State of the Art of Chemosensors in a Biomedical Context
Abstract
:1. Introduction
2. Chemosensor Types
2.1. State-of-the-Art of Plasmonic Sensors
2.2. Future Direction of Research in Plasmonic Sensors
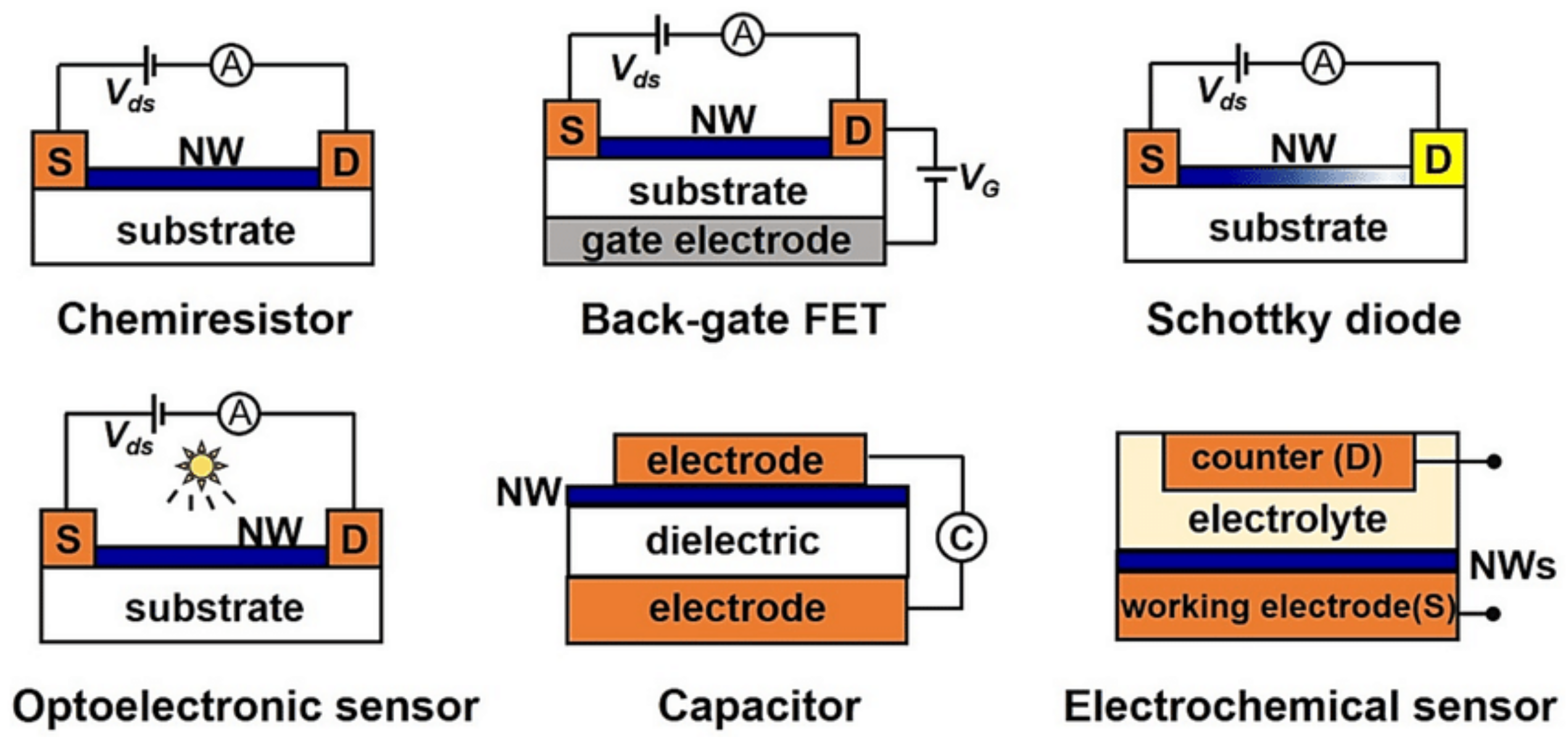
2.3. State of the Art of Field Effect Transistor (FET) Type Sensors
2.4. Future Direction of Research in FET Type Sensors
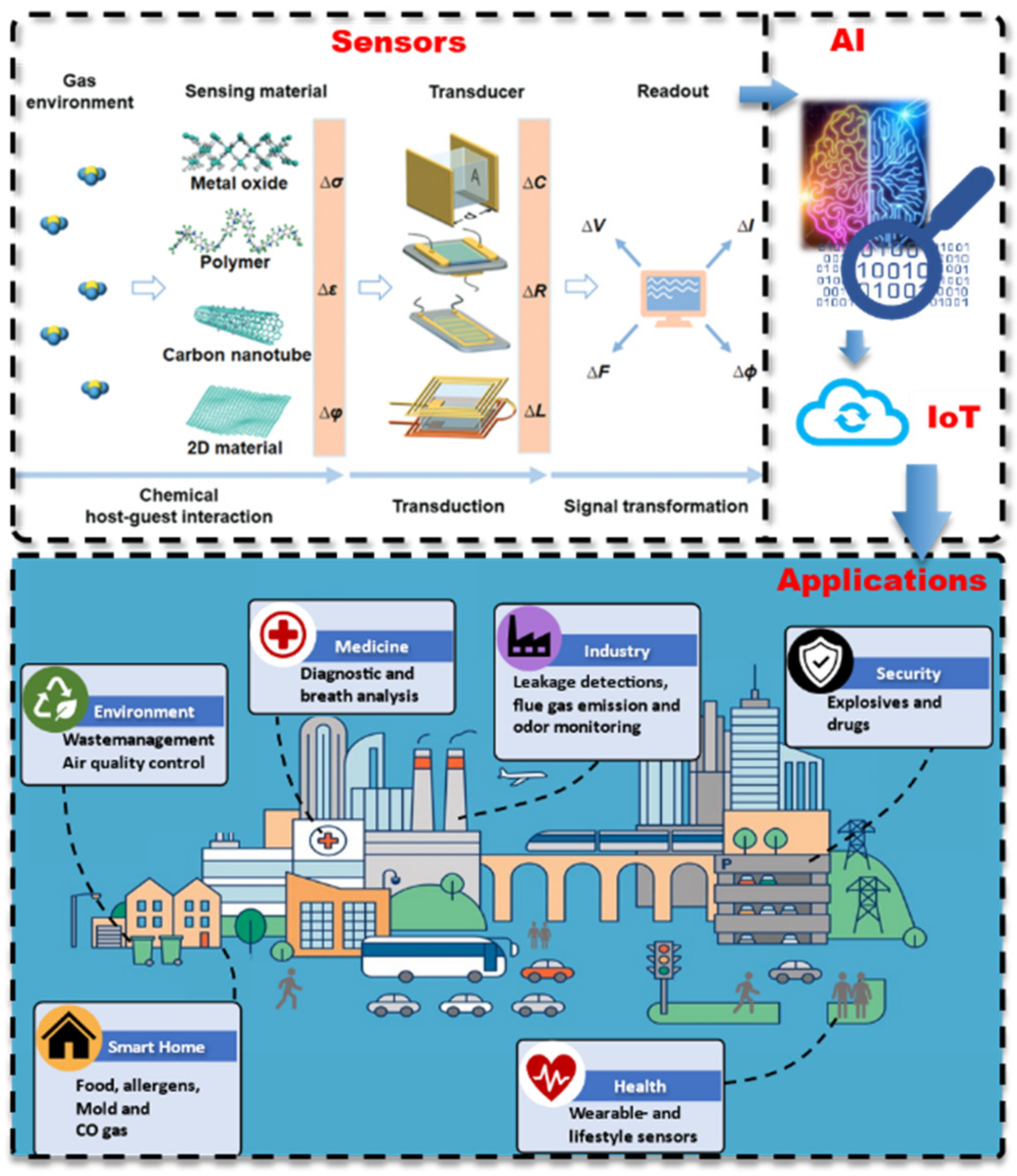
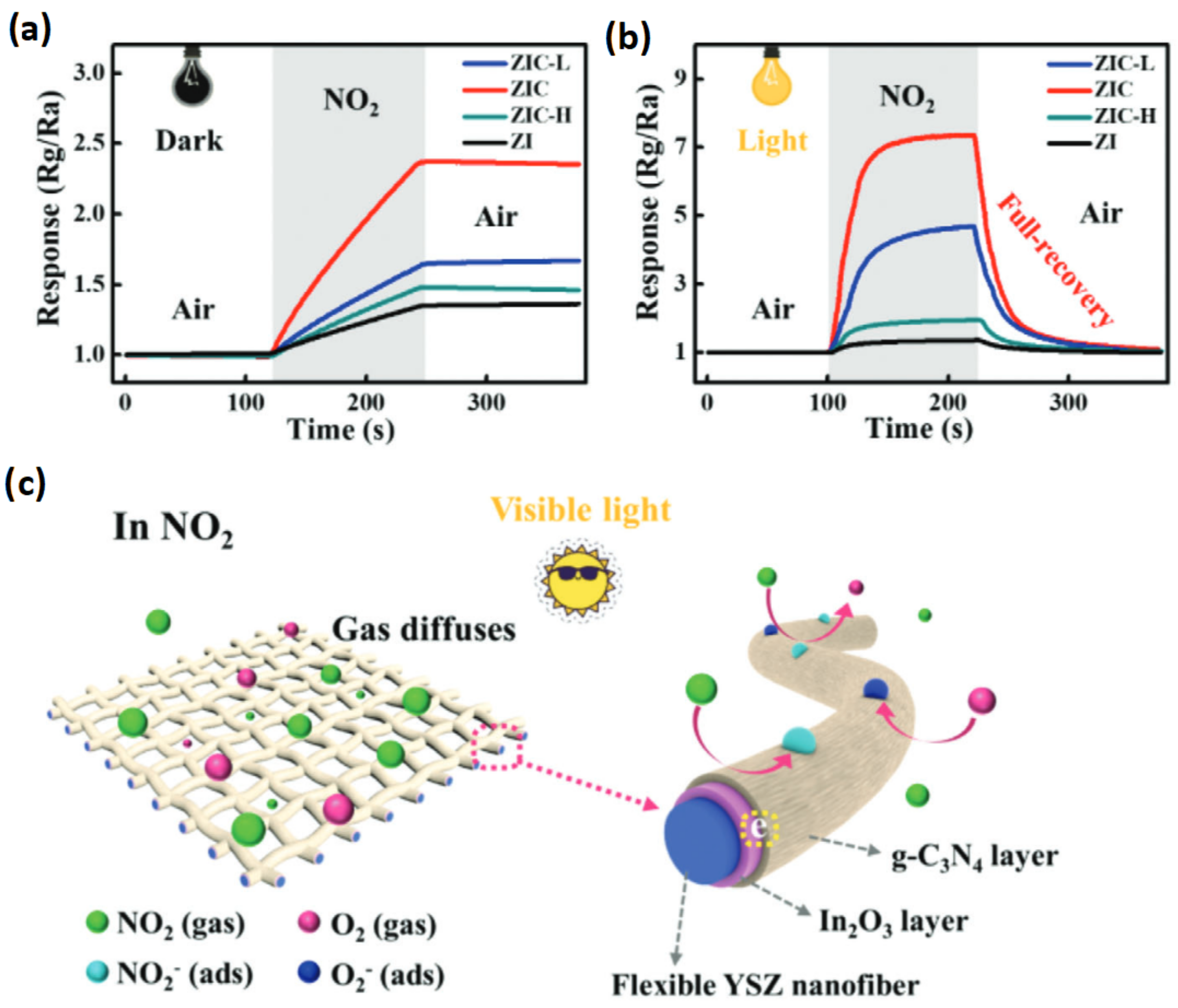
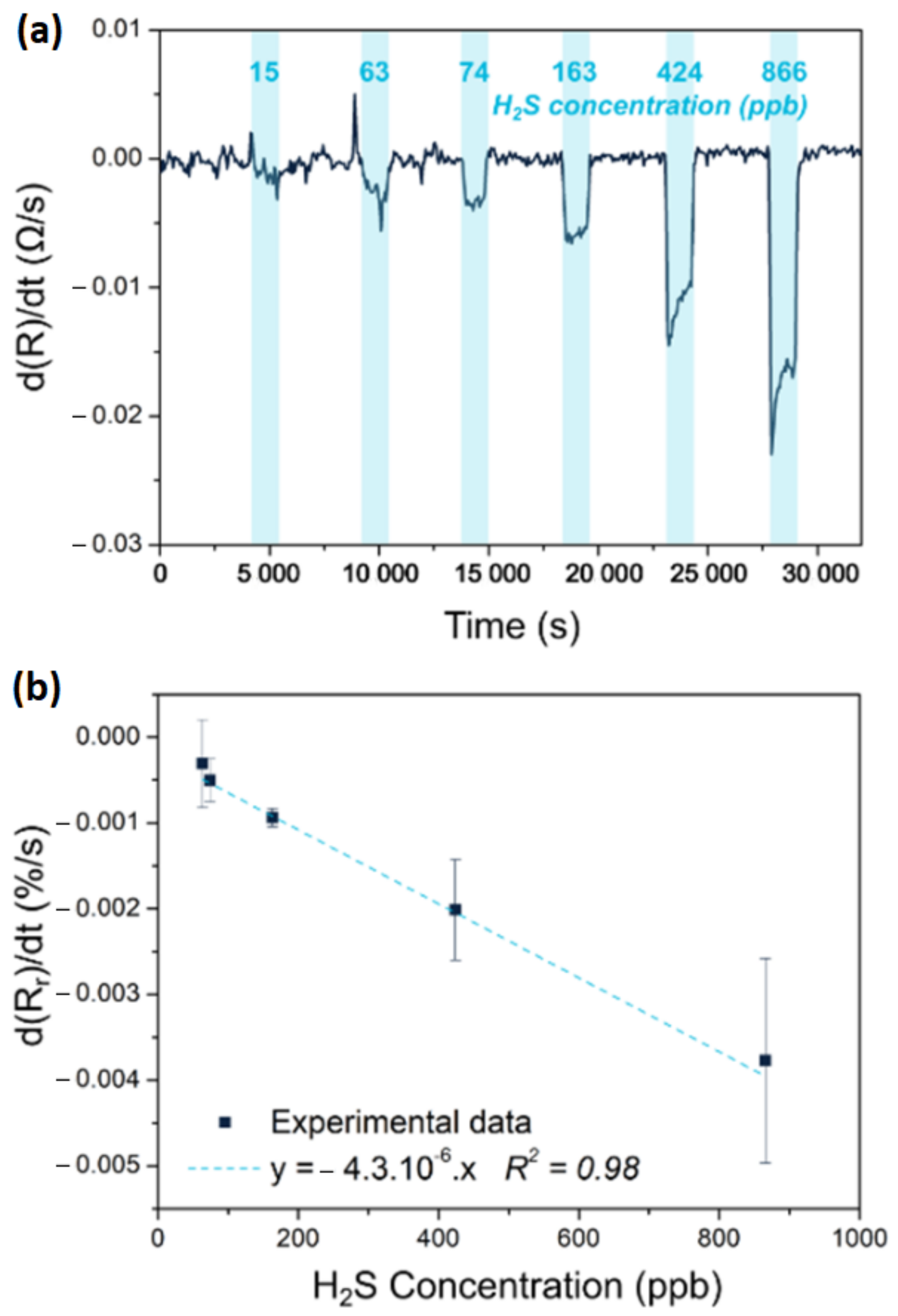
2.5. State of the Art of Chemiresistive Sensors
2.6. Future Direction of Research in Chemiresistive Sensors
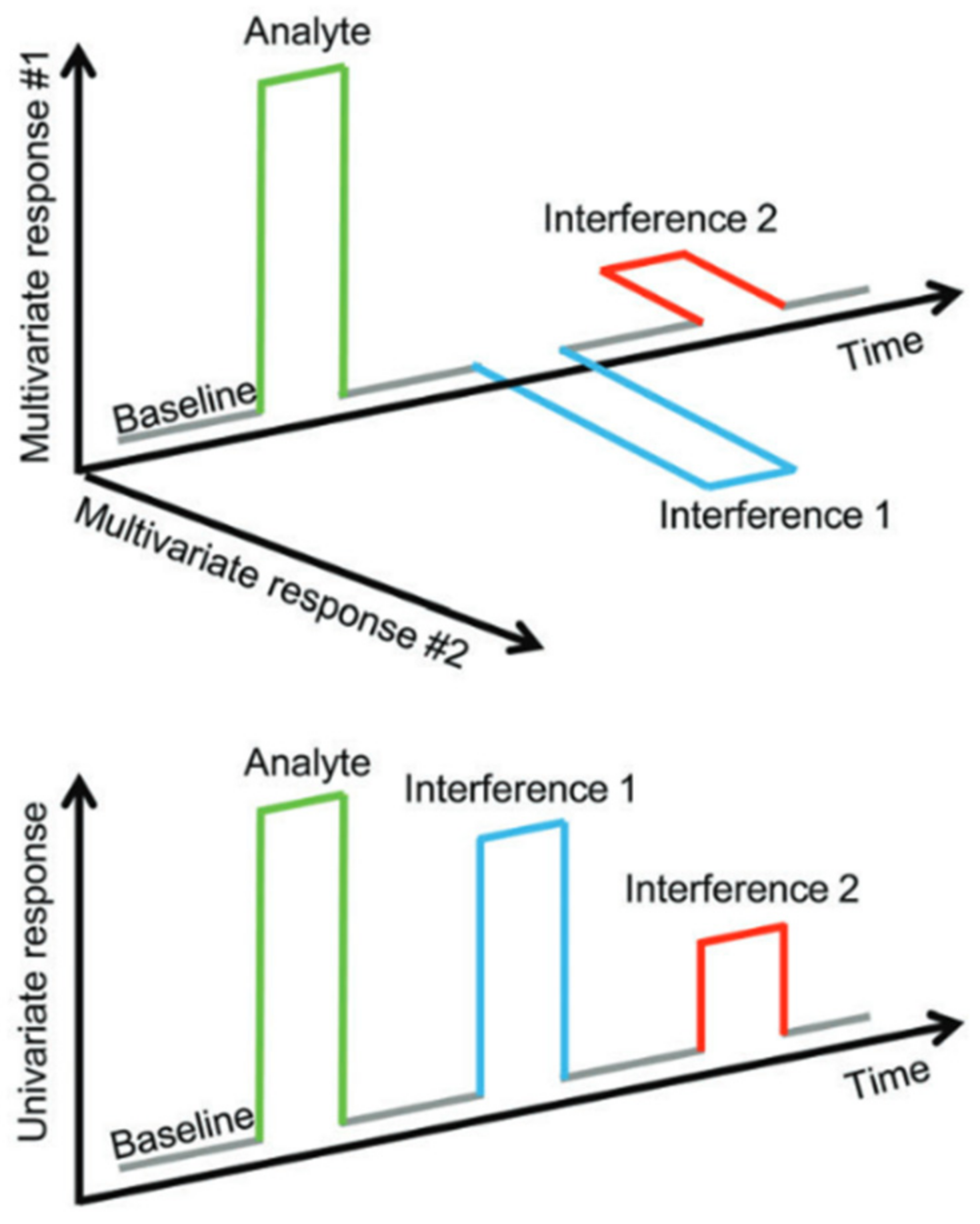
2.7. State of the Art of Sensor Arrays
2.8. Future Direction of Research in Sensor Arrays
3. Applications of Chemosensors in BioMedical Context
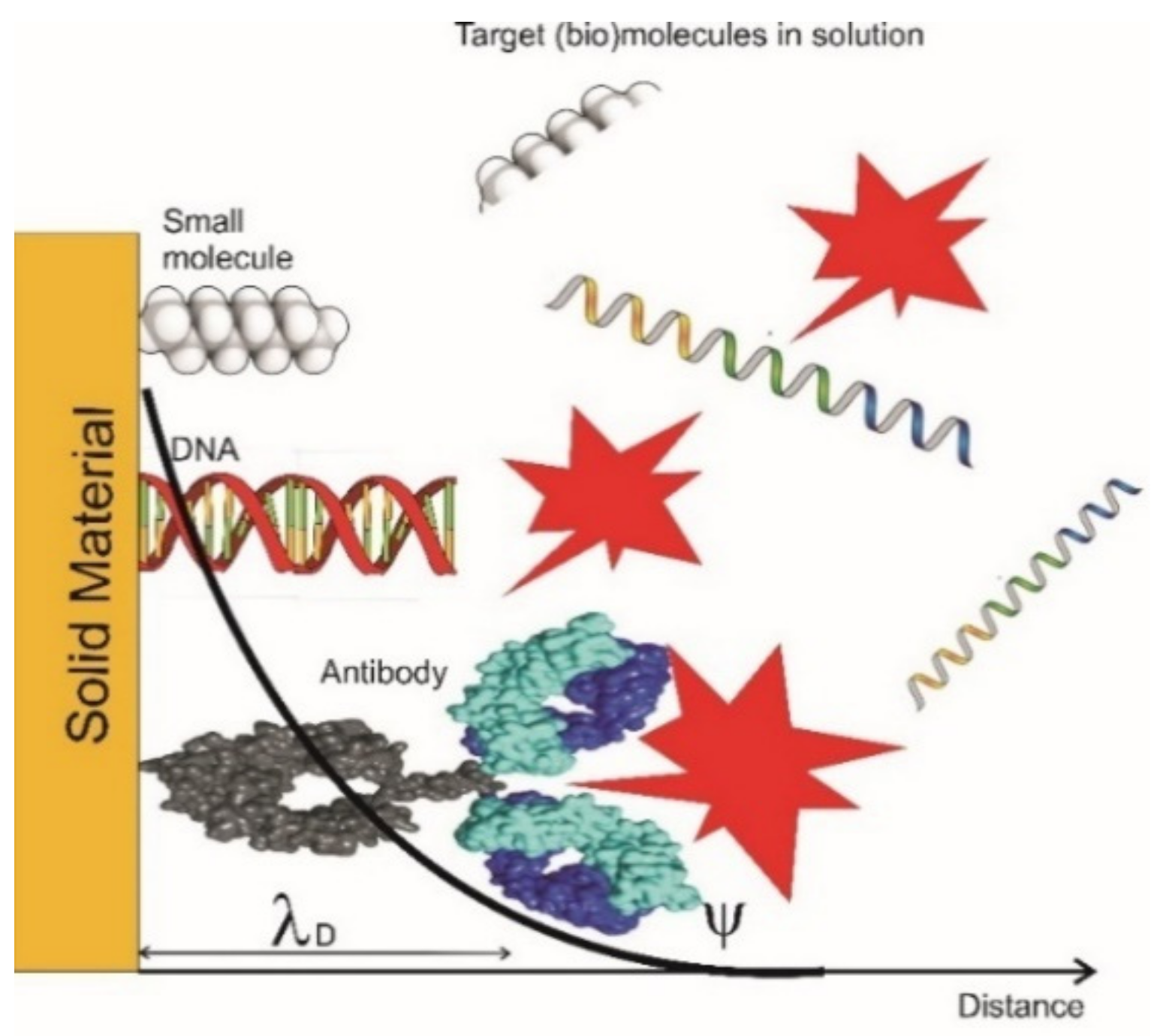
3.1. State-of-the-Art of Single Molecule Detection Technologies
3.2. Future Direction of Research Single Molecule Detection
3.3. State of the Art in Biomarkers
3.4. Future Direction of Research in Biomarkers
3.5. State of the Art in Personalized Medicine and Liquid Biopsy
3.6. Future Direction of Research in Personalized Medicine and Liquid Biopsy
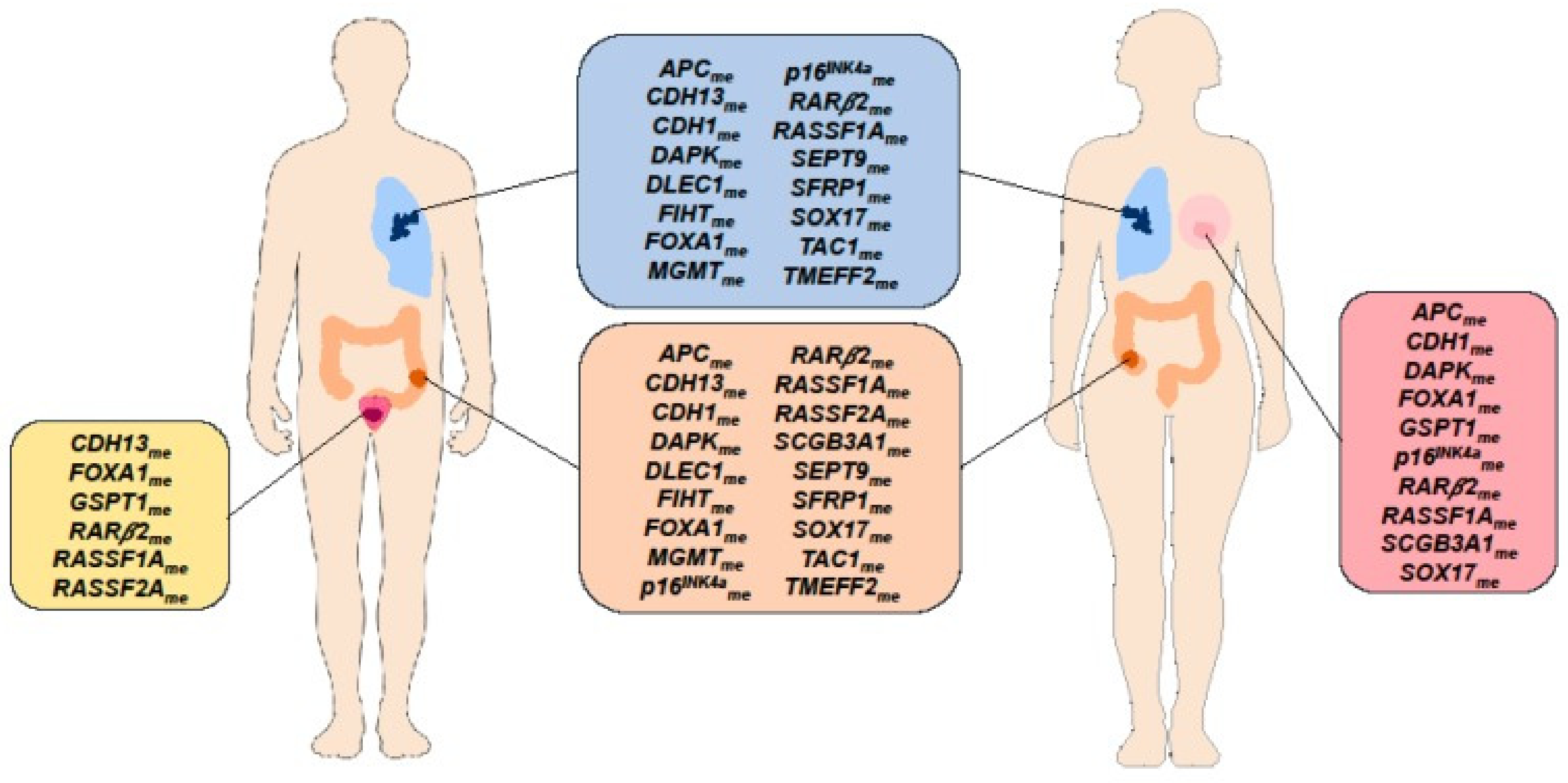
3.7. State of the Art in DNA Methylation Biomarkers for Disease Diagnosis and Therapy Stratification
3.8. Future Direction of Research in DNA Methylation Biomarkers for Disease Diagnosis and Therapy Stratification
3.9. State of the Art in Auto-Antibody (AAb) Biomarkers for Early Diagnosis
3.10. Future Direction of Research in Autoantibody Biomarkers for Early Diagnosis
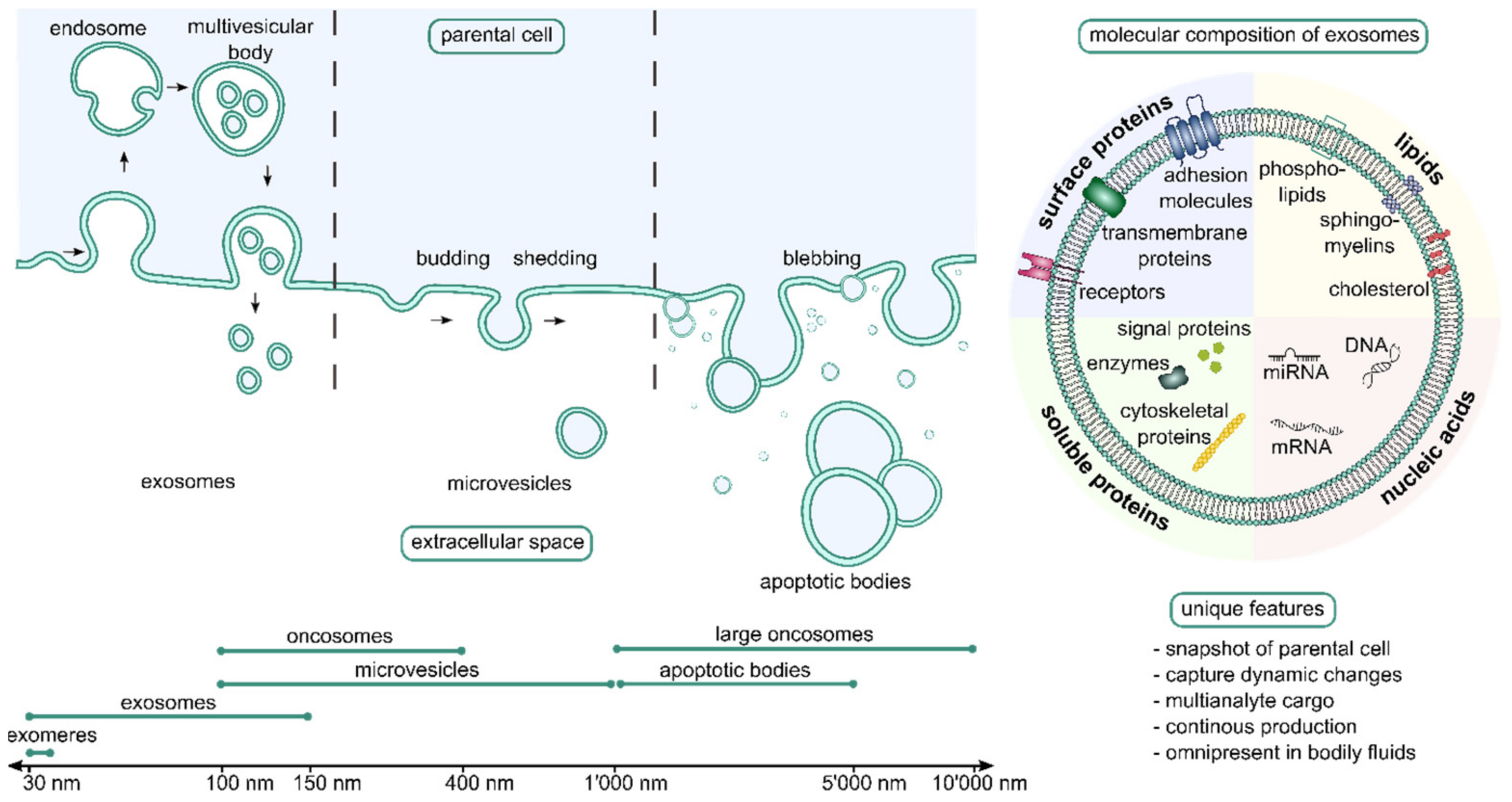
3.11. State of the Art in Extracellular Vesicles/Exosomes
3.12. Future Direction of Research in EVs
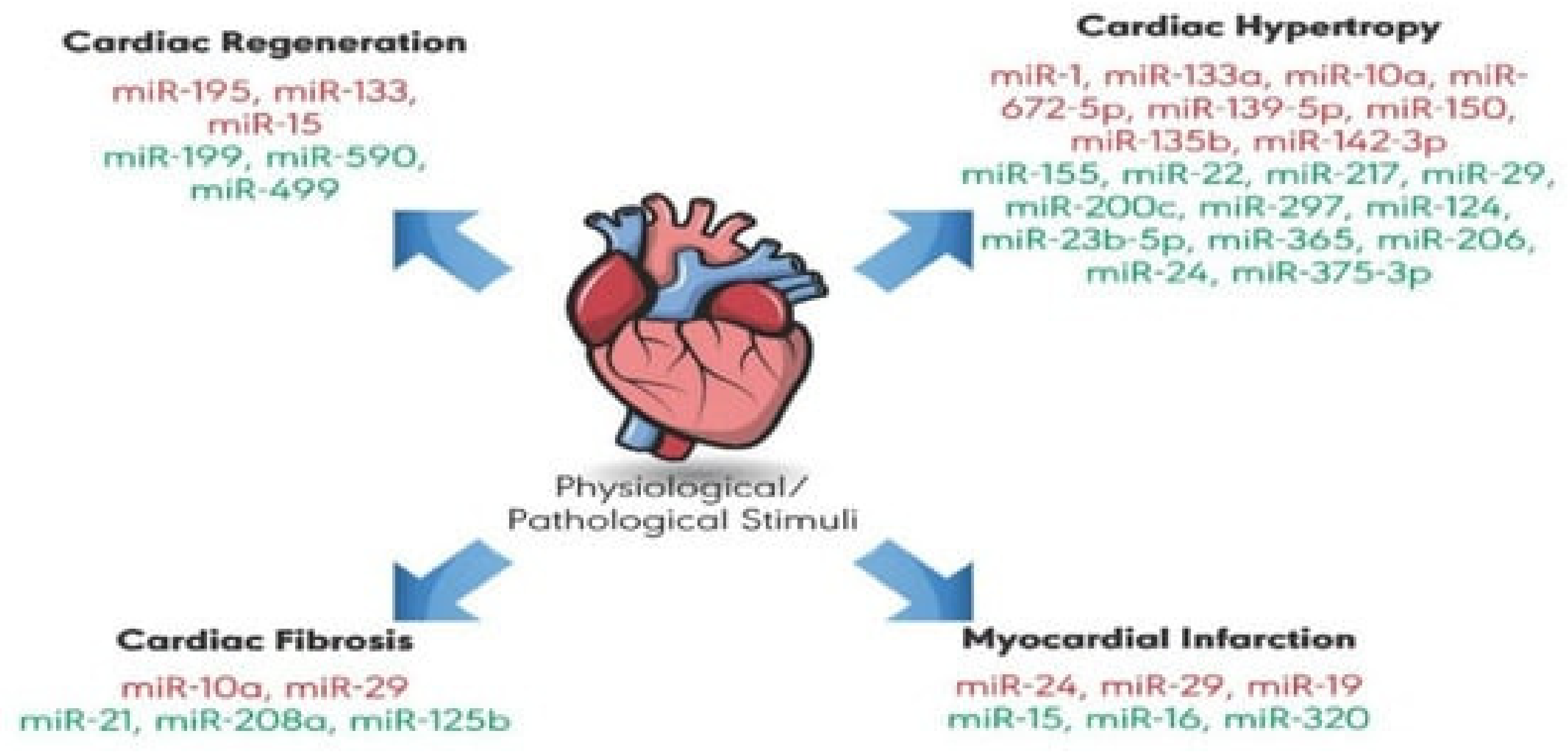
3.13. State of the Art of microRNA (miRNA) Biomarkers
3.14. Future Direction of Research in Small RNA Biomarkers
3.15. State of the Art of Saliva Biopsy
3.16. Future Direction of Research Needs in Saliva Biopsy
3.17. State of the Art in Breath Biopsy
3.18. Future Direction of Research in Breath Biopsy
3.19. State of the Art in Wearable Sweat-Based Diagnostic Textile-Based Sensors
3.20. Future Direction of Research in Textile-Based Sensors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schoepp, N.G.; Schlappi, T.S.; Curtis, M.S.; Butkovich, S.S.; Miller, S.; Humphries, R.M.; Ismagilov, R.F. Rapid Pathogen-Specific Phenotypic Antibiotic Susceptibility Testing Using Digital LAMP Quantification in Clinical Samples. Sci. Transl. Med. 2017, 9, eaal3693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rissin, D.M.; López-Longarela, B.; Pernagallo, S.; Ilyine, H.; Vliegenthart, A.D.B.; Dear, J.W.; Díaz-Mochón, J.J.; Duffy, D.C. Polymerase-Free Measurement of MicroRNA-122 with Single Base Specificity Using Single Molecule Arrays: Detection of Drug-Induced Liver Injury. PLoS ONE 2017, 12, e0179669. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.; Mishyn, V.; Leroux, Y.R.; Butruille, L.; Woitrain, E.; Barras, A.; Aspermair, P.; Happy, H.; Kleber, C.; Boukherroub, R.; et al. Highly Performing Graphene-Based Field Effect Transistor for the Differentiation between Mild-Moderate-Severe Myocardial Injury. Nano Today 2022, 43, 101391. [Google Scholar] [CrossRef]
- Sengupta, J.; Hussain, C.M. Graphene-Based Field-Effect Transistor Biosensors for the Rapid Detection and Analysis of Viruses: A Perspective in View of COVID-19. Carbon Trends 2021, 2, 100011. [Google Scholar] [CrossRef]
- Kwong Hong Tsang, D.; Lieberthal, T.J.; Watts, C.; Dunlop, I.E.; Ramadan, S.; del Rio Hernandez, A.E.; Klein, N. Chemically Functionalised Graphene FET Biosensor for the Label-Free Sensing of Exosomes. Sci. Rep. 2019, 9, 13946. [Google Scholar] [CrossRef] [Green Version]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [Green Version]
- Schuller, J.A.; Barnard, E.S.; Cai, W.; Jun, Y.C.; White, J.S.; Brongersma, M.L. Plasmonics for Extreme Light Concentration and Manipulation. Nat. Mater. 2010, 9, 193–204. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Plasmonics in Biology and Plasmon-Controlled Fluorescence. Plasmonics 2006, 1, 5–33. [Google Scholar] [CrossRef]
- Bauch, M.; Toma, K.; Toma, M.; Zhang, Q.; Dostalek, J. Plasmon-Enhanced Fluorescence Biosensors: A Review. Plasmonics 2014, 9, 781–799. [Google Scholar] [CrossRef] [Green Version]
- Lal, S.; Grady, N.K.; Kundu, J.; Levin, C.S.; Lassiter, J.B.; Halas, N.J. Tailoring Plasmonic Substrates for Surface Enhanced Spectroscopies. Chem. Soc. Rev. 2008, 37, 898. [Google Scholar] [CrossRef]
- Sharma, B.; Frontiera, R.R.; Henry, A.-I.; Ringe, E.; Van Duyne, R.P. SERS: Materials, Applications, and the Future. Mater. Today 2012, 15, 16–25. [Google Scholar] [CrossRef]
- Kinkhabwala, A.; Yu, Z.; Fan, S.; Avlasevich, Y.; Müllen, K.; Moerner, W.E. Large Single-Molecule Fluorescence Enhancements Produced by a Bowtie Nanoantenna. Nat. Photon. 2009, 3, 654–657. [Google Scholar] [CrossRef]
- Höppener, C.; Novotny, L. Exploiting the Light–Metal Interaction for Biomolecular Sensing and Imaging. Quart. Rev. Biophys. 2012, 45, 209–255. [Google Scholar] [CrossRef]
- Masson, J.-F.; Murray-Méthot, M.-P.; Live, L.S. Nanohole Arrays in Chemical Analysis: Manufacturing Methods and Applications. Analyst 2010, 135, 1483. [Google Scholar] [CrossRef]
- Xie, F.; Pang, J.S.; Centeno, A.; Ryan, M.P.; Riley, D.J.; Alford, N.M. Nanoscale Control of Ag Nanostructures for Plasmonic Fluorescence Enhancement of Near-Infrared Dyes. Nano Res. 2013, 6, 496–510. [Google Scholar] [CrossRef]
- Zhang, W.; Ding, F.; Li, W.-D.; Wang, Y.; Hu, J.; Chou, S.Y. Giant and Uniform Fluorescence Enhancement over Large Areas Using Plasmonic Nanodots in 3D Resonant Cavity Nanoantenna by Nanoimprinting. Nanotechnology 2012, 23, 225301. [Google Scholar] [CrossRef]
- Bauch, M.; Dostalek, J. Collective Localized Surface Plasmons for High Performance Fluorescence Biosensing. Opt. Express 2013, 21, 20470. [Google Scholar] [CrossRef]
- Kern, A.M.; Martin, O.J.F. Excitation and Reemission of Molecules near Realistic Plasmonic Nanostructures. Nano Lett. 2011, 11, 482–487. [Google Scholar] [CrossRef] [Green Version]
- Guillot, N.; de la Chapelle, M.L. The Electromagnetic Effect in Surface Enhanced Raman Scattering: Enhancement Optimization Using Precisely Controlled Nanostructures. J. Quant. Spectrosc. Radiat. Transf. 2012, 113, 2321–2333. [Google Scholar] [CrossRef]
- Colas, F.J.; Cottat, M.; Gillibert, R.; Guillot, N.; Djaker, N.; Lidgi-Guigui, N.; Toury, T.; Barchiesi, D.; Toma, A.; Di Fabrizio, E.; et al. Red-Shift Effects in Surface Enhanced Raman Spectroscopy: Spectral or Intensity Dependence of the Near-Field? J. Phys. Chem. C 2016, 120, 13675–13683. [Google Scholar] [CrossRef] [Green Version]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, R.; Breveglieri, G.; Zanoli, L.M.; Borgatti, M.; Spoto, G.; Gambari, R. Direct Detection of Point Mutations in Nonamplified Human Genomic DNA. Anal. Chem. 2011, 83, 8711–8717. [Google Scholar] [CrossRef] [PubMed]
- De la Rica, R.; Stevens, M.M. Plasmonic ELISA for the Ultrasensitive Detection of Disease Biomarkers with the Naked Eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.B.; Zijlstra, P. Single-Molecule Plasmon Sensing: Current Status and Future Prospects. ACS Sens. 2017, 2, 1103–1122. [Google Scholar] [CrossRef] [Green Version]
- Sheehan, P.E.; Whitman, L.J. Detection Limits for Nanoscale Biosensors. Nano Lett. 2005, 5, 803–807. [Google Scholar] [CrossRef]
- Špringer, T.; Krejčík, Z.; Homola, J. Detecting Attomolar Concentrations of MicroRNA Related to Myelodysplastic Syndromes in Blood Plasma Using a Novel Sandwich Assay with Nanoparticle Release. Biosens. Bioelectron. 2021, 194, 113613. [Google Scholar] [CrossRef]
- Riedel, T.; Hageneder, S.; Surman, F.; Pop-Georgievski, O.; Noehammer, C.; Hofner, M.; Brynda, E.; Rodriguez-Emmenegger, C.; Dostálek, J. Plasmonic Hepatitis B Biosensor for the Analysis of Clinical Saliva. Anal. Chem. 2017, 89, 2972–2977. [Google Scholar] [CrossRef]
- Finotti, A.; Allegretti, M.; Gasparello, J.; Giacomini, P.; Spandidos, D.; Spoto, G.; Gambari, R. Liquid Biopsy and PCR-Free Ultrasensitive Detection Systems in Oncology (Review). Int. J. Oncol. 2018, 53, 1395–1434. [Google Scholar] [CrossRef]
- Altug, H.; Oh, S.-H.; Maier, S.A.; Homola, J. Advances and Applications of Nanophotonic Biosensors. Nat. Nanotechnol. 2022, 17, 5–16. [Google Scholar] [CrossRef]
- Shrivastav, A.M.; Cvelbar, U.; Abdulhalim, I. A Comprehensive Review on Plasmonic-Based Biosensors Used in Viral Diagnostics. Commun. Biol. 2021, 4, 70. [Google Scholar] [CrossRef]
- Wang, Y.; Duan, L.; Deng, Z.; Liao, J. Electrically Transduced Gas Sensors Based on Semiconducting Metal Oxide Nanowires. Sensors 2020, 20, 6781. [Google Scholar] [CrossRef]
- Van Hal, R.E.G.; Eijkel, J.C.T.; Bergveld, P. A Novel Description of ISFET Sensitivity with the Buffer Capacity and Double-Layer Capacitance as Key Parameters. Sens. Actuators B Chem. 1995, 24, 201–205. [Google Scholar] [CrossRef] [Green Version]
- Yates, D.E.; Levine, S.; Healy, T.W. Site-Binding Model of the Electrical Double Layer at the Oxide/Water Interface. J. Chem. Soc. Faraday Trans. 1974, 70, 1807. [Google Scholar] [CrossRef]
- Da Silva Rodrigues, B.; Morimoto, N.; Sagazan, O.; Crand, S.; Le Bihan, F.; Mohammed-Brahim, T.; Bonnaud, O. Sensitive Continuous Monitoring of PH Thanks to Matrix of Several Suspended Gate Field Effect Transistors. ECS Trans. 2009, 23, 203–209. [Google Scholar]
- Ingebrandt, S.; Han, Y.; Nakamura, F.; Poghossian, A.; Schöning, M.J.; Offenhäusser, A. Label-Free Detection of Single Nucleotide Polymorphisms Utilizing the Differential Transfer Function of Field-Effect Transistors. Biosens. Bioelectron. 2007, 22, 2834–2840. [Google Scholar] [CrossRef]
- Kamahori, M.; Ishige, Y.; Shimoda, M. DNA Detection by an Extended-Gate FET Sensor with a High-Frequency Voltage Superimposed onto a Reference Electrode. Anal. Sci. 2007, 23, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Zeng, W.; Chen, W.; Xu, L.; Kumar, R.; Umar, A. High Sensitive and Low-Concentration Sulfur Dioxide (SO2) Gas Sensor Application of Heterostructure NiO-ZnO Nanodisks. Sens. Actuators B Chem. 2019, 298, 126870. [Google Scholar] [CrossRef]
- Capone, S.; Zuppa, M.; Presicce, D.S.; Francioso, L.; Casino, F.; Siciliano, P. Metal Oxide Gas Sensor Array for the Detection of Diesel Fuel in Engine Oil. Sens. Actuators B Chem. 2008, 131, 125–133. [Google Scholar] [CrossRef]
- Fleischer, M. Advances in Application Potential of Adsorptive-Type Solid State Gas Sensors: High-Temperature Semiconducting Oxides and Ambient Temperature GasFET Devices. Meas. Sci. Technol. 2008, 19, 042001. [Google Scholar] [CrossRef]
- Korotcenkov, G. Gas Response Control through Structural and Chemical Modification of Metal Oxide Films: State of the Art and Approaches. Sens. Actuators B Chem. 2005, 107, 209–232. [Google Scholar] [CrossRef]
- Korotcenkov, G. Metal Oxides for Solid-State Gas Sensors: What Determines Our Choice? Mater. Sci. Eng. B 2007, 139, 1–23. [Google Scholar] [CrossRef]
- Meixner, H.; Gerblinger, J.; Lampe, U.; Fleischer, M. Thin-Film Gas Sensors Based on Semiconducting Metal Oxides. Sens. Actuators B Chem. 1995, 23, 119–125. [Google Scholar] [CrossRef]
- Ng, K.; Boussaid, F.; Bermak, A. A CMOS Single-Chip Gas Recognition Circuit for Metal Oxide Gas Sensor Arrays. Circuits Syst. I Regul. Pap. IEEE Trans. 2011, 58, 1569–1580. [Google Scholar] [CrossRef]
- Wang, C.; Yin, L.; Zhang, L.; Xiang, D.; Gao, R. Metal Oxide Gas Sensors: Sensitivity and Influencing Factors. Sensors 2010, 10, 2088–2106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartlett, P.N.; Ling-Chung, S.K. Conducting Polymer Gas Sensors Part III: Results for Four Different Polymers and Five Different Vapours. Sens. Actuators 1989, 20, 287–292. [Google Scholar] [CrossRef]
- Han, S.; Huang, W.; Shi, W.; Yu, J. Performance Improvement of Organic Field-Effect Transistor Ammonia Gas Sensor Using ZnO/PMMA Hybrid as Dielectric Layer. Sens. Actuators B Chem. 2014, 203, 9–16. [Google Scholar] [CrossRef]
- Shi, W.; Yu, X.; Zheng, Y.; Yu, J. DNA Based Chemical Sensor for the Detection of Nitrogen Dioxide Enabled by Organic Field-Effect Transistor. Sens. Actuators B Chem. 2016, 222, 1003–1011. [Google Scholar] [CrossRef]
- Huang, W.; Diallo, A.K.; Dailey, J.L.; Besar, K.; Katz, H.E. Electrochemical Processes and Mechanistic Aspects of Field-Effect Sensors for Biomolecules. J. Mater. Chem. C 2015, 3, 6445–6470. [Google Scholar] [CrossRef]
- Liao, L.; Zhang, Z.; Yan, B.; Zheng, Z.; Bao, Q.L.; Wu, T.; Li, C.M.; Shen, Z.X.; Zhang, J.X.; Gong, H.; et al. Multifunctional CuO Nanowire Devices: P-Type Field Effect Transistors and CO Gas Sensors. Nanotechnology 2009, 20, 085203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Liu, Z.; Li, C.; Tang, T.; Liu, X.; Han, S.; Lei, B.; Zhou, C. Detection of NO2 down to Ppb Levels Using Individual and Multiple In2O3 Nanowire Devices. Nano Lett. 2004, 4, 1919–1924. [Google Scholar] [CrossRef]
- McAlpine, M.C.; Ahmad, H.; Wang, D.; Heath, J.R. Highly Ordered Nanowire Arrays on Plastic Substrates for Ultrasensitive Flexible Chemical Sensors. Nat. Mater. 2007, 6, 379–384. [Google Scholar] [CrossRef]
- Mabeck, J.T.; Malliaras, G.G. Chemical and Biological Sensors Based on Organic Thin-Film Transistors. Anal. Bioanal. Chem. 2005, 384, 343–353. [Google Scholar] [CrossRef]
- Ohno, Y.; Maehashi, K.; Matsumoto, K. Chemical and Biological Sensing Applications Based on Graphene Field-Effect Transistors. Biosens. Bioelectron. 2010, 26, 1727–1730. [Google Scholar] [CrossRef]
- Sarkar, D.; Liu, W.; Xie, X.; Anselmo, A.C.; Mitragotri, S.; Banerjee, K. MoS2 Field-Effect Transistor for Next-Generation Label-Free Biosensors. ACS Nano 2014, 8, 3992–4003. [Google Scholar] [CrossRef]
- Patolsky, F.; Lieber, C.M. Nanowire Nanosensors. Mater. Today 2005, 8, 20–28. [Google Scholar] [CrossRef]
- Mishyn, V.; Hugo, A.; Rodrigues, T.; Aspermair, P.; Happy, H.; Marques, L.; Hurot, C.; Othmen, R.; Bouchiat, V.; Boukherroub, R.; et al. The Holy Grail of Pyrene-Based Surface Ligands on the Sensitivity of Graphene-Based Field Effect Transistors. Sens. Diagn. 2022, 1, 235–244. [Google Scholar] [CrossRef]
- Chia, J.S.Y.; Tan, M.T.T.; Khiew, P.S.; Chin, J.K.; Siong, C.W. A Bio-Electrochemical Sensing Platform for Glucose Based on Irreversible, Non-Covalent Pi–Pi Functionalization of Graphene Produced via a Novel, Green Synthesis Method. Sens. Actuators B Chem. 2015, 210, 558–565. [Google Scholar] [CrossRef]
- Streifer, J.A.; Kim, H.; Nichols, B.M.; Hamers, R.J. Covalent Functionalization and Biomolecular Recognition Properties of DNA-Modified Silicon Nanowires. Nanotechnology 2005, 16, 1868–1873. [Google Scholar] [CrossRef]
- Wenga, G.; Jacques, E.; Salaün, A.-C.; Rogel, R.; Pichon, L.; Geneste, F. Step-Gate Polysilicon Nanowires Field Effect Transistor Compatible with CMOS Technology for Label-Free DNA Biosensor. Biosens. Bioelectron. 2013, 40, 141–146. [Google Scholar] [CrossRef] [Green Version]
- Maki, W.C.; Mishra, N.N.; Cameron, E.G.; Filanoski, B.; Rastogi, S.K.; Maki, G.K. Nanowire-Transistor Based Ultra-Sensitive DNA Methylation Detection. Biosens. Bioelectron. 2008, 23, 780–787. [Google Scholar] [CrossRef]
- Singh, S.; Zack, J.; Kumar, D.; Srivastava, S.; Gupta, G.; Saluja, D.; Khan, M.A.; Singh, P. DNA Hybridization on Silicon Nanowires. Thin Solid Films 2010, 519, 1151–1155. [Google Scholar] [CrossRef]
- Zhang, G.-J.; Chua, J.H.; Chee, R.-E.; Agarwal, A.; Wong, S.M.; Buddharaju, K.D.; Balasubramanian, N. Highly Sensitive Measurements of PNA-DNA Hybridization Using Oxide-Etched Silicon Nanowire Biosensors. Biosens. Bioelectron. 2008, 23, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Khamis, S.; Jones, R.; Johnson, A.T.C.; Preti, G.; Kwak, J.; Gelperin, A. DNA-Decorated Carbon Nanotube-Based FETs as Ultrasensitive Chemical Sensors: Discrimination of Homologues, Structural Isomers, and Optical Isomers. AIP Adv. 2012, 2, 022110. [Google Scholar] [CrossRef]
- Kong, T.; Su, R.; Zhang, B.; Zhang, Q.; Cheng, G. CMOS-Compatible, Label-Free Silicon-Nanowire Biosensors to Detect Cardiac Troponin I for Acute Myocardial Infarction Diagnosis. Biosens. Bioelectron. 2012, 34, 267–272. [Google Scholar] [CrossRef]
- Zheng, G.; Patolsky, F.; Cui, Y.; Wang, W.U.; Lieber, C.M. Multiplexed Electrical Detection of Cancer Markers with Nanowire Sensor Arrays. Nat. Biotechnol. 2005, 23, 1294–1301. [Google Scholar] [CrossRef]
- Villamizar, R.A.; Maroto, A.; Rius, F.X.; Inza, I.; Figueras, M.J. Fast Detection of Salmonella Infantis with Carbon Nanotube Field Effect Transistors. Biosens. Bioelectron. 2008, 24, 279–283. [Google Scholar] [CrossRef]
- Mishra, N.N.; Maki, W.C.; Cameron, E.; Nelson, R.; Winterrowd, P.; Rastogi, S.K.; Filanoski, B.; Maki, G.K. Ultra-Sensitive Detection of Bacterial Toxin with Silicon Nanowire Transistor. Lab Chip 2008, 8, 868. [Google Scholar] [CrossRef]
- Patolsky, F.; Zheng, G.; Hayden, O.; Lakadamyali, M.; Zhuang, X.; Lieber, C.M. Electrical Detection of Single Viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 14017–14022. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.-J.; Zhang, L.; Huang, M.; Luo, Z.; Tay, G.; Lim, E.-J.; Kang, T.; Chen, Y. Silicon Nanowire Biosensor for Highly Sensitive and Rapid Detection of Dengue Virus. Sens. Actuators B Chem. 2010, 146, 138–144. [Google Scholar] [CrossRef]
- Juang, D.S.; Lin, C.-H.; Huo, Y.-R.; Tang, C.-Y.; Cheng, C.-R.; Wu, H.-S.; Huang, S.-F.; Kalnitsky, A.; Lin, C.-C. Proton-ELISA: Electrochemical Immunoassay on a Dual-Gated ISFET Array. Biosens. Bioelectron. 2018, 117, 175–182. [Google Scholar] [CrossRef]
- Mulla, Y.; Tuccori, E.; Magliulo, M.; Lattanzi, G.; Palazzo, G.; Persaud, K.; Torsi, L. Capacitance-Modulated Transistor Detects Odorant Binding Protein Chiral Interactions. Nat. Commun. 2015, 6, 6010. [Google Scholar] [CrossRef] [Green Version]
- Schöning, M.; Poghossian, A. Recent Advances in Biologically Sensitive Field-Effect Transistors (BioFETs). Analyst 2002, 127, 1137–1151. [Google Scholar] [CrossRef] [Green Version]
- Hanif, A.; Farooq, R.; Rehman, M.U.; Khan, R.; Majid, S.; Ganaie, M.A. Aptamer Based Nanobiosensors: Promising Healthcare Devices. Saudi Pharm. J. 2019, 27, 312–319. [Google Scholar] [CrossRef]
- Hayasaka, T.; Lin, A.; Copa, V.C.; Lopez, L.P.; Loberternos, R.A.; Ballesteros, L.I.M.; Kubota, Y.; Liu, Y.; Salvador, A.A.; Lin, L. An Electronic Nose Using a Single Graphene FET and Machine Learning for Water, Methanol, and Ethanol. Microsyst. Nanoeng. 2020, 6, 50. [Google Scholar] [CrossRef]
- Leu, M.; Doll, T.; Flietner, B.; Lechner, J.; Eisele, I. Evaluation of Gas Mixtures with Different Sensitive Layers Incorporated in Hybrid FET Structures. Sens. Actuators B Chem. 1994, 19, 678–681. [Google Scholar] [CrossRef]
- Li, L.; Lu, F.; Wang, C.; Zhang, F.; Liang, W.; Kuga, S.; Dong, Z.; Zhao, Y.; Huang, Y.; Wu, M. Flexible Double-Cross-Linked Cellulose-Based Hydrogel and Aerogel Membrane for Supercapacitor Separator. J. Mater. Chem. A 2018, 6, 24468–24478. [Google Scholar] [CrossRef]
- Macchia, E.; Sarcina, L.; Driescher, C.; Gounani, Z.; Tewari, A.; Osterbacka, R.; Palazzo, G.; Tricase, A.; Kovacs Vajna, Z.M.; Viola, F.; et al. Single-Molecule Bioelectronic Label-Free Assay of Both Protein and Genomic Markers of Pancreatic Mucinous Cysts’ in Whole Blood Serum. Adv. Electron. Mater. 2021, 7, 2100304. [Google Scholar] [CrossRef]
- Lee, K.; Yoo, Y.K.; Chae, M.-S.; Hwang, K.S.; Lee, J.; Kim, H.; Hur, D.; Lee, J.H. Highly Selective Reduced Graphene Oxide (RGO) Sensor Based on a Peptide Aptamer Receptor for Detecting Explosives. Sci. Rep. 2019, 9, 10297. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Ogbeide, O.; Macadam, N.; Sun, Q.; Yu, W.; Li, Y.; Su, B.-L.; Hasan, T.; Huang, X.; Huang, W. Printed Gas Sensors. Chem. Soc. Rev. 2020, 49, 1756–1789. [Google Scholar] [CrossRef]
- Scaccabarozzi, A.D.; Basu, A.; Aniés, F.; Liu, J.; Zapata-Arteaga, O.; Warren, R.; Firdaus, Y.; Nugraha, M.I.; Lin, Y.; Campoy-Quiles, M.; et al. Doping Approaches for Organic Semiconductors. Chem. Rev. 2021, 122, 4420–4492. [Google Scholar] [CrossRef]
- Tyagi, P.; Sharma, A.; Tomar, M.; Gupta, V. A Comparative Study of RGO-SnO2 and MWCNT-SnO2 Nanocomposites Based SO2 Gas Sensors. Sens. Actuators B Chem. 2017, 248, 980–986. [Google Scholar] [CrossRef]
- Palimar, S.; Kaushik, S.D.; Siruguri, V.; Swain, D.; Viegas, A.E.; Narayana, C.; Sundaram, N.G. Investigation of Ca Substitution on the Gas Sensing Potential of LaFeO3 Nanoparticles towards Low Concentration SO2 Gas. Dalton Trans. 2016, 45, 13547–13555. [Google Scholar] [CrossRef] [PubMed]
- Septiani, N.L.W.; Saputro, A.G.; Kaneti, Y.V.; Maulana, A.L.; Fathurrahman, F.; Lim, H.; Yuliarto, B.; Nugraha, M.I.; Dipojono, H.K.; Golberg, D.; et al. Hollow Zinc Oxide Microsphere–Multiwalled Carbon Nanotube Composites for Selective Detection of Sulfur Dioxide. ACS Appl. Nano Mater. 2020, 3, 8982–8996. [Google Scholar] [CrossRef]
- Joshi, N.; Hayasaka, T.; Liu, Y.; Liu, H.; Oliveira, O.N.; Lin, L. A Review on Chemiresistive Room Temperature Gas Sensors Based on Metal Oxide Nanostructures, Graphene and 2D Transition Metal Dichalcogenides. Microchim. Acta 2018, 185, 213. [Google Scholar] [CrossRef]
- Han, C.; Li, X.; Liu, Y.; Tang, Y.; Liu, M.; Li, X.; Shao, C.; Ma, J.; Liu, Y. Flexible All-Inorganic Room-Temperature Chemiresistors Based on Fibrous Ceramic Substrate and Visible-Light-Powered Semiconductor Sensing Layer. Adv. Sci. 2021, 8, 2102471. [Google Scholar] [CrossRef]
- Gholami, P.; Rashidi, A.; Khaleghi Abbasabadi, M.; Pourkhalil, M.; Jahangiri, M.; Izadi, N. Synthesis and Characterization of ZnO-Functionalized Multiwall Carbon Nanotubes Nanocomposite as NOx Gas Sensor. Res. Chem. Intermed. 2020, 46, 3911–3927. [Google Scholar] [CrossRef]
- Shetty, S.S.; Jayarama, A.; Bhat, S.; Karunasagar, I.; Pinto, R. A Review on Metal-Oxide Based Trace Ammonia Sensor for Detection of Renal Disease by Exhaled Breath Analysis. Mater. Today Proc. 2022, 55, 113–117. [Google Scholar] [CrossRef]
- Jung, S.; Baik, K.H.; Ren, F.; Pearton, S.J.; Jang, S. AlGaN/GaN Heterostructure Based Schottky Diode Sensors with ZnO Nanorods for Environmental Ammonia Monitoring Applications. ECS J. Solid State Sci. Technol. 2018, 7, Q3020–Q3024. [Google Scholar] [CrossRef]
- Kneer, J.; Knobelspies, S.; Bierer, B.; Wöllenstein, J.; Palzer, S. New Method to Selectively Determine Hydrogen Sulfide Concentrations Using CuO Layers. Sens. Actuators B Chem. 2016, 222, 625–631. [Google Scholar] [CrossRef]
- Hjiri, M.; Lassaad, E.M.; Leonardi, S.G.; Pistone, A.; Mavilia, L.; Neri, G. Al-Doped ZnO for Highly Sensitive CO Gas Sensors. Sens. Actuators B Chem. 2014, 196, 413–420. [Google Scholar] [CrossRef]
- Koralli, P.; Petropoulou, G.; Mouzakis, D.; Mousdis, G.; Kompitsas, M. Efficient CO Sensing by a CuO: Au Nanocomposite Thin Film Deposited by PLD on a Pyrex Tube. Sens. Actuators A Phys. 2021, 332, 113120. [Google Scholar] [CrossRef]
- Xiao, T.; Wang, X.-Y.; Zhao, Z.-H.; Li, L.; Zhang, L.; Yao, H.-C.; Wang, J.-S.; Li, Z.-J. Highly Sensitive and Selective Acetone Sensor Based on C-Doped WO3 for Potential Diagnosis of Diabetes Mellitus. Sens. Actuators B Chem. 2014, 199, 210–219. [Google Scholar] [CrossRef]
- Lee, J.E.; Lim, C.K.; Park, H.J.; Song, H.; Choi, S.-Y.; Lee, D.-S. ZnO–CuO Core-Hollow Cube Nanostructures for Highly Sensitive Acetone Gas Sensors at the Ppb Level. ACS Appl. Mater. Interfaces 2020, 12, 35688–35697. [Google Scholar] [CrossRef] [PubMed]
- Nakate, U.T.; Patil, P.; Nakate, Y.T.; Na, S.-I.; Yu, Y.; Hahn, Y.-B. Ultrathin Ternary Metal Oxide Bi2MoO6 Nanosheets for High Performance Asymmetric Supercapacitor and Gas Sensor Applications. Appl. Surf. Sci. 2021, 551, 149422. [Google Scholar] [CrossRef]
- Rafiee, Z.; Roshan, H.; Sheikhi, M.H. Low Concentration Ethanol Sensor Based on Graphene/ZnO Nanowires. Ceram. Int. 2021, 47, 5311–5317. [Google Scholar] [CrossRef]
- Lou, C.; Lei, G.; Liu, X.; Xie, J.; Li, Z.; Zheng, W.; Goel, N.; Kumar, M.; Zhang, J. Design and Optimization Strategies of Metal Oxide Semiconductor Nanostructures for Advanced Formaldehyde Sensors. Coord. Chem. Rev. 2022, 452, 214280. [Google Scholar] [CrossRef]
- Jo, Y.K.; Jeong, S.-Y.; Moon, Y.K.; Jo, Y.-M.; Yoon, J.-W.; Lee, J.-H. Exclusive and Ultrasensitive Detection of Formaldehyde at Room Temperature Using a Flexible and Monolithic Chemiresistive Sensor. Nat. Commun. 2021, 12, 4955. [Google Scholar] [CrossRef]
- Moon, Y.K.; Jeong, S.-Y.; Jo, Y.-M.; Jo, Y.K.; Kang, Y.C.; Lee, J.-H. Highly Selective Detection of Benzene and Discrimination of Volatile Aromatic Compounds Using Oxide Chemiresistors with Tunable Rh-TiO2 Catalytic Overlayers. Adv. Sci. 2021, 8, 2004078. [Google Scholar] [CrossRef]
- Park, Y.; Yoo, R.; Ryull Park, S.; Lee, J.H.; Jung, H.; Lee, H.-S.; Lee, W. Highly Sensitive and Selective Isoprene Sensing Performance of ZnO Quantum Dots for a Breath Analyzer. Sens. Actuators B Chem. 2019, 290, 258–266. [Google Scholar] [CrossRef]
- Herrera-Rivera, R.; Olvera, M.; Maldonado, A. Propane Sensor Pellets Based on Nanopowders Cu-Doped ZnO. In Proceedings of the 2017 14th International Conference on Electrical Engineering, Computing Science and Automatic Control (CCE), Mexico City, Mexico, 20–22 October 2017; p. 5. [Google Scholar]
- Kim, J.-H.; Lee, J.-H.; Park, Y.; Kim, J.-Y.; Mirzaei, A.; Kim, H.W.; Kim, S.S. Toluene-and Benzene-Selective Gas Sensors Based on Pt-and Pd-Functionalized ZnO Nanowires in Self-Heating Mode. Sens. Actuators B Chem. 2019, 294, 78–88. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Qin, L.; Liu, D.; Liu, Y.; Liu, F.; Song, H.; Wang, Y.; Lu, G. Preparation of Au-Loaded TiO2 Pecan-Kernel-like and Its Enhanced Toluene Sensing Performance. Sens. Actuators B Chem. 2018, 255, 2240–2247. [Google Scholar] [CrossRef]
- Gao, H.; Wei, D.; Lin, P.; Liu, C.; Sun, P.; Shimanoe, K.; Yamazoe, N.; Lu, G. The Design of Excellent Xylene Gas Sensor Using Sn-Doped NiO Hierarchical Nanostructure. Sens. Actuators B Chem. 2017, 253, 1152–1162. [Google Scholar] [CrossRef]
- Moon, Y.K.; Jeong, S.-Y.; Kang, Y.C.; Lee, J.-H. Metal Oxide Gas Sensors with Au Nanocluster Catalytic Overlayer: Toward Tuning Gas Selectivity and Response Using a Novel Bilayer Sensor Design. ACS Appl. Mater. Interfaces 2019, 11, 32169–32177. [Google Scholar] [CrossRef]
- Wu, H.; Zhou, Y.; Guo, J.; Zhao, L.; Wang, T.; Yan, X.; Wang, C.; Liu, F.; Sun, P.; Lu, G. Highly Sensitive and Selective Xylene Sensor Based on Pp Heterojunctions Composites Derived from Off-Stoichiometric Cobalt Tungstate. Sens. Actuators B Chem. 2022, 351, 130973. [Google Scholar] [CrossRef]
- Saito, N.; Haneda, H.; Watanabe, K.; Shimanoe, K.; Sakaguchi, I. Highly Sensitive Isoprene Gas Sensor Using Au-Loaded Pyramid-Shaped ZnO Particles. Sens. Actuators B Chem. 2021, 326, 128999. [Google Scholar] [CrossRef]
- Poloju, M.; Jayababu, N.; Rao, E.V.; Rao, R.G.; Reddy, M.R. Development of CdO/ZnO Nanocomposites for the Rapid Detection and Discrimination of n-Butanol. Surf. Interfaces 2020, 20, 100586. [Google Scholar] [CrossRef]
- Srinivasan, P.; Mehtre, S. Zinc Oxide Nanoparticles from Coriandrum Sativum as Sensor for Detection of N-Butanol and Nitric Oxide Gas. Mater. Today Proc. 2022, 51, 1760–1764. [Google Scholar] [CrossRef]
- Wahab, R.; Khan, F.; Ahmad, N.; Alam, M. Molybdenum Rods Assembled with Nanosheets: A High Catalytic Material for Phenol Sensing. Mater. Today Chem. 2020, 18, 100347. [Google Scholar] [CrossRef]
- Rahman, M.M.; Balkhoyor, H.B.; Asiri, A.M. Phenolic Sensor Development Based on Chromium Oxide-Decorated Carbon Nanotubes for Environmental Safety. J. Environ. Manag. 2017, 188, 228–237. [Google Scholar] [CrossRef]
- Maccato, C.; Bigiani, L.; Carraro, G.; Gasparotto, A.; Sada, C.; Comini, E.; Barreca, D. Toward the Detection of Poisonous Chemicals and Warfare Agents by Functional Mn3O4 Nanosystems. ACS Appl. Mater. Interfaces 2018, 10, 12305–12310. [Google Scholar] [CrossRef]
- Xu, X.; Arab Pour Yazdi, M.; Sanchez, J.-B.; Billard, A.; Berger, F.; Martin, N. Exploiting the Dodecane and Ozone Sensing Capabilities of Nanostructured Tungsten Oxide Films. Sens. Actuators B Chem. 2018, 266, 773–783. [Google Scholar] [CrossRef]
- Xu, X.; Yazdi, M.A.P.; Sanchez, J.-B.; Billard, A.; Berger, F.; Martin, N. Reactive Co-Sputtering of Tungsten Oxide Thin Films by Glancing Angle Deposition for Gas Sensors. Mater. Today Proc. 2019, 6, 314–318. [Google Scholar] [CrossRef]
- Moon, H.G.; Jung, Y.; Han, S.D.; Shim, Y.-S.; Jung, W.-S.; Lee, T.; Lee, S.; Park, J.H.; Baek, S.-H.; Kim, J.-S.; et al. All Villi-like Metal Oxide Nanostructures-Based Chemiresistive Electronic Nose for an Exhaled Breath Analyzer. Sens. Actuators B Chem. 2018, 257, 295–302. [Google Scholar] [CrossRef]
- Han, J.-K.; Kang, M.; Jeong, J.; Cho, I.; Yu, J.-M.; Yoon, K.-J.; Park, I.; Choi, Y.-K. Artificial Olfactory Neuron for an In-Sensor Neuromorphic Nose. Adv. Sci. 2022, 15, 2106017. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Wang, Y.; Zhang, C.; Pittman, Z.A.; Oliveira, A.M.; Fu, K.; Zhao, J.; Srivastava, R.; Willis, B.G. Classification of Tea Aromas Using Multi-Nanoparticle Based Chemiresistor Arrays. Sensors 2019, 19, 2547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bano, K.; Bajwa, S.Z.; Bassous, N.J.; Webster, T.J.; Shaheen, A.; Taj, A.; Hameed, S.; Tehseen, B.; Dai, Z.; Iqbal, M.Z.; et al. Development of Biocompatible 1D CuO Nanoneedles and Their Potential for Sensitive, Mass-Based Detection of Anti-Tuberculosis Drugs. Appl. Nanosci. 2019, 9, 1341–1351. [Google Scholar] [CrossRef]
- Alavi-Tabari, S.A.R.; Khalilzadeh, M.A.; Karimi-Maleh, H. Simultaneous Determination of Doxorubicin and Dasatinib as Two Breast Anticancer Drugs Uses an Amplified Sensor with Ionic Liquid and ZnO Nanoparticle. J. Electroanal. Chem. 2018, 811, 84–88. [Google Scholar] [CrossRef]
- Ansari, S.; Ansari, M.S.; Satsangee, S.P.; Jain, R. WO3 Decorated Graphene Nanocomposite Based Electrochemical Sensor: A Prospect for the Detection of Anti-Anginal Drug. Anal. Chim. Acta 2019, 1046, 99–109. [Google Scholar] [CrossRef]
- Gupta, V.; Karimi-Maleh, H.; Agarwal, S.; Karimi, F.; Bijad, M.; Farsi, M.; Shahidi, S.-A. Fabrication of a Food Nano-Platform Sensor for Determination of Vanillin in Food Samples. Sensors 2018, 18, 2817. [Google Scholar] [CrossRef] [Green Version]
- Gobbi, E.; Falasconi, M.; Zambotti, G.; Sberveglieri, V.; Pulvirenti, A.; Sberveglieri, G. Rapid Diagnosis of Enterobacteriaceae in Vegetable Soups by a Metal Oxide Sensor Based Electronic Nose. Sens. Actuators B Chem. 2015, 207, 1104–1113. [Google Scholar] [CrossRef]
- Konduru, T.; Rains, G.; Li, C. A Customized Metal Oxide Semiconductor-Based Gas Sensor Array for Onion Quality Evaluation: System Development and Characterization. Sensors 2015, 15, 1252–1273. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Wei, Z.; Li, Y.; Qu, J.; Zu, B.; Dou, X. Highly Sensitive and Rapid Chemiresistive Sensor towards Trace Nitro-Explosive Vapors Based on Oxygen Vacancy-Rich and Defective Crystallized In-Doped ZnO. Sens. Actuators B Chem. 2017, 244, 983–991. [Google Scholar] [CrossRef]
- Sowmya, B.; John, A.; Panda, P. A Review on Metal-Oxide Based Pn and Nn Heterostructured Nano-Materials for Gas Sensing Applications. Sens. Int. 2021, 2, 100085. [Google Scholar]
- Knoblauch, J.; Illyaskutty, N.; Kohler, H. Early Detection of Fires in Electrical Installations by Thermally Modulated SnO2/Additive-Multi Sensor Arrays. Sens. Actuators B Chem. 2015, 217, 36–40. [Google Scholar] [CrossRef]
- Khairy, M.; Ayoub, H.A.; Banks, C.E. Non-Enzymatic Electrochemical Platform for Parathion Pesticide Sensing Based on Nanometer-Sized Nickel Oxide Modified Screen-Printed Electrodes. Food Chem. 2018, 255, 104–111. [Google Scholar] [CrossRef]
- Khan, N.; Athar, T.; Fouad, H.; Umar, A.; Ansari, Z.A.; Ansari, S.G. Application of Pristine and Doped SnO2 Nanoparticles as a Matrix for Agro-Hazardous Material (Organophosphate) Detection. Sci. Rep. 2017, 7, 42510. [Google Scholar] [CrossRef] [Green Version]
- Ulloa, A.M.; Glassmaker, N.; Oduncu, M.R.; Xu, P.; Wei, A.; Cakmak, M.; Stanciu, L. Roll-to-Roll Manufactured Sensors for Nitroaromatic Organophosphorus Pesticides Detection. ACS Appl. Mater. Interfaces 2021, 13, 35961–35971. [Google Scholar] [CrossRef]
- Lange, U.; Mirsky, V.M. Chemiresistors Based on Conducting Polymers: A Review on Measurement Techniques. Anal. Chim. Acta 2011, 687, 105–113. [Google Scholar] [CrossRef]
- Yang, S.; Bintinger, J.; Park, S.; Jain, S.; Alexandrou, K.; Fruhmann, P.; Besar, K.; Katz, H.; Kymissis, I. Inexpensive, Versatile, and Robust USB-Driven Sensor Platform. IEEE Sens. Lett. 2017, 1, 5000304. [Google Scholar] [CrossRef]
- Shirakawa, H. Nobel Lecture: The Discovery of Polyacetylene Film—The Dawning of an Era of Conducting Polymers. Rev. Mod. Phys. 2001, 73, 713–718. [Google Scholar] [CrossRef] [Green Version]
- Malinauskas, M.; Žukauskas, A.; Hasegawa, S.; Hayasaki, Y.; Mizeikis, V.; Buividas, R.; Juodkazis, S. Ultrafast Laser Processing of Materials: From Science to Industry. Light Sci. Appl. 2016, 5, e16133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.-Q.; Liu, Y.; Zhong, D.; Nikzad, S.; Liu, S.; Yu, Z.; Liu, D.; Wu, H.-C.; Zhu, C.; Li, J.; et al. Monolithic Optical Microlithography of High-Density Elastic Circuits. Science 2021, 373, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Shahid, N.; Rappon, T.; Berta, W. Applications of Artificial Neural Networks in Health Care Organizational Decision-Making: A Scoping Review. PLoS ONE 2019, 14, e0212356. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, Y.; Kim, T.; Eom, T.H.; Kim, S.Y.; Jang, H.W. Chemoresistive Materials for Electronic Nose: Progress, Perspectives, and Challenges. InfoMat 2019, 1, 289–316. [Google Scholar] [CrossRef] [Green Version]
- Kwak, D.; Lei, Y.; Maric, R. Ammonia Gas Sensors: A Comprehensive Review. Talanta 2019, 204, 713–730. [Google Scholar] [CrossRef]
- Duc, C.; Boukhenane, M.L.; Fagniez, T.; Khouchaf, L.; Redon, N.; Wojkiewicz, J.-L. Conductive Polymer Composites for Hydrogen Sulphide Sensors Working at Sub-PPM Level and Room Temperature. Sensors 2021, 21, 6529. [Google Scholar] [CrossRef]
- Phillips, B.; Banerjee, S.; Tu, X.; Fang, L. Electrical Vapour Sensing with Macrocyclic Molecular Receptors. Supramol. Chem. 2020, 32, 165–177. [Google Scholar] [CrossRef]
- Park, H.; Kim, D.-H.; Ma, B.S.; Shin, E.; Kim, Y.; Kim, T.-S.; Kim, F.S.; Kim, I.-D.; Kim, B.J. High-Performance, Flexible NO2 Chemiresistors Achieved by Design of Imine-Incorporated n-Type Conjugated Polymers. Adv. Sci. 2022, 9, 2200270. [Google Scholar] [CrossRef]
- Amini, A.; Jafari, A.; Vafaei, M.; Mahmoodian, M. Stability Enhancement of Polypyrrole Thin Film Ammonia Sensor by Camphor Sulfonic Acid Dopant. J. Mater. Sci. Mater. Electron. 2022, 33, 1293–1306. [Google Scholar] [CrossRef]
- Tarek, M.; Khozemy, E.; Ghobashy, M. Radiation Synthesis of Gas Sensor Based on Polyaniline Nanoflake-Poly Vinyl Alcohol Film for Four Hazardous Gases (NH3, CO2, H2S and Phenol). Arab. J. Nucl. Sci. Appl. 2020, 53, 210–221. [Google Scholar]
- Danesh, E.; Ghaffarian, S.R.; Molla-Abbasi, P. Non-Solvent Induced Phase Separation as a Method for Making High-Performance Chemiresistors Based on Conductive Polymer Nanocomposites. Sens. Actuators B Chem. 2011, 155, 562–567. [Google Scholar] [CrossRef]
- Zamiri, G.; Haseeb, A.S.M.A. Recent Trends and Developments in Graphene/Conducting Polymer Nanocomposites Chemiresistive Sensors. Materials 2020, 13, 3311. [Google Scholar] [CrossRef]
- Yan, Y.; Yang, G.; Xu, J.-L.; Zhang, M.; Kuo, C.-C.; Wang, S.-D. Conducting Polymer-Inorganic Nanocomposite-Based Gas Sensors: A Review. Sci. Technol. Adv. Mater. 2020, 21, 768–786. [Google Scholar] [CrossRef]
- Chang, L.-Y.; Chuang, M.-Y.; Zan, H.-W.; Meng, H.-F.; Lu, C.-J.; Yeh, P.-H.; Chen, J.-N. One-Minute Fish Freshness Evaluation by Testing the Volatile Amine Gas with an Ultrasensitive Porous-Electrode-Capped Organic Gas Sensor System. ACS Sens. 2017, 2, 531–539. [Google Scholar] [CrossRef]
- Ly, T.N.; Park, S. Highly Sensitive Ammonia Sensor for Diagnostic Purpose Using Reduced Graphene Oxide and Conductive Polymer. Sci. Rep. 2018, 8, 18030. [Google Scholar] [CrossRef]
- Jisha, P.; Suma, M. Synthesis and Electrical Characterization of Protonic Acid Doped Polyaniline for Detection of Monoterpene Vapours to Diagnose Malaria. In Proceedings of the 2019 International Conference on Data Science and Communication (IconDSC), Bangalore, India, 1–2 March 2019; pp. 1–5. [Google Scholar]
- Pirovano, P.; Dorrian, M.; Shinde, A.; Donohoe, A.; Brady, A.J.; Moyna, N.M.; Wallace, G.; Diamond, D.; McCaul, M. A Wearable Sensor for the Detection of Sodium and Potassium in Human Sweat during Exercise. Talanta 2020, 219, 121145. [Google Scholar] [CrossRef]
- Wilson, A.D.; Lester, D.G.; Oberle, C.S. Development of Conductive Polymer Analysis for the Rapid Detection and Identification of Phytopathogenic Microbes. Phytopathology 2004, 94, 419–431. [Google Scholar] [CrossRef]
- Matindoust, S.; Farzi, G.; Nejad, M.B.; Shahrokhabadi, M.H. Polymer-Based Gas Sensors to Detect Meat Spoilage: A Review. React. Funct. Polym. 2021, 165, 104962. [Google Scholar] [CrossRef]
- Samanta, S.; Roy, P.; Kar, P. Sensing of Ethanol and Other Alcohol Contaminated Ethanol by Conducting Functional Poly (o-Phenylenediamine). Mater. Sci. Eng. B 2020, 256, 114541. [Google Scholar] [CrossRef]
- Supraja, P.; Tripathy, S.; Singh, R.; Singh, V.; Chaudhury, G.; Singh, S.G. Towards Point-of-Care Diagnosis of Alzheimer’s Disease: Multi-Analyte Based Portable Chemiresistive Platform for Simultaneous Detection of β-Amyloid (1–40) and (1–42) in Plasma. Biosens. Bioelectron. 2021, 186, 113294. [Google Scholar] [CrossRef]
- Wang, M.-H.; Ji, B.-W.; Gu, X.-W.; Tian, H.-C.; Kang, X.-Y.; Yang, B.; Wang, X.-L.; Chen, X.; Li, C.-Y.; Liu, J.-Q. Direct Electrodeposition of Graphene Enhanced Conductive Polymer on Microelectrode for Biosensing Application. Biosens. Bioelectron. 2018, 99, 99–107. [Google Scholar] [CrossRef]
- Ma, Z.; Shi, W.; Yan, K.; Pan, L.; Yu, G. Doping Engineering of Conductive Polymer Hydrogels and Their Application in Advanced Sensor Technologies. Chem. Sci. 2019, 10, 6232–6244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Li, X.; Sui, G.; Du, R.; Zhang, Q.; Fu, Q. Plasma Modification of PU Foam for Piezoresistive Sensor with High Sensitivity, Mechanical Properties and Long-Term Stability. Chem. Eng. J. 2020, 381, 122666. [Google Scholar] [CrossRef]
- Khan, Z.U.; Edberg, J.; Hamedi, M.M.; Gabrielsson, R.; Granberg, H.; Wågberg, L.; Engquist, I.; Berggren, M.; Crispin, X. Thermoelectric Polymers and Their Elastic Aerogels. Adv. Mater. 2016, 28, 4556–4562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, C.-Y.; Zhang, Z. A Novel Gas Sensor Signal Drift Adjustment Method Based on Controlled Measurement. In Proceedings of the 2018 China Semiconductor Technology International Conference (CSTIC), Shanghai, China, 11–12 March 2018; pp. 1–5. [Google Scholar]
- John, A.T.; Murugappan, K.; Nisbet, D.R.; Tricoli, A. An Outlook of Recent Advances in Chemiresistive Sensor-Based Electronic Nose Systems for Food Quality and Environmental Monitoring. Sensors 2021, 21, 2271. [Google Scholar] [CrossRef] [PubMed]
- Amrani, M.E.H.; Dowdeswell, R.M.; Payne, P.A.; Persaud, K.C. An Intelligent Gas Sensing System. Sens. Actuators B Chem. 1997, 44, 512–516. [Google Scholar] [CrossRef]
- Potyrailo, R.A. Toward High Value Sensing: Monolayer-Protected Metal Nanoparticles in Multivariable Gas and Vapor Sensors. Chem. Soc. Rev. 2017, 46, 5311–5346. [Google Scholar] [CrossRef] [PubMed]
- Potyrailo, R.A. Multivariable Sensors for Ubiquitous Monitoring of Gases in the Era of Internet of Things and Industrial Internet. Chem. Rev. 2016, 116, 11877–11923. [Google Scholar] [CrossRef]
- Gruber, J.; Nascimento, H.M.; Yamauchi, E.Y.; Li, R.W.C.; Esteves, C.H.A.; Rehder, G.P.; Gaylarde, C.C.; Shirakawa, M.A. A Conductive Polymer Based Electronic Nose for Early Detection of Penicillium Digitatum in Post-Harvest Oranges. Mater. Sci. Eng. C 2013, 33, 2766–2769. [Google Scholar] [CrossRef]
- Cordeiro, J.R.; Li, R.W.C.; Takahashi, É.S.; Rehder, G.P.; Ceccantini, G.; Gruber, J. Wood Identification by a Portable Low-Cost Polymer-Based Electronic Nose. RSC Adv. 2016, 6, 109945–109949. [Google Scholar] [CrossRef]
- Hsieh, Y.-C.; Yao, D.-J. Intelligent Gas-Sensing Systems and Their Applications. J. Micromech. Microeng. 2018, 28, 093001. [Google Scholar] [CrossRef]
- Chandler, R.; Das, A.; Gibson, T.; Dutta, R. Detection of Oil Pollution in Seawater: Biosecurity Prevention Using Electronic Nose Technology. In Proceedings of the 2015 31st IEEE International Conference on Data Engineering Workshops, Seoul, Korea, 13–17 April 2015; pp. 98–100. [Google Scholar]
- Yang, Y.; Liu, H.; Gu, Y. A Model Transfer Learning Framework with Back-Propagation Neural Network for Wine and Chinese Liquor Detection by Electronic Nose. IEEE Access 2020, 8, 105278–105285. [Google Scholar] [CrossRef]
- Tiggemann, L.; Ballen, S.; Bocalon, C.; Graboski, A.M.; Manzoli, A.; de Paula Herrmann, P.S.; Steffens, J.; Valduga, E.; Steffens, C. Low-Cost Gas Sensors with Polyaniline Film for Aroma Detection. J. Food Eng. 2016, 180, 16–21. [Google Scholar] [CrossRef]
- Tan, C.; Xie, D.; Liu, Y.; Peng, W.; Li, X.; Ai, L.; Wu, C.; Wen, C.; Huang, X.; Guo, J. Identification of Different Bile Species and Fermentation Times of Bile Arisaema Based on an Intelligent Electronic Nose and Least Squares Support Vector Machine. Anal. Chem. 2018, 90, 3460–3466. [Google Scholar] [CrossRef]
- Julian, T.; Hidayat, S.N.; Rianjanu, A.; Dharmawan, A.B.; Wasisto, H.S.; Triyana, K. Intelligent Mobile Electronic Nose System Comprising a Hybrid Polymer-Functionalized Quartz Crystal Microbalance Sensor Array. ACS Omega 2020, 5, 29492–29503. [Google Scholar] [CrossRef]
- Buck, L.; Axel, R. A Novel Multigene Family May Encode Odorant Receptors: A Molecular Basis for Odor Recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Schwarze, M.; Tietze, M.L.; Ortmann, F.; Kleemann, H.; Leo, K. Universal Limit for Air-Stable Molecular n-Doping in Organic Semiconductors. ACS Appl. Mater. Interfaces 2020, 12, 40566–40571. [Google Scholar] [CrossRef]
- Xu, Y.; Sun, H.; Liu, A.; Zhu, H.-H.; Li, W.; Lin, Y.-F.; Noh, Y.-Y. Doping: A Key Enabler for Organic Transistors. Adv. Mater. 2018, 30, 1801830. [Google Scholar] [CrossRef]
- Salzmann, I.; Heimel, G.; Oehzelt, M.; Winkler, S.; Koch, N. Molecular Electrical Doping of Organic Semiconductors: Fundamental Mechanisms and Emerging Dopant Design Rules. Acc. Chem. Res. 2016, 49, 370–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, N.; Bae, J. Biosensors—Publication Trends and Knowledge Domain Visualization. Sensors 2019, 19, 2615. [Google Scholar] [CrossRef] [Green Version]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-Molecule Enzyme-Linked Immunosorbent Assay Detects Serum Proteins at Subfemtomolar Concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Marras, S.A.E. Efficiencies of Fluorescence Resonance Energy Transfer and Contact-Mediated Quenching in Oligonucleotide Probes. Nucleic Acids Res. 2002, 30, 122e. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asanuma, D.; Sakabe, M.; Kamiya, M.; Yamamoto, K.; Hiratake, J.; Ogawa, M.; Kosaka, N.; Choyke, P.L.; Nagano, T.; Kobayashi, H.; et al. Sensitive β-Galactosidase-Targeting Fluorescence Probe for Visualizing Small Peritoneal Metastatic Tumours In Vivo. Nat. Commun. 2015, 6, 6463. [Google Scholar] [CrossRef] [Green Version]
- Tricase, A.; Imbriano, A.; Macchia, E.; Sarcina, L.; Scandurra, C.; Torricelli, F.; Cioffi, N.; Torsi, L.; Bollella, P. Enzyme Based Amperometric Wide Field Biosensors: Is Single-Molecule Detection Possible? Electrochem. Sci. Adv. 2022, e2100215. [Google Scholar] [CrossRef]
- Macchia, E.; Torricelli, F.; Bollella, P.; Sarcina, L.; Tricase, A.; Di Franco, C.; Österbacka, R.; Kovács-Vajna, Z.M.; Scamarcio, G.; Torsi, L. Large-Area Interfaces for Single-Molecule Label-Free Bioelectronic Detection. Chem. Rev. 2022, 122, 4636–4699. [Google Scholar] [CrossRef]
- Macchia, E.; Tiwari, A.; Manoli, K.; Holzer, B.; Ditaranto, N.; Picca, R.A.; Cioffi, N.; Di Franco, C.; Scamarcio, G.; Palazzo, G.; et al. Label-Free and Selective Single-Molecule Bioelectronic Sensing with a Millimeter-Wide Self-Assembled Monolayer of Anti-Immunoglobulins. Chem. Mater. 2019, 31, 6476–6483. [Google Scholar] [CrossRef]
- Macchia, E.; Manoli, K.; Holzer, B.; Franco, C.D.; Ghittorelli, M.; Torricelli, F.; Alberga, D.; Mangiatordi, G.F.; Palazzo, G.; Scamarcio, G.; et al. Single-Molecule Detection with a Millimetre-Sized Transistor. Nat. Commun. 2018, 9, 3223. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium; Whitehead Institute for Biomedical Research Center for Genome Research; Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [Green Version]
- De Wit, S.; van Dalum, G.; Lenferink, A.T.M.; Tibbe, A.G.J.; Hiltermann, T.J.N.; Groen, H.J.M.; van Rijn, C.J.M.; Terstappen, L.W.M.M. The Detection of EpCAM+ and EpCAM–Circulating Tumor Cells. Sci. Rep. 2015, 5, 12270. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.; Kinde, I.; Agrawal, N.; Bartlett, B.; Wang, H.; Luber, B.; Kinzler, K.; Vogelstein, B.; et al. Abstract 5606: Detection of Circulating Tumor DNA in Early and Late Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [Green Version]
- Nedaeinia, R.; Manian, M.; Jazayeri, M.H.; Ranjbar, M.; Salehi, R.; Sharifi, M.; Mohaghegh, F.; Goli, M.; Jahednia, S.H.; Avan, A.; et al. Circulating Exosomes and Exosomal MicroRNAs as Biomarkers in Gastrointestinal Cancer. Cancer Gene Ther. 2017, 24, 48–56. [Google Scholar] [CrossRef]
- Schübeler, D. Function and Information Content of DNA Methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Jin, Z.; Liu, Y. DNA Methylation in Human Diseases. Genes Dis. 2018, 5, 1–8. [Google Scholar] [CrossRef]
- Constâncio, V.; Nunes, S.P.; Henrique, R.; Jerónimo, C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells 2020, 9, 624. [Google Scholar] [CrossRef] [Green Version]
- Moss, J.; Zick, A.; Grinshpun, A.; Carmon, E.; Maoz, M.; Ochana, B.L.; Abraham, O.; Arieli, O.; Germansky, L.; Meir, K.; et al. Circulating Breast-Derived DNA Allows Universal Detection and Monitoring of Localized Breast Cancer. Ann. Oncol. 2020, 31, 395–403. [Google Scholar] [CrossRef] [Green Version]
- Tham, C.; Chew, M.; Soong, R.; Lim, J.; Ang, M.; Tang, C.; Zhao, Y.; Ong, S.Y.K.; Liu, Y. Postoperative Serum Methylation Levels of TAC1 and SEPT9 Are Independent Predictors of Recurrence and Survival of Patients with Colorectal Cancer. Cancer 2014, 120, 3131–3141. [Google Scholar] [CrossRef]
- Dong, D.; Zhang, J.; Zhang, R.; Li, F.; Li, Y.; Jia, Y. Multiprobe Assay for Clinical SEPT9 Methylation Based on the Carbon Dot-Modified Liquid-Exfoliated Graphene Field Effect Transistor with a Potential to Present a Methylation Panorama. ACS Omega 2020, 5, 16228–16237. [Google Scholar] [CrossRef]
- Wei, L.; Zhao, S.; Wang, G.; Zhang, S.; Luo, W.; Qin, Z.; Bi, X.; Tan, Y.; Meng, M.; Qin, J.; et al. SMAD7 Methylation as a Novel Marker in Atherosclerosis. Biochem. Biophys. Res. Commun. 2018, 496, 700–705. [Google Scholar] [CrossRef]
- Xu, M.; Li, J.; Chen, X.; Han, L.; Li, L.; Liu, Y. MTHFD1 Promoter Hypermethylation Increases the Risk of Hypertension. Clin. Exp. Hypertens. 2019, 41, 422–427. [Google Scholar] [CrossRef]
- Nakatochi, M.; Ichihara, S.; Yamamoto, K.; Naruse, K.; Yokota, S.; Asano, H.; Matsubara, T.; Yokota, M. Epigenome-Wide Association of Myocardial Infarction with DNA Methylation Sites at Loci Related to Cardiovascular Disease. Clin. Epigenet. 2017, 9, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, W.; Xia, Y.; Bell, C.G.; Yet, I.; Ferreira, T.; Ward, K.J.; Gao, F.; Loomis, A.K.; Hyde, C.L.; Wu, H.; et al. An Integrated Epigenomic Analysis for Type 2 Diabetes Susceptibility Loci in Monozygotic Twins. Nat. Commun. 2014, 5, 5719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayols-Baixeras, S.; Subirana, I.; Fernández-Sanlés, A.; Sentí, M.; Lluís-Ganella, C.; Marrugat, J.; Elosua, R. DNA Methylation and Obesity Traits: An Epigenome-Wide Association Study. The REGICOR Study. Epigenetics 2017, 12, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Arosio, B.; Falconi, A.; Micioni Di Bonaventura, M.V.; Karimi, M.; Mari, D.; Casati, M.; Maccarrone, M.; D’Addario, C. Global Changes in DNA Methylation in Alzheimer’s Disease Peripheral Blood Mononuclear Cells. Brain Behav. Immun. 2015, 45, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Masliah, E.; Dumaop, W.; Galasko, D.; Desplats, P. Distinctive Patterns of DNA Methylation Associated with Parkinson Disease: Identification of Concordant Epigenetic Changes in Brain and Peripheral Blood Leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Smith, D.; Richards, D.; et al. Sensitive and Specific Multi-Cancer Detection and Localization Using Methylation Signatures in Cell-Free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Galardi, F.; De Luca, F.; Romagnoli, D.; Biagioni, C.; Moretti, E.; Biganzoli, L.; Di Leo, A.; Migliaccio, I.; Malorni, L.; Benelli, M. Cell-Free DNA-Methylation-Based Methods and Applications in Oncology. Biomolecules 2020, 10, 1677. [Google Scholar] [CrossRef]
- Ilijazi, D.; Pulverer, W.; Ertl, I.E.; Lemberger, U.; Kimura, S.; Abufaraj, M.; D’Andrea, D.; Pradere, B.; Bruchbacher, A.; Graf, A.; et al. Discovery of Molecular DNA Methylation-Based Biomarkers through Genome-Wide Analysis of Response Patterns to BCG for Bladder Cancer. Cells 2020, 9, 1839. [Google Scholar] [CrossRef]
- Gampenrieder, S.P.; Rinnerthaler, G.; Hackl, H.; Pulverer, W.; Weinhaeusel, A.; Ilic, S.; Hufnagl, C.; Hauser-Kronberger, C.; Egle, A.; Risch, A.; et al. DNA Methylation Signatures Predicting Bevacizumab Efficacy in Metastatic Breast Cancer. Theranostics 2018, 8, 2278–2288. [Google Scholar] [CrossRef]
- Zaenker, P.; Ziman, M.R. Serologic Autoantibodies as Diagnostic Cancer Biomarkers—A Review. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2161–2181. [Google Scholar] [CrossRef] [Green Version]
- Luna Coronell, J.A.; Syed, P.; Sergelen, K.; Gyurján, I.; Weinhäusel, A. The Current Status of Cancer Biomarker Research Using Tumour-Associated Antigens for Minimal Invasive and Early Cancer Diagnostics. J. Proteom. 2012, 76, 102–115. [Google Scholar] [CrossRef]
- Luna-Coronell, J.A.; Vierlinger, K.; Gamperl, M.; Hofbauer, J.; Berger, I.; Weinhäusel, A. The Prostate Cancer Immunome: In Silico Functional Analysis of Antigenic Proteins from Microarray Profiling with IgG. Proteomics 2016, 16, 1204–1214. [Google Scholar] [CrossRef]
- Luna Coronell, J.A.; Sergelen, K.; Hofer, P.; Gyurján, I.; Brezina, S.; Hettegger, P.; Leeb, G.; Mach, K.; Gsur, A.; Weinhäusel, A. The Immunome of Colon Cancer: Functional In Silico Analysis of Antigenic Proteins Deduced from IgG Microarray Profiling. Genom. Proteom. Bioinform. 2018, 16, 73–84. [Google Scholar] [CrossRef]
- Jodeleit, H.; Milchram, L.; Soldo, R.; Beikircher, G.; Schönthaler, S.; Al-Amodi, O.; Wolf, E.; Beigel, F.; Weinhäusel, A.; Siebeck, M.; et al. Autoantibodies as Diagnostic Markers and Potential Drivers of Inflammation in Ulcerative Colitis. PLoS ONE 2020, 15, e0228615. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, F.M.; Mair, F.S.; Anderson, W.; Armory, P.; Briggs, A.; Chew, C.; Dorward, A.; Haughney, J.; Hogarth, F.; Kendrick, D.; et al. Earlier Diagnosis of Lung Cancer in a Randomised Trial of an Autoantibody Blood Test Followed by Imaging. Eur. Respir. J. 2020, 57, 2000670. [Google Scholar] [CrossRef]
- Hettegger, P.; Huber, J.; Paßecker, K.; Soldo, R.; Kegler, U.; Nöhammer, C.; Weinhäusel, A. High Similarity of IgG Antibody Profiles in Blood and Saliva Opens Opportunities for Saliva Based Serology. PLoS ONE 2019, 14, e0218456. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Špilak, A.; Brachner, A.; Kegler, U.; Neuhaus, W.; Noehammer, C. Implications and Pitfalls for Cancer Diagnostics Exploiting Extracellular Vesicles. Adv. Drug Deliv. Rev. 2021, 175, 113819. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy Delivery Tools and Biomarkers of Diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; How Huang, K.; Jen Lee, M.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Zhou, X.; Wen, W.; Shan, X.; Zhu, W.; Xu, J.; Guo, R.H.; Cheng, W.F.; Wang, F.; Qi, L.W.; Chen, Y.; et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2016, 8, 6513–6525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, H.; Stanisavljevic, L.; Storli, K.; Hestetun, K.; Dahl, O.; Myklebust, M. A four-microRNA classifier as a novel prognostic marker for tumor recurrence in stage II colon cancer. Sci. Rep. 2018, 8, 6157. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.D.; Li, Q.; Zhang, R.S.; Dai, X.L.; Chen, W.J.; Xing, D.M. Circulating microRNAs: Biomarkers of disease. Clin. Chem. 2021, 516, 46–54. [Google Scholar] [CrossRef]
- Wehbe, N.; Nasser, S.A.; Pintus, G.; Badran, A.; Eid, A.H.; Baydoun, E. MicroRNAs in Cardiac Hypertrophy. Int. J. Mol. Sci. 2019, 20, 4714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular MiRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Wang, K.-H.; Hsieh, J.-C.; Chen, C.-C.; Zan, H.-W.; Meng, H.-F.; Kuo, S.-Y.; Nguyễn, M.T.N. A Low-Cost, Portable and Easy-Operated Salivary Urea Sensor for Point-of-Care Application. Biosens. Bioelectron. 2019, 132, 352–359. [Google Scholar] [CrossRef]
- Kim, J.; Imani, S.; de Araujo, W.R.; Warchall, J.; Valdés-Ramírez, G.; Paixão, T.R.L.C.; Mercier, P.P.; Wang, J. Wearable Salivary Uric Acid Mouthguard Biosensor with Integrated Wireless Electronics. Biosens. Bioelectron. 2015, 74, 1061–1068. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.S.; Dighe, K.; Wang, Z.; Srivastava, I.; Daza, E.; Schwartz-Dual, A.S.; Ghannam, J.; Misra, S.K.; Pan, D. Detection of Prostate Specific Antigen (PSA) in Human Saliva Using an Ultra-Sensitive Nanocomposite of Graphene Nanoplatelets with Diblock-Co-Polymers and Au Electrodes. Analyst 2018, 143, 1094–1103. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Wu, C.-C.; Peng, Y.-S.; Wu, C.-W.; Chang, Y.-T.; Chang, K.-P. Detection of Anti-P53 Autoantibodies in Saliva Using Microfluidic Chips for the Rapid Screening of Oral Cancer. RSC Adv. 2018, 8, 15513–15521. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.; Pires, N.M.M. Immunodetection of Salivary Biomarkers by an Optical Microfluidic Biosensor with Polyethylenimine-Modified Polythiophene-C70 Organic Photodetectors. Biosens. Bioelectron. 2017, 94, 321–327. [Google Scholar] [CrossRef]
- Rathnayake, N.; Gieselmann, D.-R.; Heikkinen, A.M.; Tervahartiala, T.; Sorsa, T. Salivary Diagnostics-Point-of-Care Diagnostics of MMP-8 in Dentistry and Medicine. Diagnostics 2017, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Wignarajah, S.; Suaifan, G.A.R.Y.; Bizzarro, S.; Bikker, F.J.; Kaman, W.E.; Zourob, M. Colorimetric Assay for the Detection of Typical Biomarkers for Periodontitis Using a Magnetic Nanoparticle Biosensor. Anal. Chem. 2015, 87, 12161–12168. [Google Scholar] [CrossRef]
- Bhakta, S.A.; Borba, R.; Taba, M.; Garcia, C.D.; Carrilho, E. Determination of Nitrite in Saliva Using Microfluidic Paper-Based Analytical Devices. Anal. Chim. Acta 2014, 809, 117–122. [Google Scholar] [CrossRef] [Green Version]
- Sorsa, T.; Gursoy, U.K.; Nwhator, S.; Hernandez, M.; Tervahartiala, T.; Leppilahti, J.; Gursoy, M.; Könönen, E.; Emingil, G.; Pussinen, P.J.; et al. Analysis of Matrix Metalloproteinases, Especially MMP-8, in Gingival Crevicular Fluid, Mouthrinse and Saliva for Monitoring Periodontal Diseases. Periodontol 2000 2016, 70, 142–163. [Google Scholar] [CrossRef]
- De Castro, L.F.; de Freitas, S.V.; Duarte, L.C.; de Souza, J.A.C.; Paixão, T.R.L.C.; Coltro, W.K.T. Salivary Diagnostics on Paper Microfluidic Devices and Their Use as Wearable Sensors for Glucose Monitoring. Anal. Bioanal. Chem. 2019, 411, 4919–4928. [Google Scholar] [CrossRef]
- Lee, B.H.; Kim, S.H.; Ko, Y.; Park, J.C.; Ji, S.; Gu, M.B. The Sensitive Detection of ODAM by Using Sandwich-Type Biosensors with a Cognate Pair of Aptamers for the Early Diagnosis of Periodontal Disease. Biosens. Bioelectron. 2019, 126, 122–128. [Google Scholar] [CrossRef]
- Chen, Z.; Mauk, M.G.; Wang, J.; Abrams, W.R.; Corstjens, P.L.A.M.; Niedbala, R.S.; Malamud, D.; Bau, H.H. A Microfluidic System for Saliva-Based Detection of Infectious Diseases. Ann. N. Y. Acad. Sci. 2007, 1098, 429–436. [Google Scholar] [CrossRef]
- Chen, D.; Mauk, M.; Qiu, X.; Liu, C.; Kim, J.; Ramprasad, S.; Ongagna, S.; Abrams, W.R.; Malamud, D.; Corstjens, P.L.A.M.; et al. An Integrated, Self-Contained Microfluidic Cassette for Isolation, Amplification, and Detection of Nucleic Acids. Biomed. Microdevices 2010, 12, 705–719. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Zhu, H.; Malamud, D.; Barber, C.; Ongagna Yhombi, S.; Yasmin, R.; Modak, S.; Janal, M.; Abrams, W.; Montagna, R. A Rapid, Self-Confirming Assay for HIV: Simultaneous Detection of Anti-HIV Antibodies and Viral RNA. J. AIDS Clin. Res. 2016, 7, 540. [Google Scholar] [CrossRef] [Green Version]
- Sabalza, M.; Yasmin, R.; Barber, C.A.; Castro, T.; Malamud, D.; Kim, B.J.; Zhu, H.; Montagna, R.A.; Abrams, W.R. Detection of Zika Virus Using Reverse-Transcription LAMP Coupled with Reverse Dot Blot Analysis in Saliva. PLoS ONE 2018, 13, e0192398. [Google Scholar] [CrossRef] [Green Version]
- Cabral-Miranda, G.; Cardoso, A.R.; Ferreira, L.C.S.; Sales, M.G.F.; Bachmann, M.F. Biosensor-Based Selective Detection of Zika Virus Specific Antibodies in Infected Individuals. Biosens. Bioelectron. 2018, 113, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Žukovskaja, O.; Jahn, I.; Weber, K.; Cialla-May, D.; Popp, J. Detection of Pseudomonas Aeruginosa Metabolite Pyocyanin in Water and Saliva by Employing the SERS Technique. Sensors 2017, 17, 1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oblath, E.A.; Henley, W.H.; Alarie, J.P.; Ramsey, J.M. A Microfluidic Chip Integrating DNA Extraction and Real-Time PCR for the Detection of Bacteria in Saliva. Lab Chip 2013, 13, 1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haji Mohammadi, M.; Mulder, S.; Khashayar, P.; Kalbasi, A.; Azimzadeh, M.; Aref, A.R. Saliva Lab-on-a-Chip Biosensors: Recent Novel Ideas and Applications in Disease Detection. Microchem. J. 2021, 168, 106506. [Google Scholar] [CrossRef]
- Cheng, Y.L.; Rees, T.; Wright, J. A Review of Research on Salivary Biomarkers for Oral Cancer Detection. Clin. Transl. Med. 2014, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Javaid, M.A.; Ahmed, A.S.; Durand, R.; Tran, S.D. Saliva as a Diagnostic Tool for Oral and Systemic Diseases. J. Oral Biol. Craniofac. Res. 2016, 6, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Aro, K.; Wei, F.; Wong, D.T.; Tu, M. Saliva Liquid Biopsy for Point-of-Care Applications. Front. Public Health 2017, 5, 77. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, L.L.; Pacheco, V.B.; Borges, L.; Athwal, H.K.; de Paula Eduardo, F.; Bezinelli, L.; Correa, L.; Jimenez, M.; Dame-Teixeira, N.; Lombaert, I.M.A.; et al. Saliva in the Diagnosis of COVID-19: A Review and New Research Directions. J. Dent. Res. 2020, 99, 1435–1443. [Google Scholar] [CrossRef]
- Isho, B.; Abe, K.T.; Zuo, M.; Jamal, A.J.; Rathod, B.; Wang, J.H.; Li, Z.; Chao, G.; Rojas, O.L.; Bang, Y.M.; et al. Persistence of Serum and Saliva Antibody Responses to SARS-CoV-2 Spike Antigens in COVID-19 Patients. Sci. Immunol. 2020, 5, eabe5511. [Google Scholar] [CrossRef]
- Lin, G.C.; Smajlhodzic, M.; Bandian, A.-M.; Friedl, H.-P.; Leitgeb, T.; Oerter, S.; Stadler, K.; Giese, U.; Peham, J.R.; Bingle, L.; et al. An In Vitro Barrier Model of the Human Submandibular Salivary Gland Epithelium Based on a Single Cell Clone of Cell Line HTB-41: Establishment and Application for Biomarker Transport Studies. Biomedicines 2020, 8, 302. [Google Scholar] [CrossRef]
- Lin, G.C.; Leitgeb, T.; Vladetic, A.; Friedl, H.-P.; Rhodes, N.; Rossi, A.; Roblegg, E.; Neuhaus, W. Optimization of an Oral Mucosa In Vitro Model Based on Cell Line TR146. Tissue Barriers 2020, 8, 1748459. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Lin, C.-C.; Joon, A.; Feng, Z.; Troche, G.; Lira, M.E.; Chia, D.; Mao, M.; Ho, C.-L.; Su, W.-C.; et al. Noninvasive Saliva-Based EGFR Gene Mutation Detection in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 190, 1117–1126. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wei, F.; Schafer, C.; Wong, D.T.W. Detection of Tumor Cell-Specific MRNA and Protein in Exosome-Like Microvesicles from Blood and Saliva. PLoS ONE 2014, 9, e110641. [Google Scholar] [CrossRef]
- Kim, N.-H.; Choi, S.-J.; Yang, D.-J.; Bae, J.; Park, J.; Kim, I.-D. Highly Sensitive and Selective Hydrogen Sulfide and Toluene Sensors Using Pd Functionalized WO3 Nanofibers for Potential Diagnosis of Halitosis and Lung Cancer. Sens. Actuators B Chem. 2014, 193, 574–581. [Google Scholar] [CrossRef]
- Wilson, A.D. Application of Electronic-Nose Technologies and VOC-Biomarkers for the Noninvasive Early Diagnosis of Gastrointestinal Diseases. Sensors 2018, 18, 2613. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.-W.; Lee, J.-H. Toward Breath Analysis on a Chip for Disease Diagnosis Using Semiconductor-Based Chemiresistors: Recent Progress and Future Perspectives. Lab Chip 2017, 17, 3537–3557. [Google Scholar] [CrossRef]
- Queralto, N.; Berliner, A.N.; Goldsmith, B.; Martino, R.; Rhodes, P.; Lim, S.H. Detecting Cancer by Breath Volatile Organic Compound Analysis: A Review of Array-Based Sensors. J. Breath Res. 2014, 8, 027112. [Google Scholar] [CrossRef]
- Shirasu, M.; Touhara, K. The Scent of Disease: Volatile Organic Compounds of the Human Body Related to Disease and Disorder. J. Biochem. 2011, 150, 257–266. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.-Y.; Wang, Y.-C.; Peng, H.-Y.; Huang, C.-H. Breath Biopsy of Breast Cancer Using Sensor Array Signals and Machine Learning Analysis. Sci. Rep. 2021, 11, 103. [Google Scholar] [CrossRef]
- Bos, L.D.; Sterk, P.J.; Fowler, S.J. Breathomics in the Setting of Asthma and Chronic Obstructive Pulmonary Disease. J. Allergy Clin. Immunol. 2016, 138, 970–976. [Google Scholar] [CrossRef] [Green Version]
- Van der Schee, M.; Pinheiro, H.; Gaude, E. Breath Biopsy for Early Detection and Precision Medicine in Cancer. Ecancermedicalscience 2018, 12, ed84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, X.; Monge, M.E.; McCarty, N.A.; Stecenko, A.A.; Fernández, F.M. Feasibility of Early Detection of Cystic Fibrosis Acute Pulmonary Exacerbations by Exhaled Breath Condensate Metabolomics: A Pilot Study. J. Proteome Res. 2017, 16, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Montuschi, P.; Paris, D.; Melck, D.; Lucidi, V.; Ciabattoni, G.; Raia, V.; Calabrese, C.; Bush, A.; Barnes, P.J.; Motta, A. NMR Spectroscopy Metabolomic Profiling of Exhaled Breath Condensate in Patients with Stable and Unstable Cystic Fibrosis. Thorax 2012, 67, 222–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trefz, P.; Obermeier, J.; Lehbrink, R.; Schubert, J.K.; Miekisch, W.; Fischer, D.-C. Exhaled Volatile Substances in Children Suffering from Type 1 Diabetes Mellitus: Results from a Cross-Sectional Study. Sci. Rep. 2019, 9, 15707. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, A.; Hrinczenko, B.; Swain, G.; Peteu, S. Exhaled Breath Biomarker Sensing. Biosens. Bioelectron. 2021, 182, 113193. [Google Scholar] [CrossRef]
- Hatamie, A.; Angizi, S.; Kumar, S.; Pandey, C.M.; Simchi, A.; Willander, M.; Malhotra, B.D. Review—Textile Based Chemical and Physical Sensors for Healthcare Monitoring. J. Electrochem. Soc. 2020, 167, 037546. [Google Scholar] [CrossRef]




| Name | Manufacturer | Biomarker(s) | Biosample | Application | Sensitivity (%) | Specifity (%) |
|---|---|---|---|---|---|---|
| Cologuard | Exact Sciences Corp. | NDRG4, BMP3 | Stool | CRC early detection | 92 | 87 |
| Epi proColon 2.0 | Epigenomics GA | SEPT9 | Plasma | CRC early detection | 81 | 97 |
| Epi proLung | Epigenomics GA | PTGER4, SHOX2 | Plasma | Lung cancer detection | 90 | 73 |
| Cervi-M | Epigene, iStat Biomedical Co. | ZNF582, PAX1 | Cervical brush | Cervical cancer detection | 77 *, 70 ** | 87 *, 82 ** |
| Oral-M | Epigene, iStat Biomedical Co. | ZNF582, PAX1 | Oral swab | Oral cancer detection | 85 *, 72 ** | 87 *, 86 ** |
| Assure MDx | MDxHealth | TWIST1, ONECUT2, OTX1 (+FGFR3, TERT, HRAS mutations) | Urine | Bladder cancer detection | 97 | 83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kny, E.; Reiner-Rozman, C.; Dostalek, J.; Hassel, A.-W.; Nöhammer, C.; Pfaffeneder-Mantai, F.; Szunerits, S.; Weber, V.; Knoll, W.; Kleber, C. State of the Art of Chemosensors in a Biomedical Context. Chemosensors 2022, 10, 199. https://doi.org/10.3390/chemosensors10060199
Kny E, Reiner-Rozman C, Dostalek J, Hassel A-W, Nöhammer C, Pfaffeneder-Mantai F, Szunerits S, Weber V, Knoll W, Kleber C. State of the Art of Chemosensors in a Biomedical Context. Chemosensors. 2022; 10(6):199. https://doi.org/10.3390/chemosensors10060199
Chicago/Turabian StyleKny, Erich, Ciril Reiner-Rozman, Jakub Dostalek, Achim-Walter Hassel, Christa Nöhammer, Florian Pfaffeneder-Mantai, Sabine Szunerits, Viktoria Weber, Wolfgang Knoll, and Christoph Kleber. 2022. "State of the Art of Chemosensors in a Biomedical Context" Chemosensors 10, no. 6: 199. https://doi.org/10.3390/chemosensors10060199
APA StyleKny, E., Reiner-Rozman, C., Dostalek, J., Hassel, A.-W., Nöhammer, C., Pfaffeneder-Mantai, F., Szunerits, S., Weber, V., Knoll, W., & Kleber, C. (2022). State of the Art of Chemosensors in a Biomedical Context. Chemosensors, 10(6), 199. https://doi.org/10.3390/chemosensors10060199








