors10040128







