Microfluidic-Enabled Multi-Cell-Densities-Patterning and Culture Device for Characterization of Yeast Strains’ Growth Rates under Mating Pheromone
Abstract
:1. Introduction
2. Materials and Methods
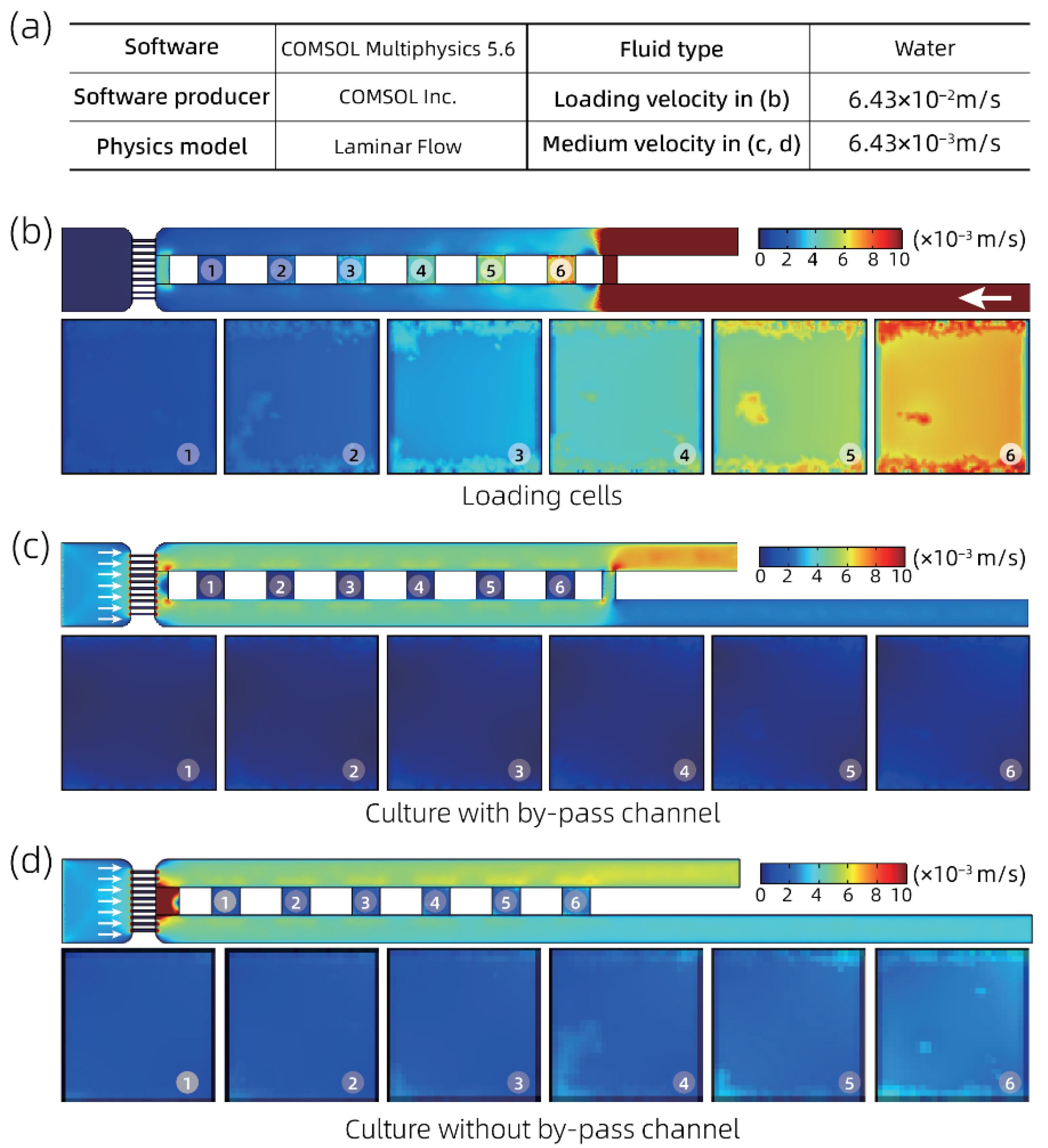
2.1. System Setup and Microfluidic Chip Design
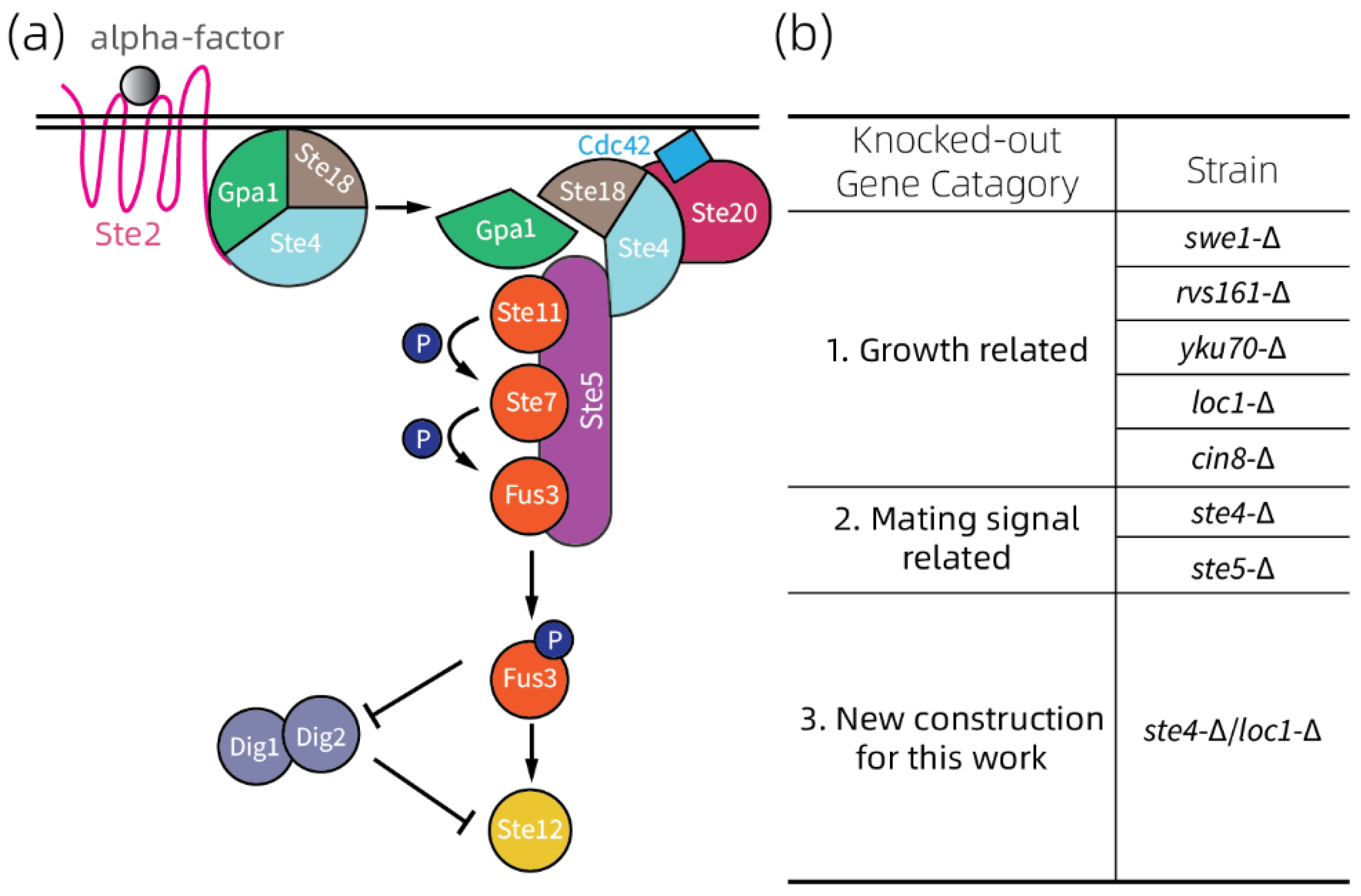
2.2. Selection of Yeast Strains
2.3. Experiment Workflow and Image Acquisition
3. Results
3.1. Validation on Function of Loading Yeasts with a Multi-Cell-Density Pattern
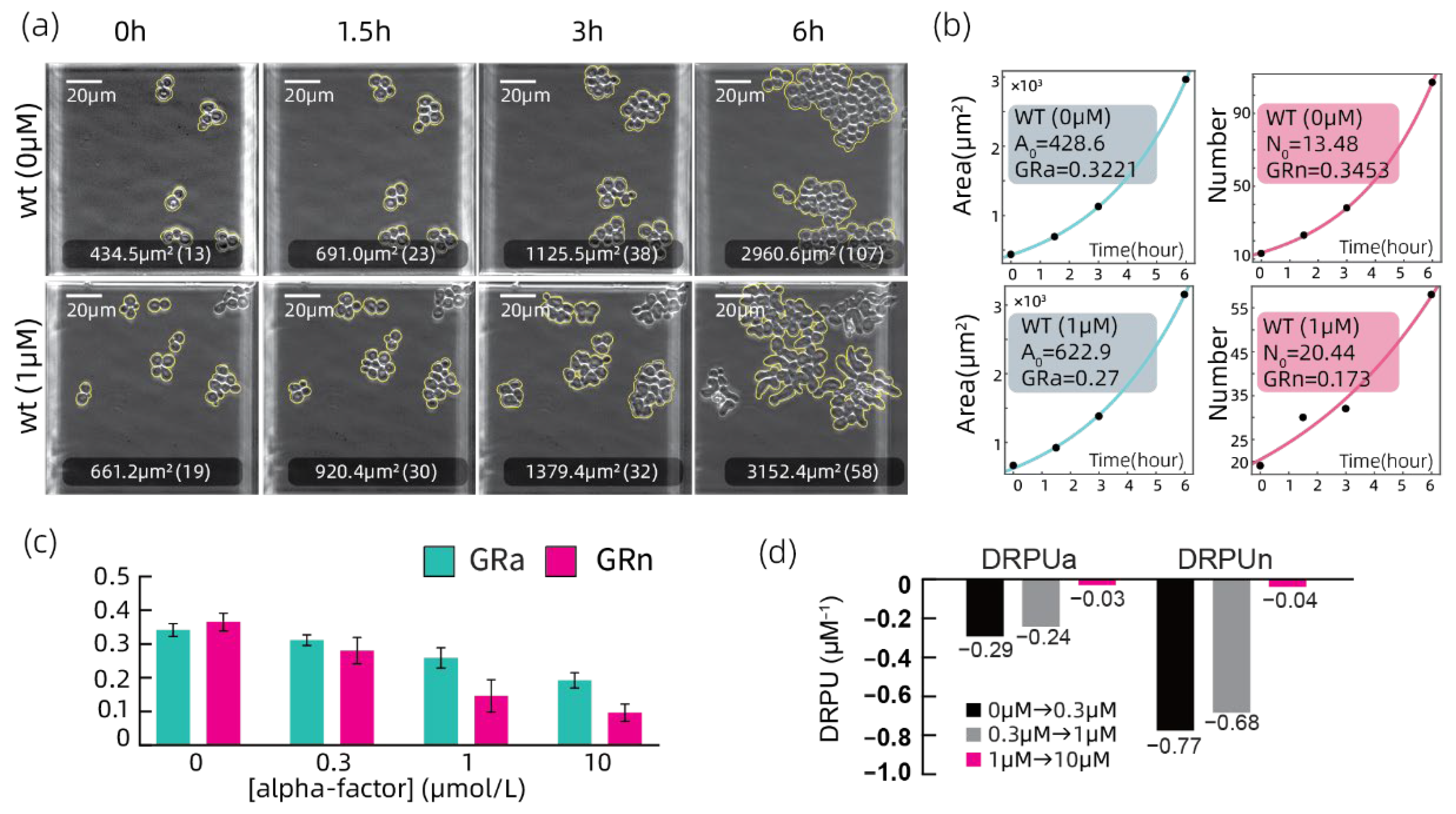
3.2. Growth Rate Analyzation on the Wild Type Strain
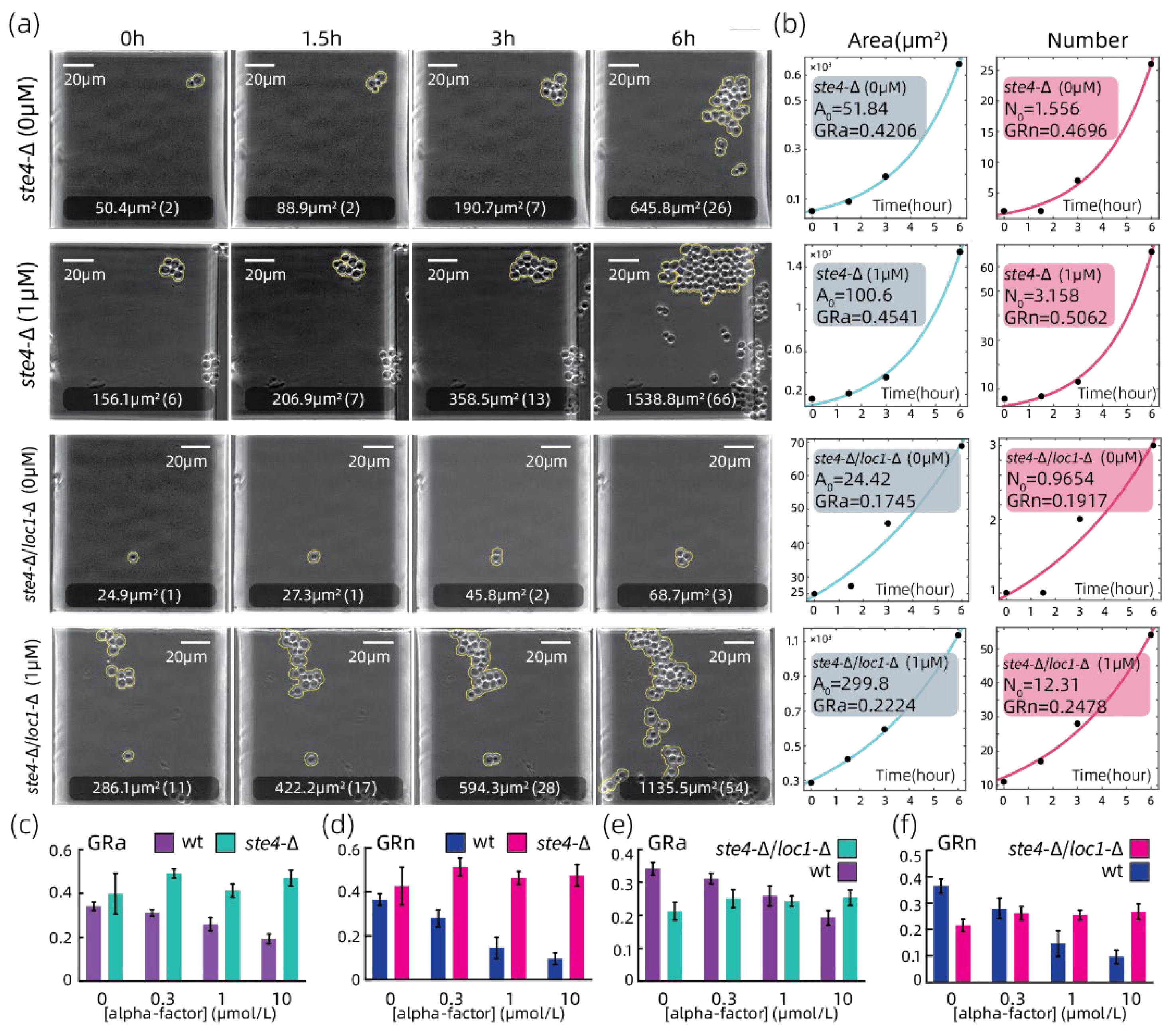
3.3. Growth Rate Analyzation on Strains with Growth-Related Genes Absent
3.4. Growth Rate Analyzation on Strains with Defective Mating Pheromone Response Pathway
3.5. Landscape View of Eight Gene Knock-out Strains under Four Levels of Mating Pheromone A-Factor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poulsen, A.K.; Petersen, M.O.; Olsen, L.F. Single Cell Studies and Simulation of Cell-Cell Interactions Using Oscillating Glycolysis in Yeast Cells. Biophys. Chem. 2007, 125, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Kortmann, H.; Blank, L.M.; Schmid, A. Single Cell Analysis Reveals Unexpected Growth Phenotype of S. Cerevisiae. Cytometry 2009, 75, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Puchkov, E.O. Single Yeast Cell Vacuolar Milieu Viscosity Assessment by Fluorescence Polarization Microscopy with Computer Image Analysis. Yeast 2012, 29, 185–190. [Google Scholar] [CrossRef]
- Durandau, E.; Aymoz, D.; Pelet, S. Dynamic Single Cell Measurements of Kinase Activity by Synthetic Kinase Activity Relocation Sensors. BMC Biol. 2015, 13, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, Y.T.; Koh, J.L.; Friesen, H.; Duffy, S.K.; Cox, M.J.; Moses, A.; Moffat, J.; Boone, C.; Andrews, B.J. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell 2015, 161, 1413–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.B.; Wang, Q.Y.; Ke, Y.X.; Liu, S.Y.; Ju, J.Q.; Lim, W.A.; Tang, C.; Wei, P. Design of Tunable Oscillatory Dynamics in a Synthetic Nf-Kappab Signaling Circuit. Cell Syst. 2017, 5, 460–470.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.Y.; Li, P.C. A Three-Dimensional Flow Control Concept for Single-Cell Experiments on a Microchip. 2. Fluorescein Diacetate Metabolism and Calcium Mobilization in a Single Yeast Cell as Stimulated by Glucose and pH Changes. Anal. Chem. 2004, 76, 5282–5292. [Google Scholar] [CrossRef]
- Liti, G.; Schacherer, J. The Rise of Yeast Population Genomics. Comptes Rendus Biol. 2011, 334, 612–619. [Google Scholar] [CrossRef]
- Chen, M.T.; Weiss, R. Artificial Cell-Cell Communication in Yeast Saccharomyces Cerevisiae Using Signaling Elements from Arabidopsis Thaliana. Nat. Biotechnol. 2005, 23, 1551–1555. [Google Scholar] [CrossRef]
- Regot, S.; Macia, J.; Conde, N.; Furukawa, K.; Kjellen, J.; Peeters, T.; Hohmann, S.; de Nadal, E.; Posas, F.; Sole, R. Distributed Biological Computation with Multicellular Engineered Networks. Nature 2011, 469, 207–211. [Google Scholar] [CrossRef]
- Vachova, L.; Palkova, Z. Aging and Longevity of Yeast Colony Populations: Metabolic Adaptation and Differentiation. Biochem. Soc. Trans. 2011, 39, 1471–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billerbeck, S.; Brisbois, J.; Agmon, N.; Jimenez, M.; Temple, J.; Shen, M.; Boeke, J.D.; Cornish, V.W. A Scalable Peptide-Gpcr Language for Engineering Multicellular Communication. Nat. Commun. 2018, 9, 5057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossouw, D.; Meiring, S.P.; Bauer, F.F. Modifying Saccharomyces Cerevisiae Adhesion Properties Regulates Yeast Ecosystem Dynamics. mSphere 2018, 3, e00383-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Huang, Y.; Wu, Z. Simulation of Yeast Cooperation in 2D. Bull. Math. Biol. 2016, 78, 531–555. [Google Scholar] [CrossRef]
- Wloch-Salamon, D.M.; Fisher, R.M.; Regenberg, B. Division of Labour in the Yeast: Saccharomyces Cerevisiae. Yeast 2017, 34, 399–406. [Google Scholar] [CrossRef] [Green Version]
- Kurth, F.; Schumann, C.A.; Blank, L.M.; Schmid, A.; Manz, A.; Dittrich, P.S. Bilayer Microfluidic Chip for Diffusion-Controlled Activation of Yeast Species. J. Chromatogr. A 2008, 1206, 77–82. [Google Scholar] [CrossRef]
- Kang, X.; Jiang, L.; Chen, X.; Yuan, H.; Luo, C.; Ouyang, Q. Pump-Free Multi-Well-Based Microfluidic System for High-Throughput Analysis of Size-Control Relative Genes in Budding Yeast. Integr. Biol. 2014, 6, 685–693. [Google Scholar] [CrossRef]
- Yuan, H.; Zhang, R.; Shao, B.; Wang, X.; Ouyang, Q.; Hao, N.; Luo, C. Protein Expression Patterns of the Yeast Mating Response. Integr. Biol. 2016, 8, 712–719. [Google Scholar] [CrossRef]
- Shao, B.; Yuan, H.; Zhang, R.; Wang, X.; Zhang, S.; Ouyang, Q.; Hao, N.; Luo, C. Reconstructing the Regulatory Circuit of Cell Fate Determination in Yeast Mating Response. PLoS Comput. Biol. 2017, 13, e1005671. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Kang, Y.; Pan, X.; Yu, J.; Ouyang, Q.; Luo, C. Studies of the Drug Resistance Response of Sensitive and Drug-Resistant Strains in a Microfluidic System. Integr. Biol. 2014, 6, 143–151. [Google Scholar] [CrossRef]
- Ran, M.; Wang, Y.; Wang, S.; Luo, C. Pump-Free Gradient-Based Micro-Device Enables Quantitative and High-Throughput Bacterial Growth Inhibition Analysis. Biomed. Microdevices 2015, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Quan, Q.; Zhang, S.; Wang, X.; Ouyang, Q.; Wang, Y.; Yang, G.; Luo, C. A Parallel and Quantitative Cell Migration Assay Using a Novel Multi-Well-Based Device. Biomed. Microdevices 2016, 18, 99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, R.; Wei, M.; Zhang, Y. A Novel Method of Cell Culture Based on the Microfluidic Chip for Regulation of Cell Density. Biotechnol. Bioeng. 2020, 118, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yuan, H.; Wang, S.; Ouyang, Q.; Chen, Y.; Hao, N.; Luo, C. High-Throughput Single-Cell Analysis for the Proteomic Dynamics Study of the Yeast Osmotic Stress Response. Sci. Rep. 2017, 7, 42200. [Google Scholar] [CrossRef] [PubMed]
- Hulme, S.E.; Shevkoplyas, S.S.; Apfeld, J.; Fontana, W.; Whitesides, G.M. A Microfabricated Array of Clamps for Immobilizing and Imaging C. Elegans. Lab Chip 2007, 7, 1515–1523. [Google Scholar] [CrossRef]
- Bardwell, L. A Walk-through of the Yeast Mating Pheromone Response Pathway. Peptides 2005, 26, 339–350. [Google Scholar] [CrossRef]
- Parker, L.L.; Kurutz, J.W.; Kent, S.B.; Kron, S.J. Control of the Yeast Cell Cycle with a Photocleavable Alpha-Factor Analogue. Angew. Chem. Int. Ed. 2006, 45, 6322–6325. [Google Scholar] [CrossRef]
- Zhang, N.N.; Dudgeon, D.D.; Paliwal, S.; Levchenko, A.; Grote, E.; Cunningham, K.W. Multiple Signaling Pathways Regulate Yeast Cell Death During the Response to Mating Pheromones. Mol. Biol. Cell 2006, 17, 3409–3422. [Google Scholar] [CrossRef]
- Goranov, A.I.; Cook, M.; Ricicova, M.; Ben-Ari, G.; Gonzalez, C.; Hansen, C.; Tyers, M.; Amon, A. The Rate of Cell Growth Is Governed by Cell Cycle Stage. Genes Dev. 2009, 23, 1408–1422. [Google Scholar] [CrossRef] [Green Version]
- Michaelis, S.; Barrowman, J. Biogenesis of the Saccharomyces Cerevisiae Pheromone a-Factor, from Yeast Mating to Human Disease. Microbiol. Mol. Biol. Rev. 2012, 76, 626–651. [Google Scholar] [CrossRef] [Green Version]
- Hao, N.; Nayak, S.; Behar, M.; Shanks, R.H.; Nagiec, M.J.; Errede, B.; Hasty, J.; Elston, T.C.; Dohlman, H.G. Regulation of Cell Signaling Dynamics by the Protein Kinase-Scaffold Ste5. Mol. Cell 2008, 30, 649–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorer, R.; Pryciak, P.M.; Hartwell, L.H. Saccharomyces Cerevisiae Cells Execute a Default Pathway to Select a Mate in the Absence of Pheromone Gradients. J. Cell Biol. 1995, 131, 845–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erdman, S.; Snyder, M. A Filamentous Growth Response Mediated by the Yeast Mating Pathway. Genetics 2001, 159, 919–928. [Google Scholar] [CrossRef] [PubMed]
- Segall, J.E. Polarization of Yeast Cells in Spatial Gradients of Alpha Mating Factor. Proc. Natl. Acad. Sci. USA 1993, 90, 8332–8336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paliwal, S.; Iglesias, P.A.; Campbell, K.; Hilioti, Z.; Groisman, A.; Levchenko, A. Mapk-Mediated Bimodal Gene Expression and Adaptive Gradient Sensing in Yeast. Nature 2007, 446, 46–51. [Google Scholar] [CrossRef]
- Molk, J.N.; Bloom, K. Microtubule Dynamics in the Budding Yeast Mating Pathway. J. Cell Sci. 2006, 119, 3485–3490. [Google Scholar] [CrossRef] [Green Version]
- Shao, D.; Zheng, W.; Qiu, W.; Ouyang, Q.; Tang, C. Dynamic Studies of Scaffold-Dependent Mating Pathway in Yeast. Biophys. J. 2006, 91, 3986–4001. [Google Scholar] [CrossRef] [Green Version]
- McClure, A.W.; Jacobs, K.C.; Zyla, T.R.; Lew, D.J. Mating in Wild Yeast: Delayed Interest in Sex after Spore Germination. Mol. Biol. Cell 2018, 29, 3119–3127. [Google Scholar] [CrossRef]
- Shellhammer, J.P.; Pomeroy, A.E.; Li, Y.; Dujmusic, L.; Elston, T.C.; Hao, N.; Dohlman, H.G. Quantitative Analysis of the Yeast Pheromone Pathway. Yeast 2019, 36, 495–518. [Google Scholar] [CrossRef]
- Doyle, T.; Botstein, D. Movement of Yeast Cortical Actin Cytoskeleton Visualized in Vivo. Proc. Natl. Acad. Sci. USA 1996, 93, 3886–3891. [Google Scholar] [CrossRef] [Green Version]
- Chant, J. Cell Polarity in Yeast. Annu. Rev. Cell Dev. Biol. 1999, 15, 365–391. [Google Scholar] [CrossRef] [PubMed]
- Ghose, D.; Elston, T.; Lew, D. Orientation of Cell Polarity by Chemical Gradients. Annu. Rev. Biophys. 2022, 51. [Google Scholar] [CrossRef] [PubMed]
- Youk, H.; Lim, W.A. Secreting and Sensing the Same Molecule Allows Cells to Achieve Versatile Social Behaviors. Science 2014, 343, 1242782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer Deletion Strains Derived from Saccharomyces Cerevisiae S288c: A Useful Set of Strains and Plasmids for Pcr-Mediated Gene Disruption and Other Applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global Analysis of Protein Localization in Budding Yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-Efficiency Yeast Transformation Using the Liac/Ss Carrier DNA/Peg Method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Booher, R.N.; Deshaies, R.J.; Kirschner, M.W. Properties of Saccharomyces Cerevisiae Wee1 and Its Differential Regulation of P34CDC28 in Response to G1 and G2 Cyclins. EMBO J. 1993, 12, 3417–3426. [Google Scholar] [CrossRef]
- Urbinati, C.R.; Gonsalvez, G.B.; Aris, J.P.; Long, R.M. Loc1p Is Required for Efficient Assembly and Nuclear Export of the 60s Ribosomal Subunit. Mol. Genet. Genom. 2006, 276, 369–377. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Shen, W.; Cai, Z.; Chen, K.; Ouyang, Q.; Wei, P.; Yang, W.; Luo, C. Microfluidic-Enabled Multi-Cell-Densities-Patterning and Culture Device for Characterization of Yeast Strains’ Growth Rates under Mating Pheromone. Chemosensors 2022, 10, 141. https://doi.org/10.3390/chemosensors10040141
Zhang J, Shen W, Cai Z, Chen K, Ouyang Q, Wei P, Yang W, Luo C. Microfluidic-Enabled Multi-Cell-Densities-Patterning and Culture Device for Characterization of Yeast Strains’ Growth Rates under Mating Pheromone. Chemosensors. 2022; 10(4):141. https://doi.org/10.3390/chemosensors10040141
Chicago/Turabian StyleZhang, Jing, Wenting Shen, Zhiyuan Cai, Kaiyue Chen, Qi Ouyang, Ping Wei, Wei Yang, and Chunxiong Luo. 2022. "Microfluidic-Enabled Multi-Cell-Densities-Patterning and Culture Device for Characterization of Yeast Strains’ Growth Rates under Mating Pheromone" Chemosensors 10, no. 4: 141. https://doi.org/10.3390/chemosensors10040141
APA StyleZhang, J., Shen, W., Cai, Z., Chen, K., Ouyang, Q., Wei, P., Yang, W., & Luo, C. (2022). Microfluidic-Enabled Multi-Cell-Densities-Patterning and Culture Device for Characterization of Yeast Strains’ Growth Rates under Mating Pheromone. Chemosensors, 10(4), 141. https://doi.org/10.3390/chemosensors10040141







