Recent Advance in Nucleus-Targeted Fluorescent Probes for Bioimaging, Detection and Therapy
Abstract
:1. Introduction
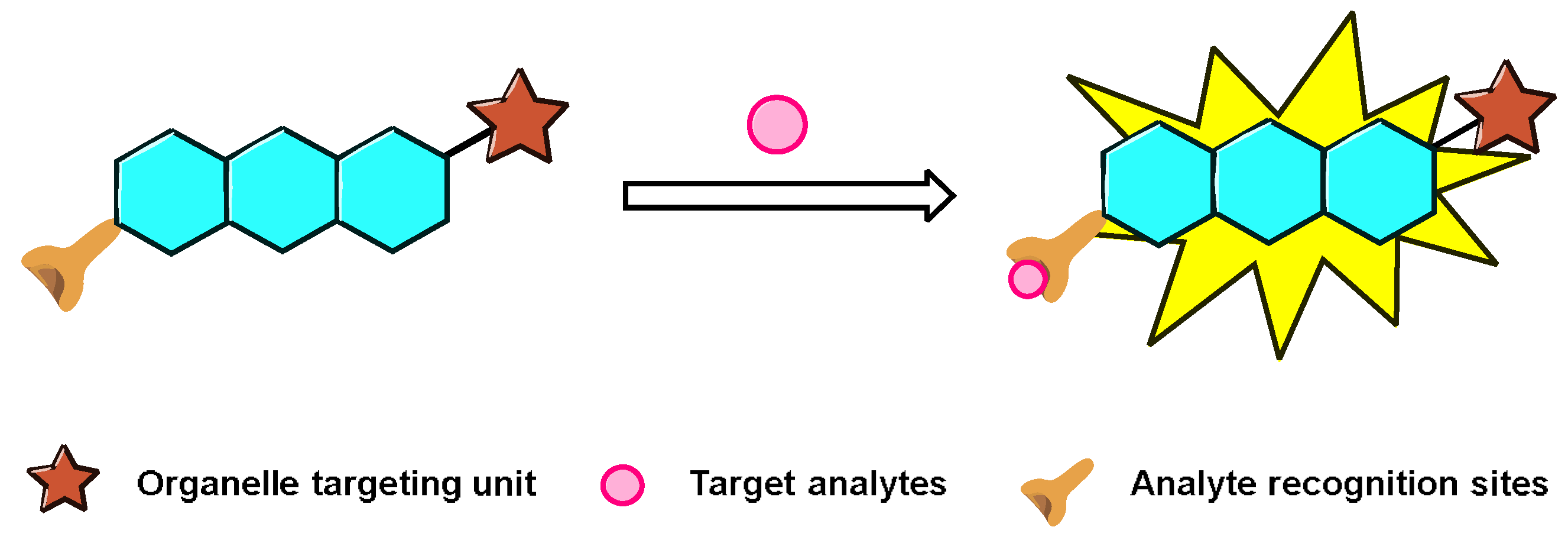
2. Representative General Strategies for Constructing Nucleus-Targeting Probes
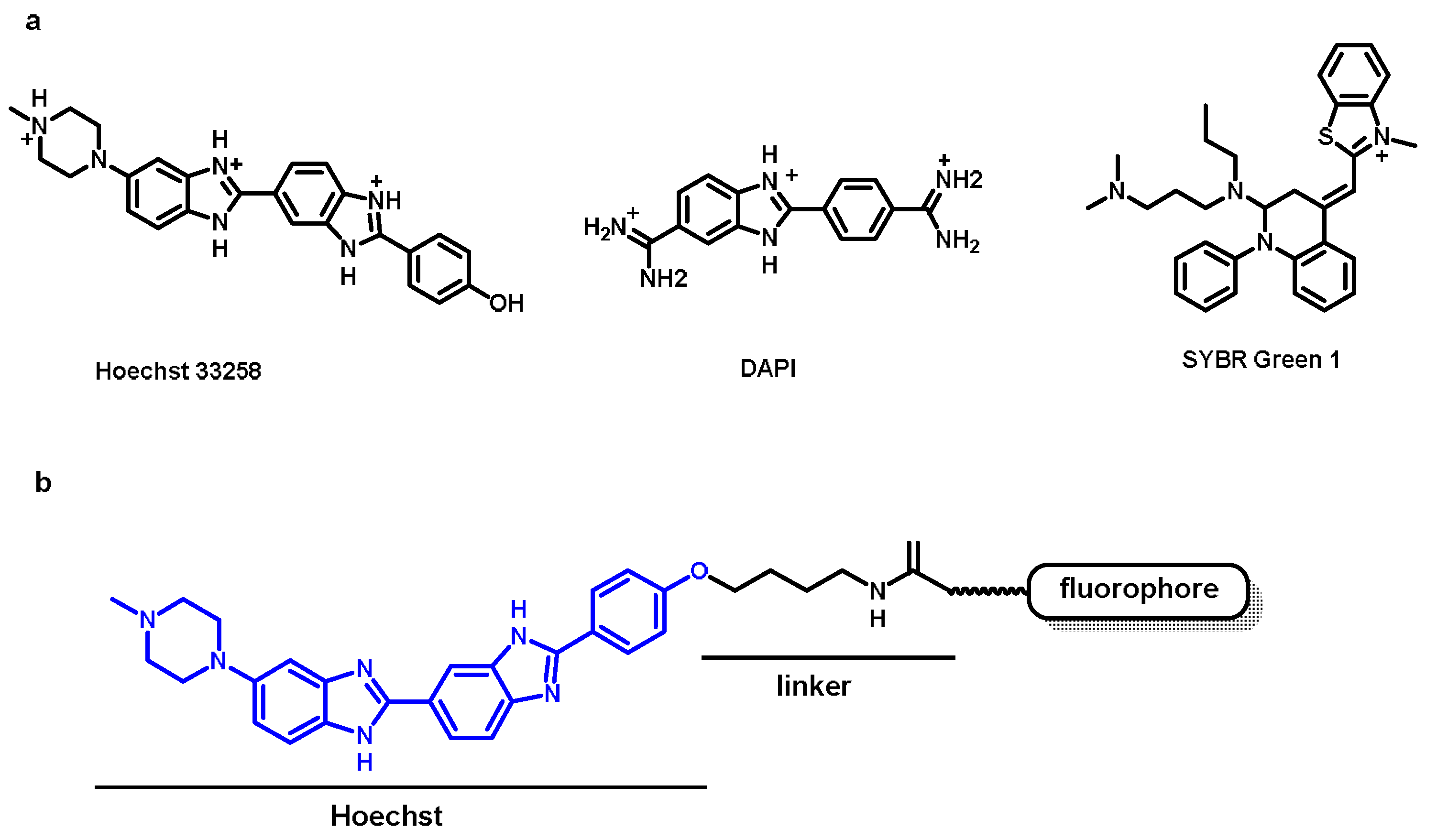
2.1. Nucleus Targeting Dye
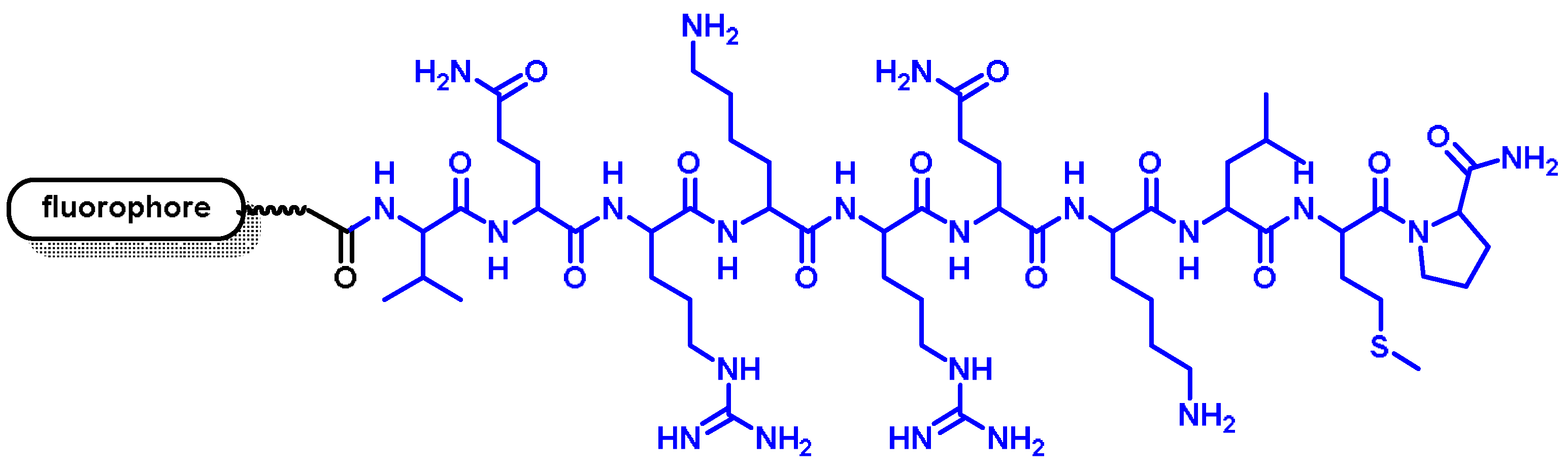
2.2. Nuclear Localization Signal (NLS)
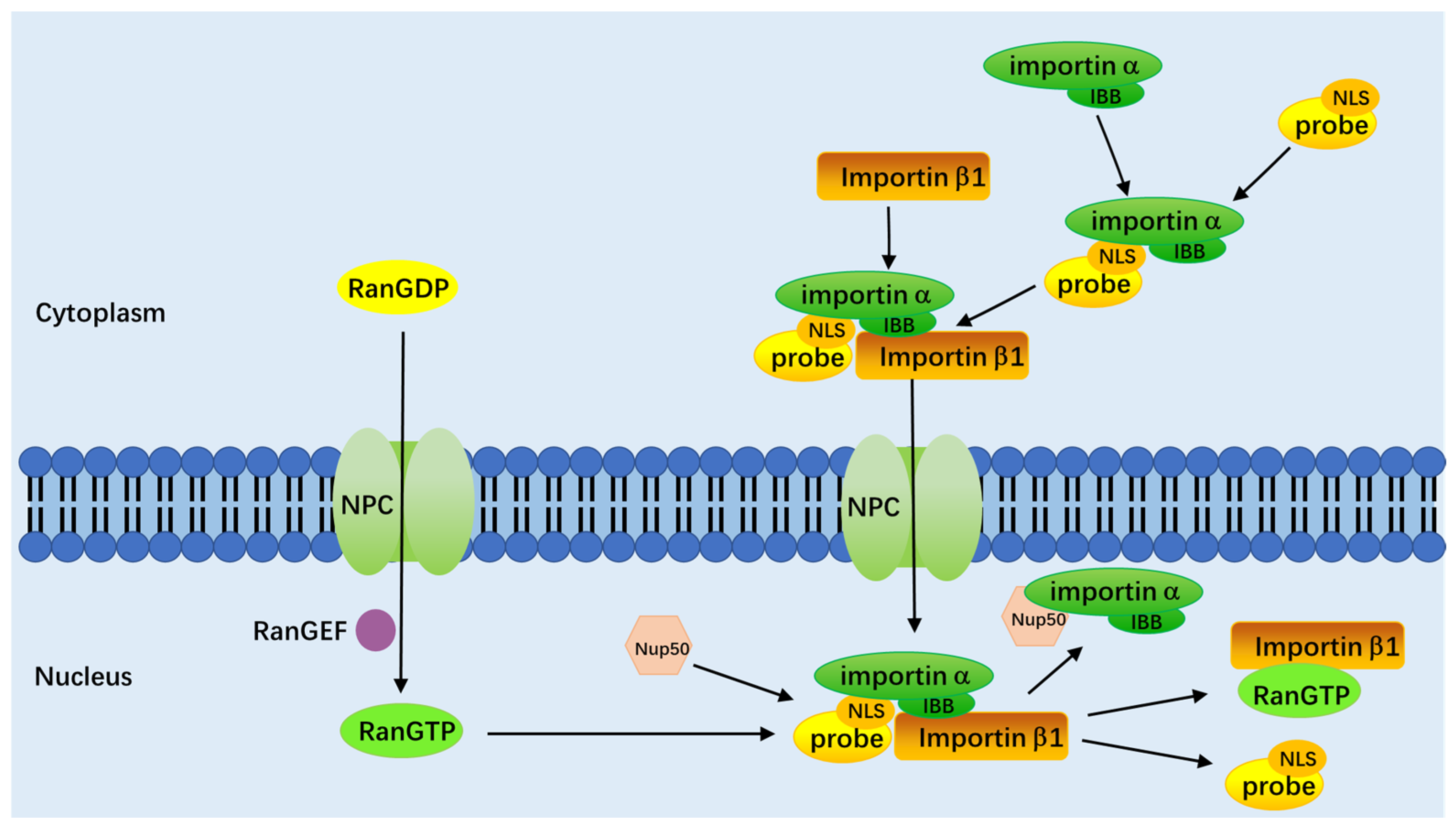
3. Mechanism of the Nucleus-Targeted Fluorescence Probe into the Nucleus
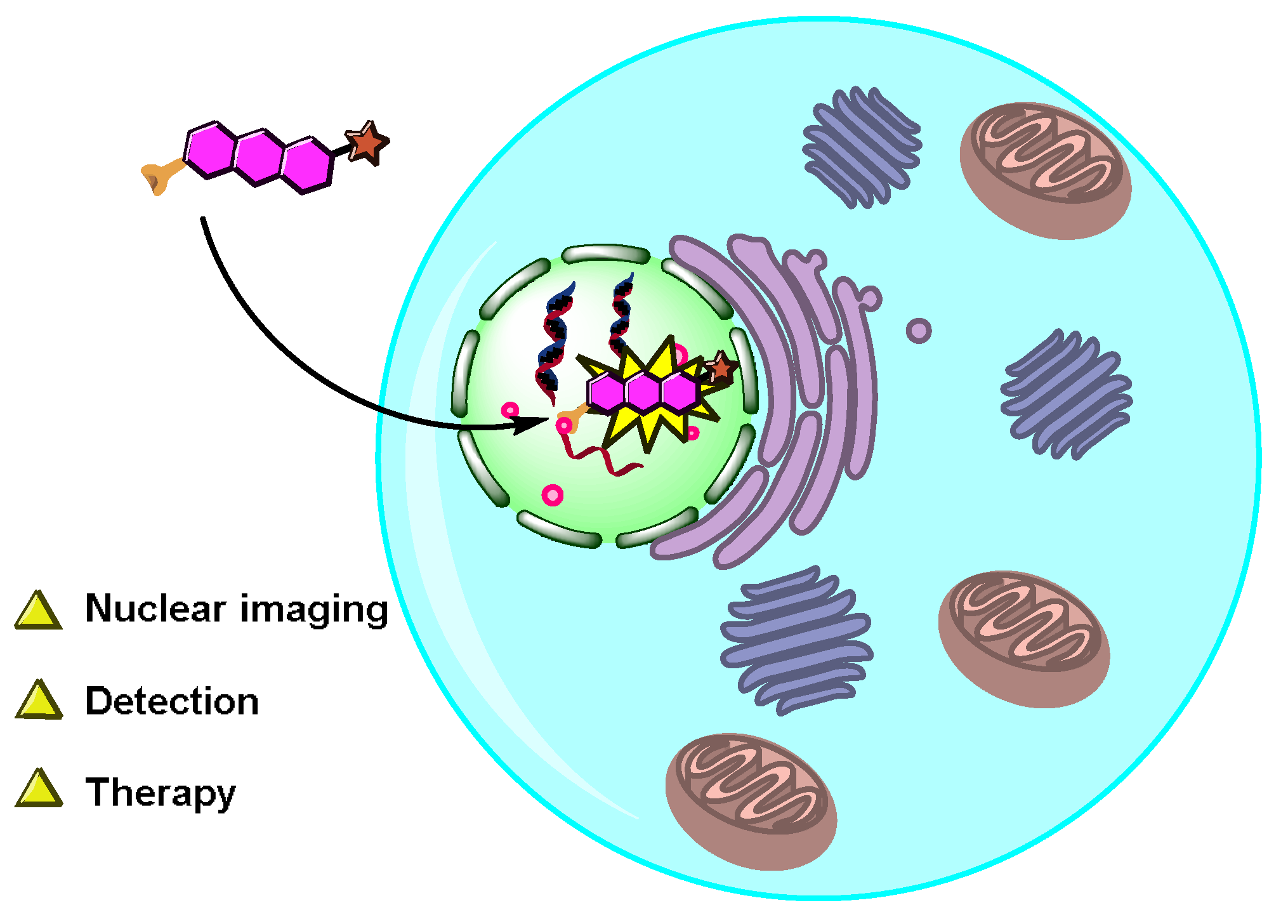
4. Applications of the Nucleus-Targeted Fluorescent Probes
4.1. Nuclear Imaging
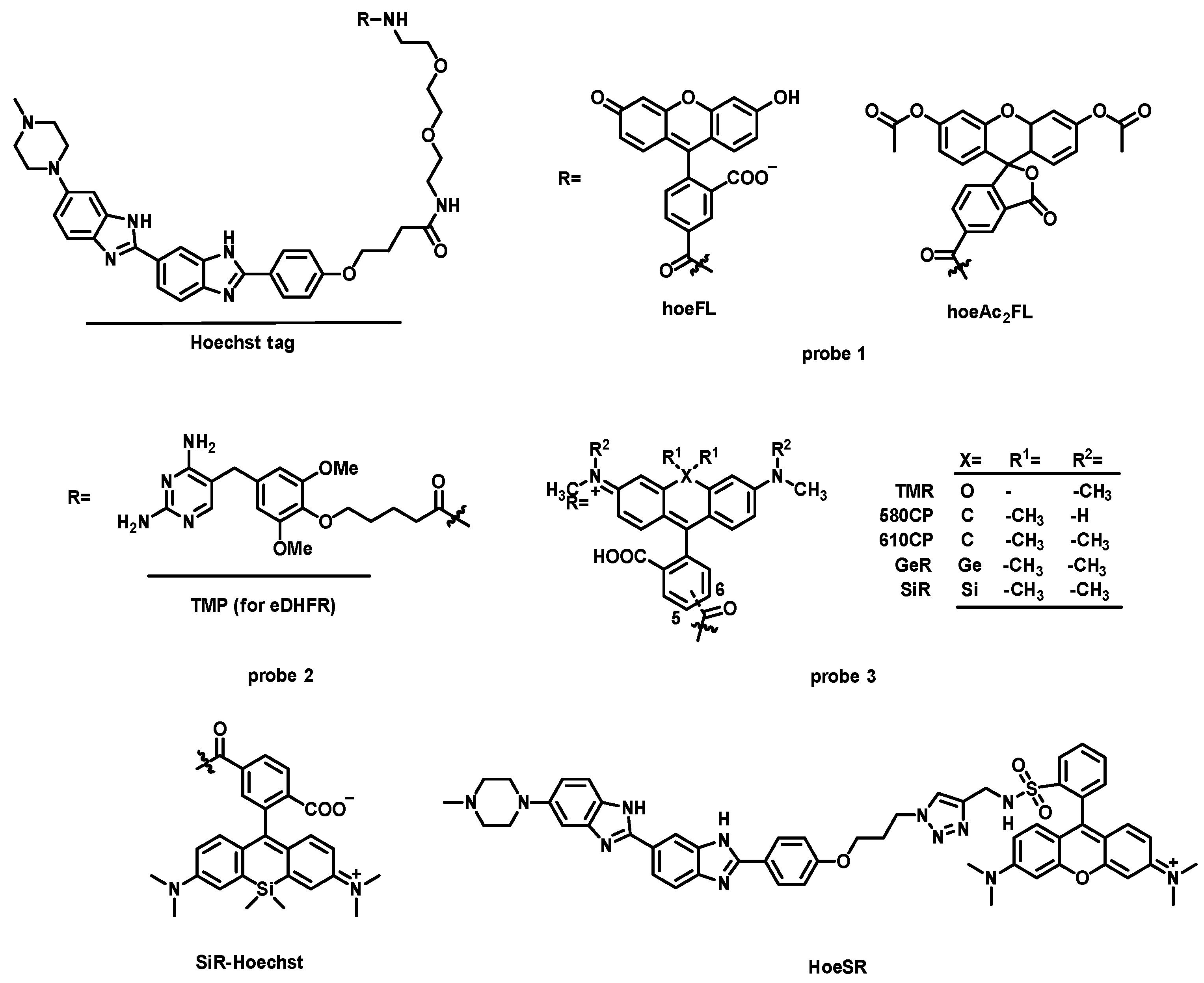
| Name [Ref.] | λex (nm) | λem (nm) | Stokes Shift (nm) | Binding Constant | Quantum Yield Bound/Unbound | Nucleic Acid Specificity |
|---|---|---|---|---|---|---|
| probe 1 [25] | 460 | 520 | 60 | KD = 2.5 × 106 M−1 | 0.0072/0.44 (hpDNA) | AT-rich DNA |
| 5-TMR (probe 3) [27] | 558 | 586 | 28 | KD = 3.65 × 107 M−1 | ~0.007/0.209 (hpDNA) | AT-rich DNA |
| 6-TMR (probe 3) | 562 | 585 | 23 | KD1 = 1.4 × 108 M−1; KD2 = 1.68 × 108 M−1 | ~0.005/0.052 (hpDNA) | AT-rich DNA |
| 5-580CP (probe 3) | 588 | 613 | 25 | KD = 1.55 × 108 M−1 | ~0.027/0.372 (hpDNA) | AT-rich DNA |
| 6-580CP (probe 3) | 594 | 619 | 25 | KD1 = 2.7 × 108 M−1; KD2 = 3.62 × 107 M−1 | ~0.011/0.124 (hpDNA) | AT-rich DNA |
| 5-610CP (probe 3) | 614 | 641 | 27 | KD = 3.47 × 107 M−1 | ~0.052/0.432 (hpDNA) | AT-rich DNA |
| 6-610CP (probe 3) | 618 | 644 | 26 | KD1 = 6.5 × 108 M−1; KD2 = 4.4 × 106 M−1 | ~0.033/0.282 (hpDNA) | AT-rich DNA |
| 5-GeR (probe 3) | 641 | 660 | 19 | KD = 4.4 × 106 M−1 | ~0.054/0.392 (hpDNA) | AT-rich DNA |
| 6-GeR (probe 3) | 643 | 662 | 19 | KD = 5.72 × 107 M−1; | ~0.033/0.207 (hpDNA) | AT-rich DNA |
| 5-SiR (probe 3) | 651 | 672 | 21 | KD = 4.8 × 106 M−1 | ~0.007/0.374 (hpDNA) | AT-rich DNA |
| 6-SiR (probe 3) | 654 | 677 | 23 | KD = 6.69 × 107 M−1 | ~0.003/0.156 (hpDNA) | AT-rich DNA |
| SiR-Hoechst [26] | 652 | 672 | 20 | KD = 8.4 × 106 M−1 | — | DNA |
| HoeSR [28] | 572 | 590 | 18 | KD = 3.5 × 106 M−1 | 0.009/0.09 (hpDNA) | DNA |
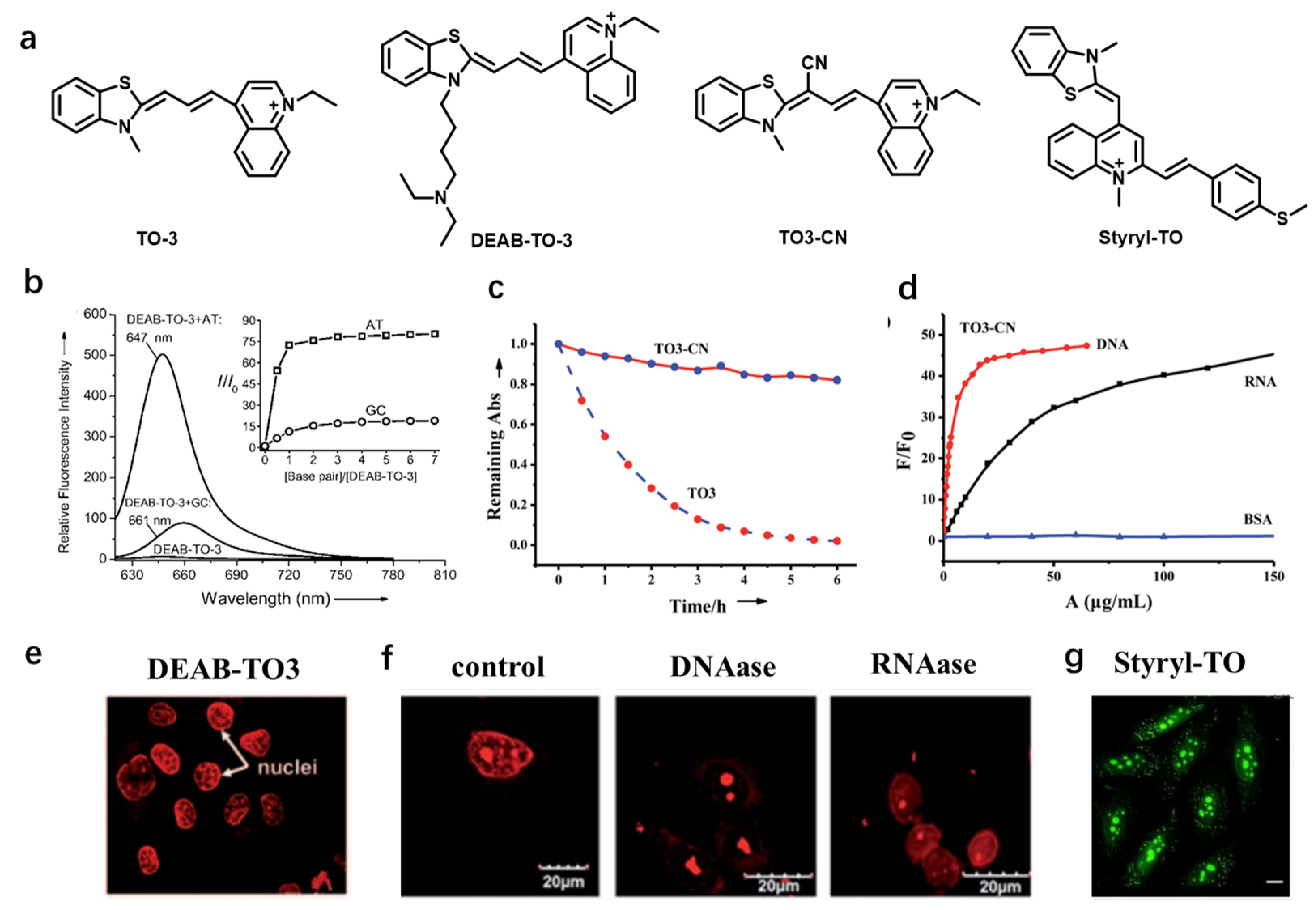
| DEAB-TO-3 [29] | 626 | 649 | 23 | — | 0.36 | AT-rich DNA |
| TO3-CN [30] | 543 | 604 | 56 (DNA); 49 (RNA) | — | 0.73 (DNA); 0.72 (RNA) | DNA and RNA |
| Styryl-TO [31] | 476 | 535 | 59 | KD = 1.23 × 106 M−1 | 0.506/0.0016 | RNA |
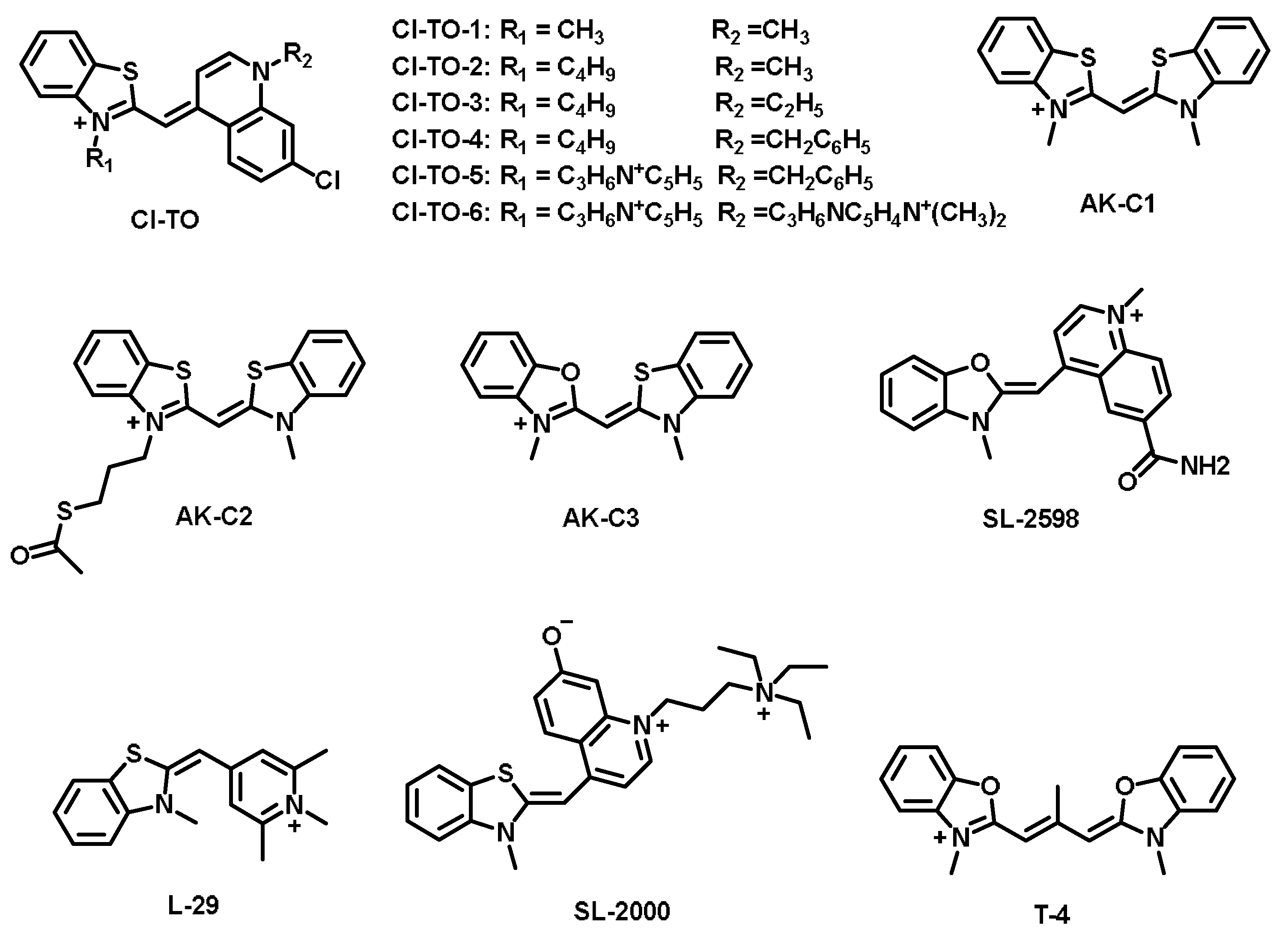
| Cl-TO-1 [32] | 508 | 534 | 26 | Ks = 2.22 × 106 M−1 | — | DNA |
| Cl-TO-2 [32] | 509 | 536 | 27 | Ks = 2.16 × 106 M−1 | — | DNA |
| Cl-TO-3 [32] | 510 | 536 | 26 | Ks = 1.26 × 106 M−1 | — | DNA |
| Cl-TO-4 [32] | 513 | 540 | 27 | Ks = 6.32 × 106 M−1 | — | DNA |
| Cl-TO-5 [32] | 511 | 538 | 27 | Ks = 4.56 × 106 M−1 | — | DNA |
| Cl-TO-6 [32] | 514 | 539 | 25 | Ks = 4.76 × 106 M−1 | — | DNA |
| AK-C1 [33] | 421 | 472 | 51 | Ks = 6.92 × 106 M−1 | 0.0005 (in water) | AU-rich RNA |
| AK-C2 [33] | 422 | 481 | 59 | Ks = 6.46 × 106 M−1 | 0.0009 (in water) | AU-rich RNA |
| AK-C3 [33] | 400 | 431 | 31 | Ks = 1.29 × 106 M−1 | 0.0031 (in water) | AU-rich RNA |
| SL-2598 [34] | 504 | 526 | 22 | — | 0.44 | RNA |
| SL-2000 [34] | 506 | 529 | 23 | — | — | RNA |
| L-29 [34] | 451 | 476 | 25 | — | — | RNA |
| T-4 [35] | 499 | 511 | 12 | — | — | RNA |
| CP [36] | 598 | 658 | 60 | — | 0.22 (in DCM) | RNA |
| CP3 [37] | 584 | 638 | 54 | — | 0.555 (in DCM) | RNA and lysosome |
| CP6 [37] | 595 | 655 | 60 | — | 0.338(in DCM) | RNA and lysosome |
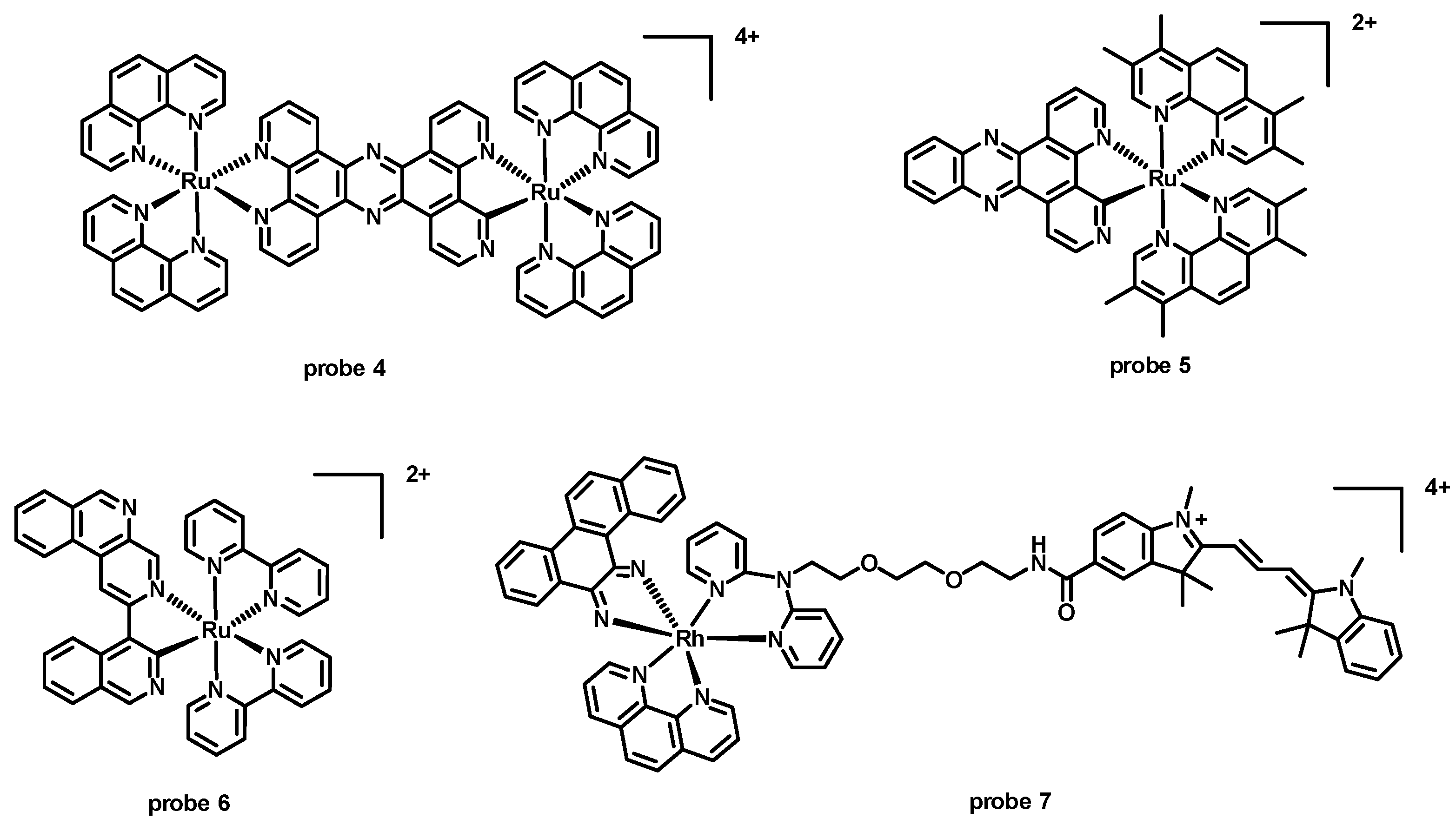
| probe 4 [24] | 450 | 658 (duplex); 631 (GQ) | 208 (duplex); 181 (GQ) | Kb > 105 (duplex); 4.4 × 106 M−1 (GQ) | — | Duplex and GQ |
| probe 5 [38] | 440 | 660 | 220 | KB = 6.8 × 104 M−1 (well-matched DNA); KB = 1.8 × 106 M−1 (mismatched DNA) | — | DNA |
| probe 6 [39] | 440 | 700 | 260 | KB = 7.3 × 103 M−1 (well-matched DNA); KB = 3.5 × 106 M−1 (mismatched DNA) | — | DNA |
| probe 7 [40] | 520 | 570 | 50 | KD = 3.2 × 107 M−1 (mismatched DNA) | — | DNA |
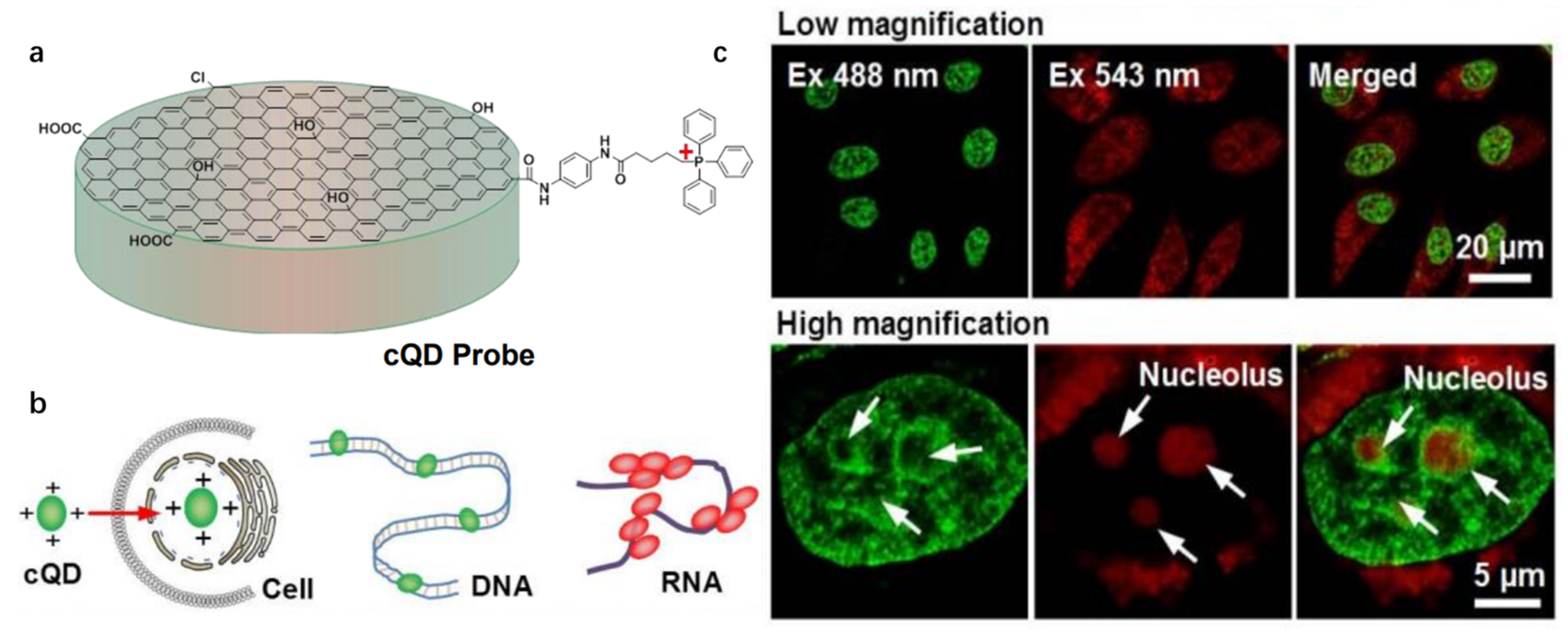
| cQD probe [44] | ~400(dsDNA); ~540(ssRNA) | ~520(dsDNA); ~620(ssRNA) | ~120 | — | ~0.080 | dsDNA; ssRNA |
4.2. Detection of Biomolecules in the Nucleus
4.2.1. Detection of DNA
4.2.2. Detection of RNA
4.2.3. Detection of the G-Quadruplex
4.2.4. Detection of DNA Conformational Changes
4.2.5. Detection of Biological Macromolecules—Histones H2B and HDACs
4.2.6. Detection of Small Molecules and Ions
- Detection of Pyrophosphate (PPi)
- b
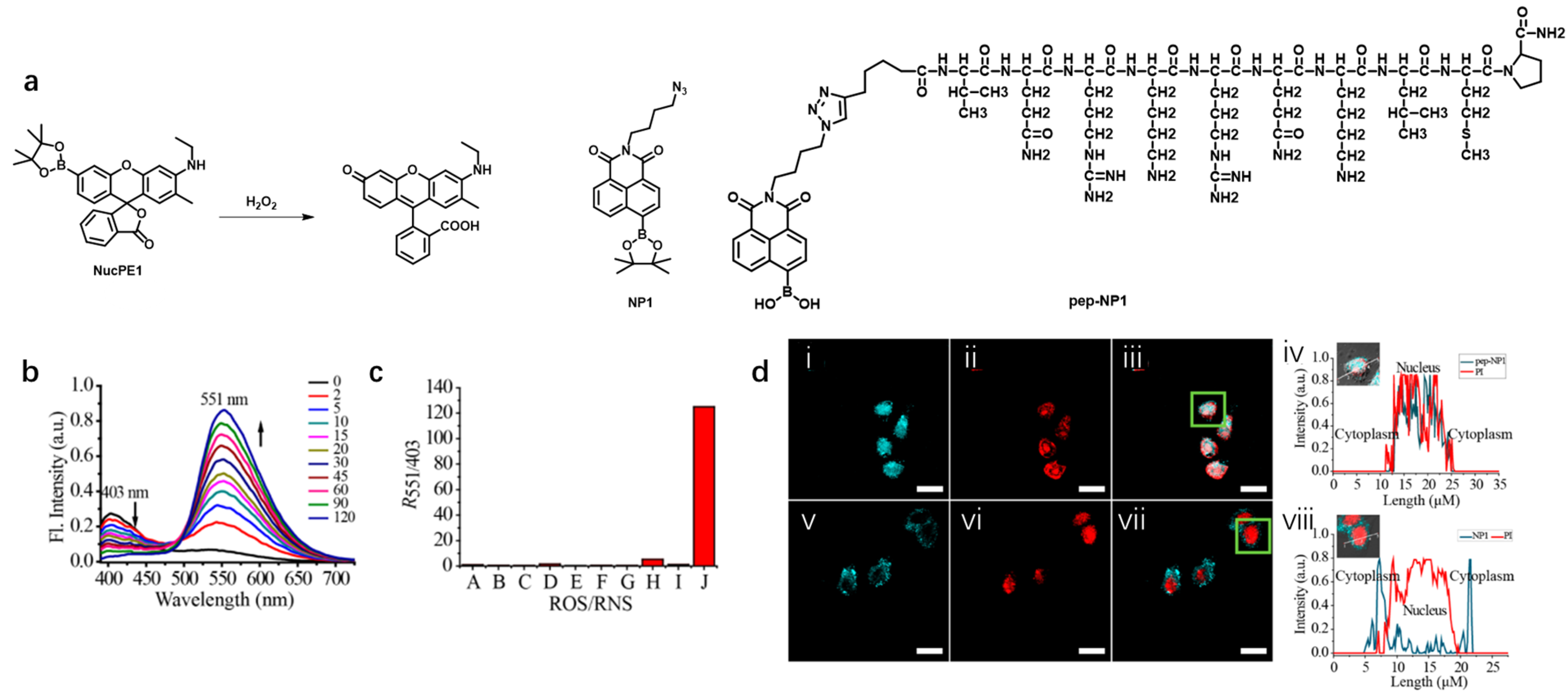
- Detection of hydrogen peroxide (H2O2)
- c
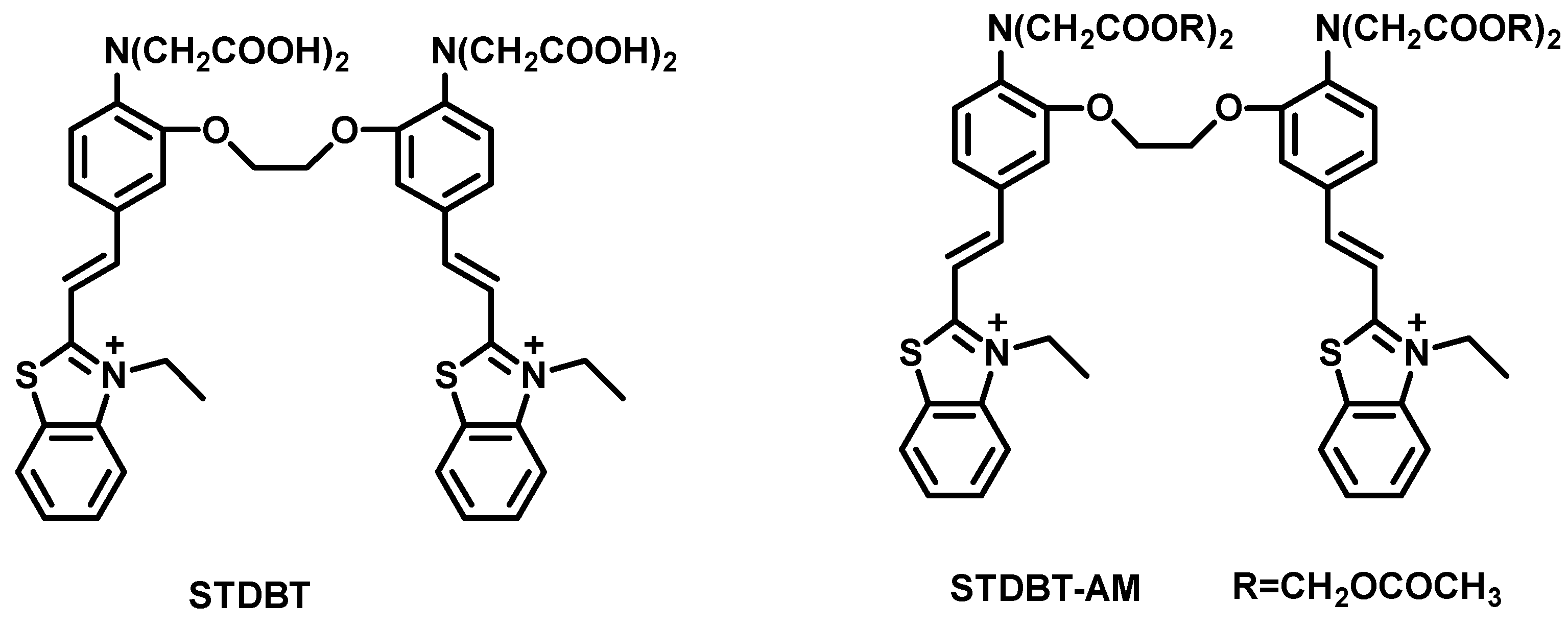
- Detection of Calcium Ions (Ca2+)
4.3. Application of Nucleus-Targeted Fluorescent Probes in Theranostics
Improved Strategies for Nucleus-Targeted Delivery Efficiency of Drugs or Probes
| Name [Ref.] | λex (nm) | λem (nm) | Stokes Shift (nm) | Binding Constant | Fluorescent Quantum Yield | 1O2 Quantum Yield | Lifetime | Extinction Coefficient |
|---|---|---|---|---|---|---|---|---|
| MeTPAE [79] | 424 | 632 | 208 | ~4.52 × 105 M−1 (dsDNA); ~1.70 × 106 M−1 (G4) | — | 0.772 | — | 39,400 M−1 cm−1 |
| Ir1-HAS [80] | 405 | 515 | 110 | — | 0.036 | 0.830 | 871.8 ns | — |
| TPCI [82] | 441 | ~580 | 139 | 5.68 × 108 M−1 | 0.03 | 0.986 | — | — |
| TPE-4EP+ [83] | 405 | 610 | 205 | — | 0.15 | — | — | — |
| Ru-tap-NLS [85] | 415 | 640 | 225 | 2.26 × 107 M−1 | 0.028 | — | 760 ns | 16,700 M−1 cm−1 |
| GTTN [92] | ~500 | ~520 | ~20 | — | 0.4179 | — | — | — |
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, Y.; Sun, C.; Ou, X.; Liu, B.; Lou, X.; Xia, F. Dual-targeted peptide-conjugated multifunctional fluorescent probe with AIEgen for efficient nucleus-specific imaging and long-term tracing of cancer cells. Chem. Sci. 2017, 8, 4571–4578. [Google Scholar] [CrossRef]
- Zhou, X.; Hao, Q.; Zhang, Q.; Liao, J.m.; Ke, J.w.; Liao, P.; Cao, B.; Lu, H. Ribosomal proteins L11 and L5 activate TAp73 by overcoming MDM2 inhibition. Cell Death Differ. 2015, 22, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Peleg, S.; Feller, C.; Ladurner, A.G.; Imhof, A. The metabolic impact on histone acetylation and transcription in ageing. Trends Biochem. Sci. 2016, 41, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Futscher, B.W.; Oshiro, M.M.; Wozniak, R.J.; Holtan, N.; Hanigan, C.L.; Duan, H.; Domann, F.E. Role for DNA methylation in the control of cell type–specific maspin expression. Nat. Genet. 2002, 31, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.J.; Ahmadzadeh, H.; Arriaga, E.A. Within the cell: Analytical techniques for subcellular analysis. Anal. Bioanal. Chem. 2005, 382, 906–917. [Google Scholar] [CrossRef]
- Lichtman, J.W.; Conchello, J.-A. Fluorescence microscopy. Nat. Meth. 2005, 2, 910–919. [Google Scholar] [CrossRef]
- Cotruvo, J.J.A.; Aron, A.T.; Ramos-Torres, K.M.; Chang, C.J. Synthetic fluorescent probes for studying copper in biological systems. Chem. Soc. Rev. 2015, 44, 4400–4414. [Google Scholar] [CrossRef]
- Alamudi, S.H.; Satapathy, R.; Kim, J.; Su, D.; Ren, H.; Das, R.; Hu, L.; Alvarado-Martínez, E.; Lee, J.Y.; Hoppmann, C.; et al. Development of background-free tame fluorescent probes for intracellular live cell imaging. Nat. Commun. 2016, 7, 11964. [Google Scholar] [CrossRef]
- Horobin, R.W.; Stockert, J.C.; Rashid-Doubell, F. Fluorescent cationic probes for nuclei of living cells: Why are they selective? A quantitative structure–activity relations analysis. Histochem. Cell Biol. 2006, 126, 165–175. [Google Scholar] [CrossRef]
- Chen, T.R. In situ detection of mycoplasma contamination in cell cultures by fluorescent Hoechst 33258 stain. Exp. Cell Res. 1977, 104, 255–262. [Google Scholar] [CrossRef]
- Kapuscinski, J. Dapi: A DNA-specific fluorescent probe. Biotech. Histochem. 1995, 70, 220–233. [Google Scholar] [CrossRef]
- Ishida, M.; Watanabe, H.; Takigawa, K.; Kurishita, Y.; Oki, C.; Nakamura, A.; Hamachi, I.; Tsukiji, S. Synthetic self-localizing ligands that control the spatial location of proteins in living cells. J. Am. Chem. Soc. 2013, 135, 12684–12689. [Google Scholar] [CrossRef] [PubMed]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin α. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef]
- Sankhala, R.S.; Lokareddy, R.K.; Begum, S.; Pumroy, R.A.; Gillilan, R.E.; Cingolani, G. Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1. Nat. Commun. 2017, 8, 979. [Google Scholar] [CrossRef]
- Lu, J.; Wu, T.; Zhang, B.; Liu, S.; Song, W.; Qiao, J.; Ruan, H. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Commun. Signal. 2021, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Atale, N.; Gupta, S.; Yadav, U.C.S.; Rani, V. Cell-death assessment by fluorescent and nonfluorescent cytosolic and nuclear staining techniques. J. Microsc. 2014, 255, 7–19. [Google Scholar] [CrossRef]
- Massignani, M.; LoPresti, C.; Blanazs, A.; Madsen, J.; Armes, S.P.; Lewis, A.L.; Battaglia, G. Controlling cellular uptake by surface chemistry, size, and surface topology at the nanoscale. Small 2009, 5, 2424–2432. [Google Scholar] [CrossRef]
- Ploeger, L.; Dullens, H.; Huisman, A.; van Diest, P. Fluorescent stains for quantification of DNA by confocal laser scanning microscopy in 3-D. Biotech. Histochem. 2008, 83, 63–69. [Google Scholar] [CrossRef]
- Yamauchi, K.; Yang, M.; Jiang, P.; Yamamoto, N.; Xu, M.; Amoh, Y.; Tsuji, K.; Bouvet, M.; Tsuchiya, H.; Tomita, K.; et al. Real-time in vivo dual-color imaging of intracapillary cancer cell and nucleus deformation and migration. Cancer Res. 2005, 65, 4246–4252. [Google Scholar] [CrossRef]
- Pfeifer, G.P.; You, Y.-H.; Besaratinia, A. Mutations induced by ultraviolet light. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2005, 571, 19–31. [Google Scholar] [CrossRef]
- Wojcik, K.; Dobrucki, J.W. Interaction of a DNA intercalator DRAQ5, and a minor groove binder SYTO17, with chromatin in live cells—Influence on chromatin organization and histone—DNA interactions. Cytometry A 2008, 73A, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yu, M.; Sun, Y.; Wu, Y.; Huang, C.; Li, F. A nonemissive iridium(iii) complex that specifically lights-up the nuclei of living cells. J. Am. Chem. Soc. 2011, 133, 11231–11239. [Google Scholar] [CrossRef]
- Liu, S.; Liang, H.; Zhang, K.Y.; Zhao, Q.; Zhou, X.; Xu, W.; Huang, W. A multifunctional phosphorescent iridium(iii) complex for specific nucleus staining and hypoxia monitoring. Chem. Commun. 2015, 51, 7943–7946. [Google Scholar] [CrossRef] [PubMed]
- Gill, M.R.; Garcia-Lara, J.; Foster, S.J.; Smythe, C.; Battaglia, G.; Thomas, J.A. A ruthenium(ii) polypyridyl complex for direct imaging of DNA structure in living cells. Nat. Chem. 2009, 1, 662–667. [Google Scholar] [CrossRef]
- Nakamura, A.; Takigawa, K.; Kurishita, Y.; Kuwata, K.; Ishida, M.; Shimoda, Y.; Hamachi, I.; Tsukiji, S. Hoechst tagging: A modular strategy to design synthetic fluorescent probes for live-cell nucleus imaging. Chem. Commun. 2014, 50, 6149–6152. [Google Scholar] [CrossRef] [PubMed]
- Lukinavičius, G.; Blaukopf, C.; Pershagen, E.; Schena, A.; Reymond, L.; Derivery, E.; Gonzalez-Gaitan, M.; D’Este, E.; Hell, S.W.; Wolfram Gerlich, D.; et al. SiR–Hoechst is a far-red DNA stain for live-cell nanoscopy. Nat. Commun. 2015, 6, 8497. [Google Scholar] [CrossRef]
- Bucevičius, J.; Keller-Findeisen, J.; Gilat, T.; Hell, S.W.; Lukinavičius, G. Rhodamine–Hoechst positional isomers for highly efficient staining of heterochromatin. Chem. Sci. 2019, 10, 1962–1970. [Google Scholar] [CrossRef]
- Zhang, X.; Ye, Z.; Zhang, X.; Man, H.; Huang, Z.; Li, N.; Xiao, Y. A targetable fluorescent probe for dSTORM super-resolution imaging of live cell nucleus DNA. Chem. Commun. 2019, 55, 1951–1954. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Wu, T.; Fan, J.; Wang, J.; Zhang, S.; Song, F.; Sun, S. An effective minor groove binder as a red fluorescent marker for live-cell DNA imaging and quantification. Angew. Chem. Int. Ed. 2011, 50, 4180–4183. [Google Scholar] [CrossRef]
- Zhang, S.; Fan, J.; Li, Z.; Hao, N.; Cao, J.; Wu, T.; Wang, J.; Peng, X. A bright red fluorescent cyanine dye for live-cell nucleic acid imaging, with high photostability and a large Stokes shift. J. Mater. Chem. B 2014, 2, 2688–2693. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Deng, Q.; Hu, D.-P.; Wang, Z.-Y.; Huang, B.-H.; Du, Z.-Y.; Fang, Y.-X.; Wong, W.-L.; Zhang, K.; Chow, C.-F. A molecular fluorescent dye for specific staining and imaging of RNA in live cells: A novel ligand integration from classical thiazole orange and styryl compounds. Chem. Commun. 2015, 51, 15241–15244. [Google Scholar] [CrossRef]
- Kurutos, A.; Ilic-Tomic, T.; Kamounah, F.S.; Vasilev, A.A.; Nikodinovic-Runic, J. Non-cytotoxic photostable monomethine cyanine platforms: Combined paradigm of nucleic acid staining and in vivo imaging. J. Photochem. Photobiol. A 2020, 397, 112598. [Google Scholar] [CrossRef]
- Kurutos, A.; Nikodinovic-Runic, J.; Veselinovic, A.; Veselinović, J.B.; Kamounah, F.S.; Ilic-Tomic, T. RNA-targeting low-molecular-weight fluorophores for nucleoli staining: Synthesis, in silico modelling and cellular imaging. New J. Chem. 2021, 45, 12818–12829. [Google Scholar] [CrossRef]
- Aristova, D.; Kosach, V.; Chernii, S.; Slominsky, Y.; Balanda, A.; Filonenko, V.; Yarmoluk, S.; Rotaru, A.; Özkan, H.G.; Mokhir, A.; et al. Monomethine cyanine probes for visualization of cellular RNA by fluorescence microscopy. Methods Appl. Fluoresc. 2021, 9, 045002. [Google Scholar] [CrossRef]
- Aristova, D.; Selin, R.; Heil, H.S.; Kosach, V.; Slominsky, Y.; Yarmoluk, S.; Pekhnyo, V.; Kovalska, V.; Henriques, R.; Mokhir, A.; et al. Trimethine cyanine dyes as NA-sensitive probes for visualization of cell compartments in fluorescence microscopy. ACS Omega 2022, 7, 47734–47746. [Google Scholar] [CrossRef]
- Zhou, B.; Liu, W.; Zhang, H.; Wu, J.; Liu, S.; Xu, H.; Wang, P. Imaging of nucleolar RNA in living cells using a highly photostable deep-red fluorescent probe. Biosens. Bioelectron. 2015, 68, 189–196. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, B.; Niu, G.; Ge, J.; Wu, J.; Zhang, H.; Xu, H.; Wang, P. Deep-red emissive crescent-shaped fluorescent dyes: Substituent effect on live cell imaging. ACS Appl. Mater. Interfaces 2015, 7, 7421–7427. [Google Scholar] [CrossRef] [PubMed]
- Boynton, A.N.; Marcélis, L.; Barton, J.K. [Ru(Me4phen)2dppz]2+, a light switch for DNA mismatches. J. Am. Chem. Soc. 2016, 138, 5020–5023. [Google Scholar] [CrossRef] [PubMed]
- Boynton, A.N.; Marcélis, L.; McConnell, A.J.; Barton, J.K. A ruthenium(ii) complex as a luminescent probe for DNA mismatches and abasic sites. Inorg. Chem. 2017, 56, 8381–8389. [Google Scholar] [CrossRef] [PubMed]
- Nano, A.; Boynton, A.N.; Barton, J.K. A rhodium-cyanine fluorescent probe: Detection and signaling of mismatches in DNA. J. Am. Chem. Soc. 2017, 139, 17301–17304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Lei, Y. Fluorescent carbon dots and their sensing applications. TrAC Trends Anal. Chem. 2017, 89, 163–180. [Google Scholar] [CrossRef]
- Hong, G.; Diao, S.; Antaris, A.L.; Dai, H. Carbon nanomaterials for biological imaging and nanomedicinal therapy. Chem. Rev. 2015, 115, 10816–10906. [Google Scholar] [CrossRef]
- Serdiuk, T.; Lysenko, V.; Mognetti, B.; Skryshevsky, V.; Géloën, A. Impact of cell division on intracellular uptake and nuclear targeting with fluorescent SiC-based nanoparticles. J. Biophotonics 2013, 6, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Zhao, J.; Zhang, R.; Tian, X.; Liu, Z.; Wang, A.; Liu, R.; Liu, B.; Han, M.-Y.; Gao, X.; et al. Membrane-penetrating carbon quantum dots for imaging nucleic acid structures in live organisms. Angew. Chem. Int. Ed. 2019, 58, 7087–7091. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.J.; Wu, P.L.; Bolze, F.; Leung, H.W.C.; Li, K.F.; Mak, N.K.; Kwong, D.W.J.; Nicoud, J.-F.; Cheah, K.W.; Wong, M.S. Cyanines as new fluorescent probes for DNA detection and two-photon excited bioimaging. Org. Lett. 2010, 12, 2194–2197. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-C.; Zheng, M.-L.; Chen, S.; Zhao, Z.-S.; Duan, X.-M. Biscarbazolylmethane-based cyanine: A two-photon excited fluorescent probe for DNA and selective cell imaging. J. Mater. Chem. B 2014, 2, 2301–2310. [Google Scholar] [CrossRef] [PubMed]
- Allain, C.; Schmidt, F.; Lartia, R.; Bordeau, G.; Fiorini-Debuisschert, C.; Charra, F.; Tauc, P.; Teulade-Fichou, M.-P. Vinyl-pyridinium triphenylamines: Novel far-red emitters with high photostability and two-photon absorption properties for staining DNA. Chembiochem 2007, 8, 424–433. [Google Scholar] [CrossRef]
- Gaur, P.; Kumar, A.; Dey, G.; Kumar, R.; Bhattacharyya, S.; Ghosh, S. Selenium incorporated cationic organochalcogen: Live cell compatible and highly photostable molecular stain for imaging and localization of intracellular DNA. ACS Appl. Mater. Interfaces 2016, 8, 10690–10699. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zou, Y.; Li, C.; Sicking, W.; Piantanida, I.; Yi, T.; Schmuck, C. A molecular peptide beacon for the ratiometric sensing of nucleic acids. J. Am. Chem. Soc. 2012, 134, 1958–1961. [Google Scholar] [CrossRef]
- Liu, L.-Y.; Zhao, Y.; Zhang, N.; Wang, K.-N.; Tian, M.; Pan, Q.; Lin, W. Ratiometric fluorescence imaging for the distribution of nucleic acid content in living cells and human tissue sections. Anal. Chem. 2021, 93, 1612–1619. [Google Scholar] [CrossRef]
- Suseela, Y.V.; Narayanaswamy, N.; Pratihar, S.; Govindaraju, T. Far-red fluorescent probes for canonical and non-canonical nucleic acid structures: Current progress and future implications. Chem. Soc. Rev. 2018, 47, 1098–1131. [Google Scholar] [CrossRef]
- Yao, Q.; Li, H.; Xian, L.; Xu, F.; Xia, J.; Fan, J.; Du, J.; Wang, J.; Peng, X. Differentiating RNA from DNA by a molecular fluorescent probe based on the “door-bolt” mechanism biomaterials. Biomaterials 2018, 177, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, Y.; Sato, Y.; Nishizawa, S. Deep-red light-up signaling of benzo[c,d]indole–quinoline monomethine cyanine for imaging of nucleolar RNA in living cells and for sequence-selective RNA analysis. Anal. Chem. 2019, 91, 14254–14260. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Wei, P.; Li, R.; Zhong, Y.; Li, X.; Xue, F.; Shi, Y.; Yi, T. Ribosomal RNA-selective light-up fluorescent probe for rapidly imaging the nucleolus in live cells. ACS Sens. 2019, 4, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Niu, J.; Wang, W.; Lin, W. Tracking of mitochondrial endogenous ribonucleic acid in the cancer cells and macrophages using a novel small-molecular fluorescent probe. Anal. Chem. 2019, 91, 1715–1718. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, W.; Sun, Y.; Song, G.; Miao, F.; Guo, F.; Tian, M.; Yu, X.; Sun, J.Z. Two-photon fluorescence imaging of RNA in nucleoli and cytoplasm in living cells based on low molecular weight probes. Dyes Pigm. 2014, 103, 191–201. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, S.; Zhang, C.; Dong, J.; Shen, C.; Zhu, J.; Xu, H.; Fu, M.; Yang, G.; Zhang, X. A water-soluble two-photon ratiometric triarylboron probe with nucleolar targeting by preferential RNA binding. Chem. Commun. 2017, 53, 11476–11479. [Google Scholar] [CrossRef]
- Lu, Y.-J.; Deng, Q.; Hou, J.-Q.; Hu, D.-P.; Wang, Z.-Y.; Zhang, K.; Luyt, L.G.; Wong, W.-L.; Chow, C.-F. Molecular engineering of thiazole orange dye: Change of fluorescent signaling from universal to specific upon binding with nucleic acids in bioassay. ACS Chem. Biol. 2016, 11, 1019–1029. [Google Scholar] [CrossRef]
- Grande, V.; Doria, F.; Freccero, M.; Würthner, F. An aggregating amphiphilic squaraine: A light-up probe that discriminates parallel G-quadruplexes. Angew. Chem. Int. Ed. 2017, 56, 7520–7524. [Google Scholar] [CrossRef]
- Grande, V.; Shen, C.-A.; Deiana, M.; Dudek, M.; Olesiak-Banska, J.; Matczyszyn, K.; Würthner, F. Selective parallel G-quadruplex recognition by a NIR-to-NIR two-photon squaraine. Chem. Sci. 2018, 9, 8375–8381. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zuo, X.; Fan, C.; Zhang, X.-E. Biomacromolecular nanostructures-based interfacial engineering: From precise assembly to precision biosensing. Natl. Sci. Rev. 2018, 5, 740–755. [Google Scholar] [CrossRef]
- Choi, J.; Majima, T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011, 40, 5893–5909. [Google Scholar] [CrossRef] [PubMed]
- Stelson, A.C.; Liu, M.; Little, C.A.E.; Long, C.J.; Orloff, N.D.; Stephanopoulos, N.; Booth, J.C. Label-free detection of conformational changes in switchable DNA nanostructures with microwave microfluidics. Nat. Commun. 2019, 10, 1174. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Huang, Q.; Qu, Z.; Li, C.; Li, Q.; Shi, J.; Fan, C.; Wang, L.; Zuo, X.; Shen, J.; et al. Probing transient DNA conformation changes with an intercalative fluorescent excimer. Angew. Chem. Int. Ed. 2021, 60, 6624–6630. [Google Scholar] [CrossRef]
- Ye, Z.; Yu, H.; Yang, W.; Zheng, Y.; Li, N.; Bian, H.; Wang, Z.; Liu, Q.; Song, Y.; Zhang, M.; et al. Strategy to lengthen the on-time of photochromic rhodamine spirolactam for super-resolution photoactivated localization microscopy. J. Am. Chem. Soc. 2019, 141, 6527–6536. [Google Scholar] [CrossRef]
- Tsankova, N.; Renthal, W.; Kumar, A.; Nestler, E.J. Epigenetic regulation in psychiatric disorders. Nat. Rev. Neurosci. 2007, 8, 355–367. [Google Scholar] [CrossRef]
- Minoshima, M.; Matsumoto, T.; Kikuchi, K. Development of a fluorogenic probe based on a DNA staining dye for continuous monitoring of the histone deacetylase reaction. Anal. Chem. 2014, 86, 7925–7930. [Google Scholar] [CrossRef]
- Rooker, D.R.; Buccella, D. Real-time detection of histone deacetylase activity with a small molecule fluorescent and spectrophotometric probe. Chem. Sci. 2015, 6, 6456–6461. [Google Scholar] [CrossRef]
- Chao, D.; Ni, S. Nanomolar pyrophosphate detection and nucleus staining in living cells with simple terpyridine–Zn(ii) complexes. Sci. Rep. 2016, 6, 26477. [Google Scholar] [CrossRef] [Green Version]
- Clayson, D.B.; Mehta, R.; Iverson, F. Oxidative DNA damage—The effects of certain genotoxic and operationally non-genotoxic carcinogens. Mutat. Res./Rev. Gene. Toxic. 1994, 317, 25–42. [Google Scholar] [CrossRef] [PubMed]
- Loft, S.; Poulsen, H.E. Cancer risk and oxidative DNA damage in man. J. Mol. Med. 1996, 74, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.C.; Tang, Y.; Chang, Z.; Chang, C.J. A nuclear-localized fluorescent hydrogen peroxide probe for monitoring sirtuin-mediated oxidative stress responses in vivo. Chem. Biol. 2011, 18, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Liu, K.; Yang, H.; Li, Y.; Lan, H.; Liu, Y.; Zhang, X.; Yi, T. A highly sensitive ratiometric fluorescent probe for the detection of cytoplasmic and nuclear hydrogen peroxide. Anal. Chem. 2014, 86, 9970–9976. [Google Scholar] [CrossRef]
- Ozmen, B.; Akkaya, E.U. Infrared fluorescence sensing of submicromolar calcium: Pushing the limits of photoinduced electron transfer. Tetrahedron Lett. 2000, 41, 9185–9188. [Google Scholar] [CrossRef]
- Zhu, B.; Jia, H.; Zhang, X.; Chen, Y.; Liu, H.; Tan, W. Engineering a subcellular targetable, red-emitting, and ratiometric fluorescent probe for Ca2+ and its bioimaging applications. Anal. Bioanal. Chem. 2010, 397, 1245–1250. [Google Scholar] [CrossRef]
- Li, W.-H.; Fraser, S.E.; Meade, T.J. A calcium-sensitive magnetic resonance imaging contrast agent. J. Am. Chem. Soc. 1999, 121, 1413–1414. [Google Scholar] [CrossRef]
- Tatay, S.; Gaviña, P.; Coronado, E.; Palomares, E. Optical mercury sensing using a benzothiazolium hemicyanine dye. Org. Lett. 2006, 8, 3857–3860. [Google Scholar] [CrossRef]
- Wang, K.-N.; Liu, L.-Y.; Mao, D.; Hou, M.-X.; Tan, C.-P.; Mao, Z.-W.; Liu, B. A nuclear-targeted AIE photosensitizer for enzyme inhibition and photosensitization in cancer cell ablation. Angew. Chem. Int. Ed. 2022, 61, e202114600. [Google Scholar]
- Zhang, P.; Huang, H.; Banerjee, S.; Clarkson, G.J.; Ge, C.; Imberti, C.; Sadler, P.J. Nucleus-targeted organoiridium–albumin conjugate for photodynamic cancer therapy. Angew. Chem. Int. Ed. 2019, 58, 2350–2354. [Google Scholar] [CrossRef]
- Sharma, B.R. Infection in patients with severe burns: Causes and prevention thereof. Infect. Dis. Clin. N. Am. 2007, 21, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, X.; He, X.; He, Z.; Yang, X.; Tian, S.; Meng, F.; Ding, D.; Luo, L.; Tang, B.Z. A dual-functional photosensitizer for ultraefficient photodynamic therapy and synchronous anticancer efficacy monitoring. Adv. Funct. Mater. 2019, 29, 1902673. [Google Scholar] [CrossRef]
- Zhang, T.; Li, Y.; Zheng, Z.; Ye, R.; Zhang, Y.; Kwok, R.T.K.; Lam, J.W.Y.; Tang, B.Z. In situ monitoring apoptosis process by a self-reporting photosensitizer. J. Am. Chem. Soc. 2019, 141, 5612–5616. [Google Scholar] [CrossRef] [PubMed]
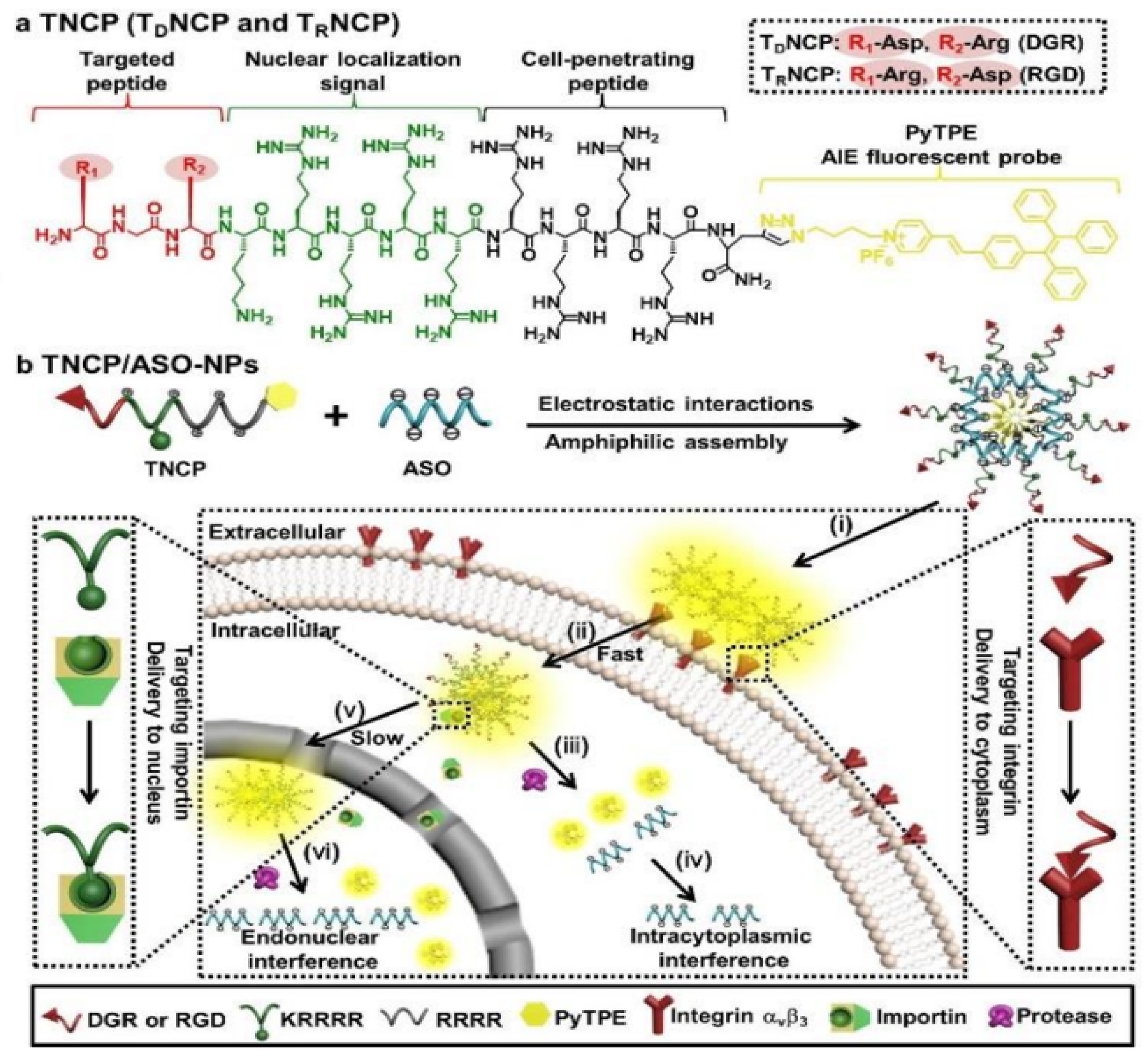
- Cheng, Y.; Sun, C.; Liu, R.; Yang, J.; Dai, J.; Zhai, T.; Lou, X.; Xia, F. A multifunctional peptide-conjugated AIEgen for efficient and sequential targeted gene delivery into the nucleus. Angew. Chem. Int. Ed. 2019, 58, 5049–5053. [Google Scholar] [CrossRef]
- Burke, C.S.; Byrne, A.; Keyes, T.E. Targeting photoinduced DNA destruction by Ru(ii) tetraazaphenanthrene in live cells by signal peptide. J. Am. Chem. Soc. 2018, 140, 6945–6955. [Google Scholar] [CrossRef]
- Song, H.; Allison, S.J.; Brabec, V.; Bridgewater, H.E.; Kasparkova, J.; Kostrhunova, H.; Novohradsky, V.; Phillips, R.M.; Pracharova, J.; Rogers, N.J.; et al. Glycoconjugated metallohelices have improved nuclear delivery and suppress tumour growth in vivo. Angew. Chem. Int. Ed. 2020, 59, 14677–14685. [Google Scholar] [CrossRef]
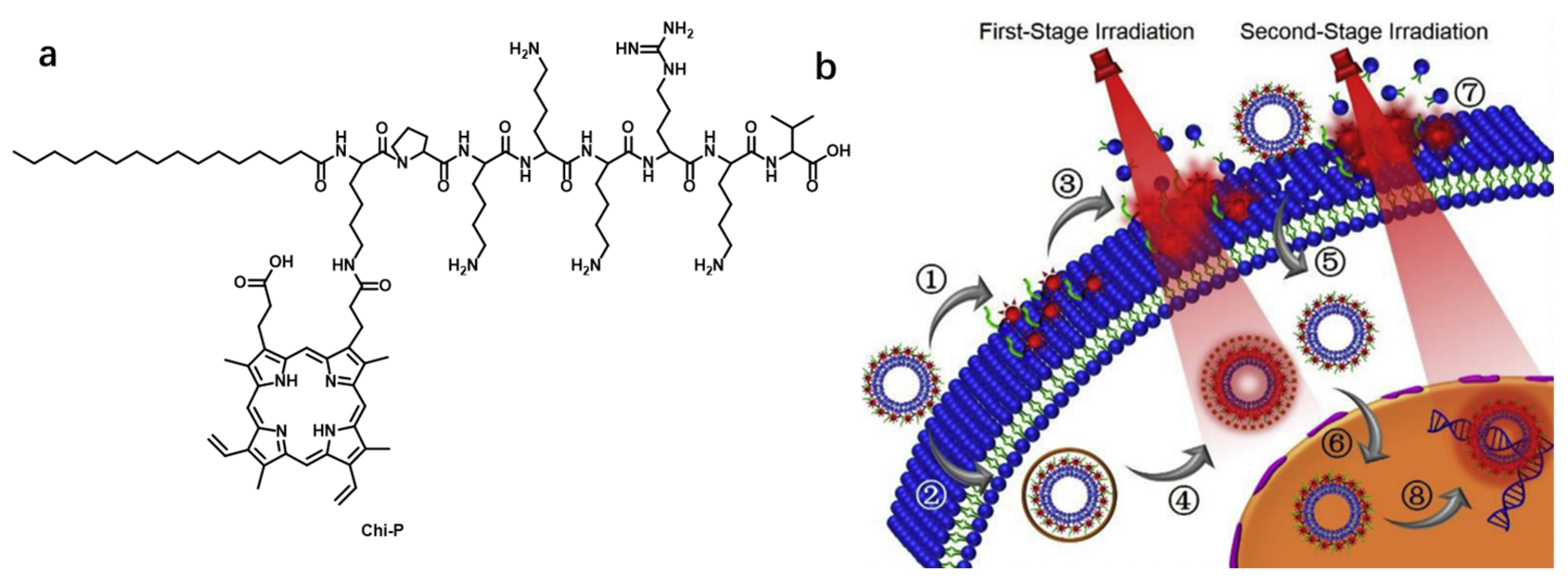
- Cheng, H.; Fan, J.-H.; Zhao, L.-P.; Fan, G.-L.; Zheng, R.-R.; Qiu, X.-Z.; Yu, X.-Y.; Li, S.-Y.; Zhang, X.-Z. Chimeric peptide engineered exosomes for dual-stage light guided plasma membrane and nucleus targeted photodynamic therapy. Biomaterials 2019, 211, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; He, Q.; Liu, J.; Chen, Y.; Ma, M.; Zhang, L.; Shi, J. Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J. Am. Chem. Soc. 2012, 134, 5722–5725. [Google Scholar] [CrossRef]
- KC, R.B.; Thapa, B.; Xu, P. pH and redox dual responsive nanoparticle for nuclear targeted drug delivery. Mol. Pharm. 2012, 9, 2719–2729. [Google Scholar]
- Yang, Y.; Zhu, W.; Feng, L.; Chao, Y.; Yi, X.; Dong, Z.; Yang, K.; Tan, W.; Liu, Z.; Chen, M. G-quadruplex-based nanoscale coordination polymers to modulate tumor hypoxia and achieve nuclear-targeted drug delivery for enhanced photodynamic therapy. Nano Lett. 2018, 18, 6867–6875. [Google Scholar] [CrossRef]
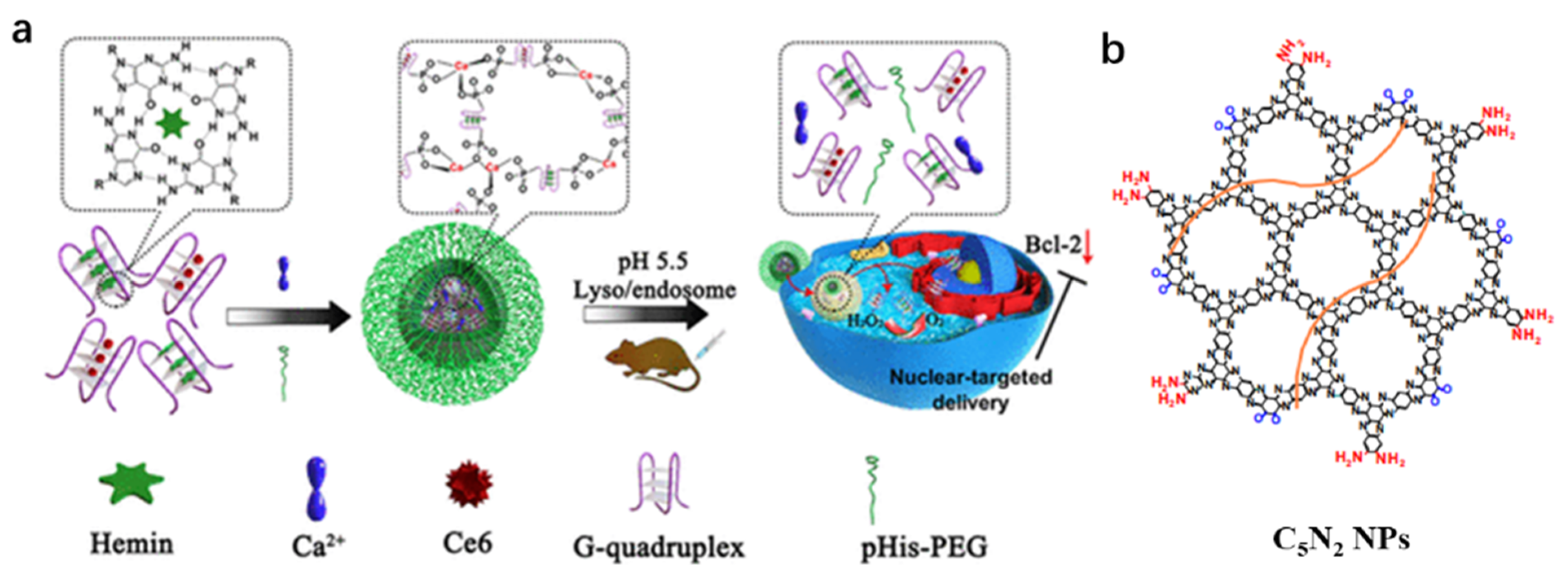
- Chen, W.; Liu, J.; Wang, Y.; Jiang, C.; Yu, B.; Sun, Z.; Lu, L. A C5N2 nanoparticle based direct nucleus delivery platform for synergistic cancer therapy. Angew. Chem. Int. Ed. 2019, 58, 6290–6294. [Google Scholar] [CrossRef]
- Lei, Z.; Ding, L.; Yao, C.; Mo, F.; Li, C.; Huang, Y.; Yin, X.; Li, M.; Liu, J.; Zhang, Y.; et al. A highly efficient tumor-targeting nanoprobe with a novel cell membrane permeability mechanism. Adv. Mater. 2019, 31, 1807456. [Google Scholar] [CrossRef]




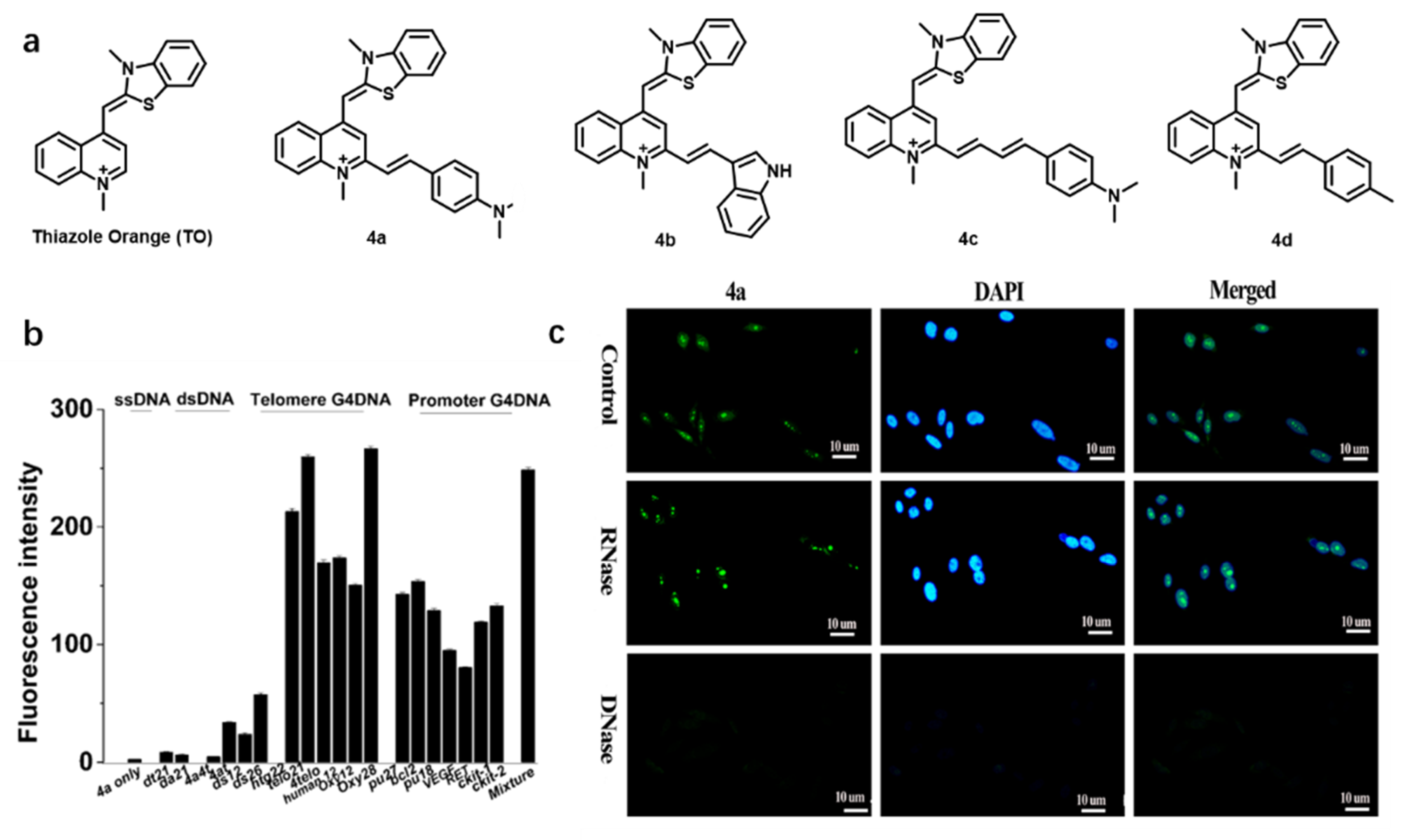
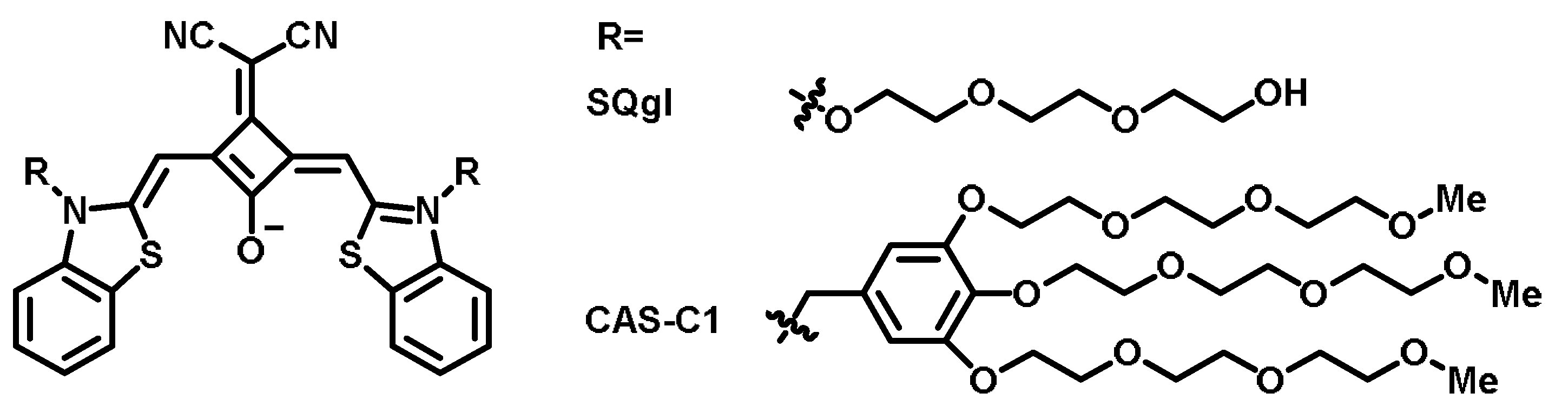
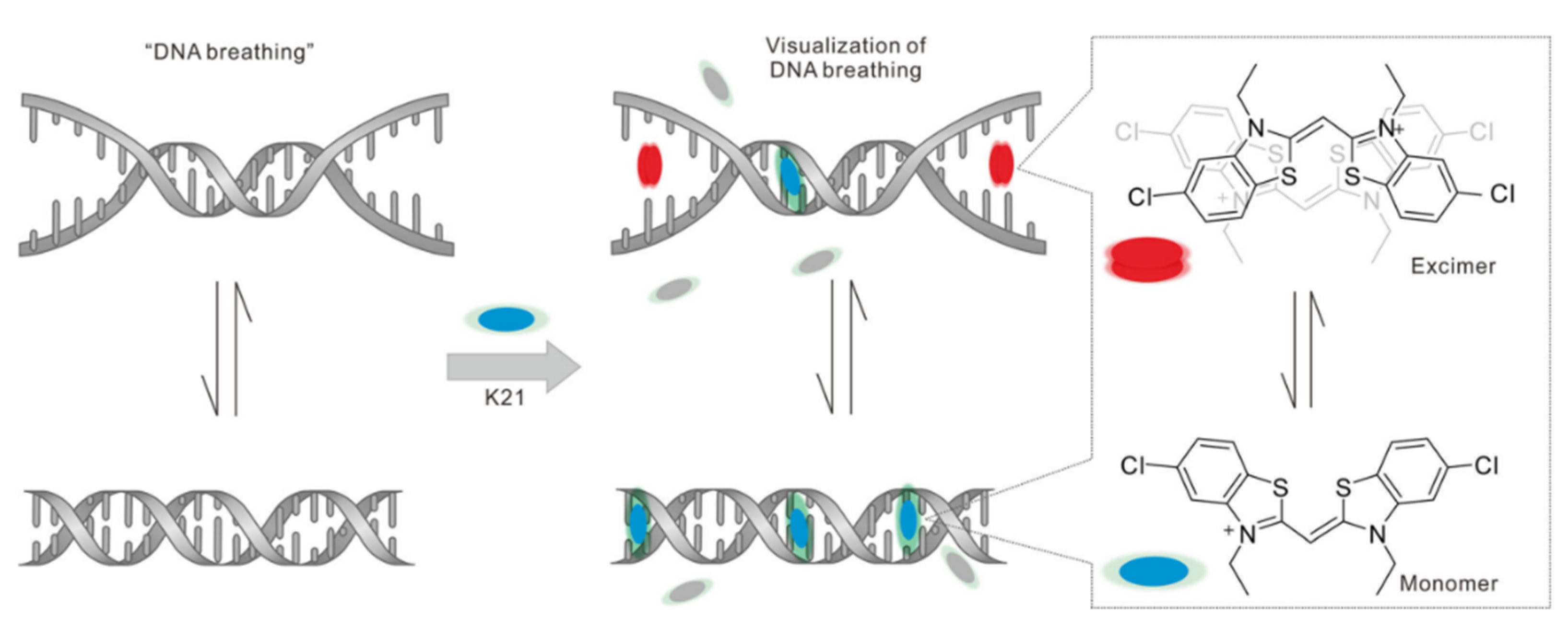

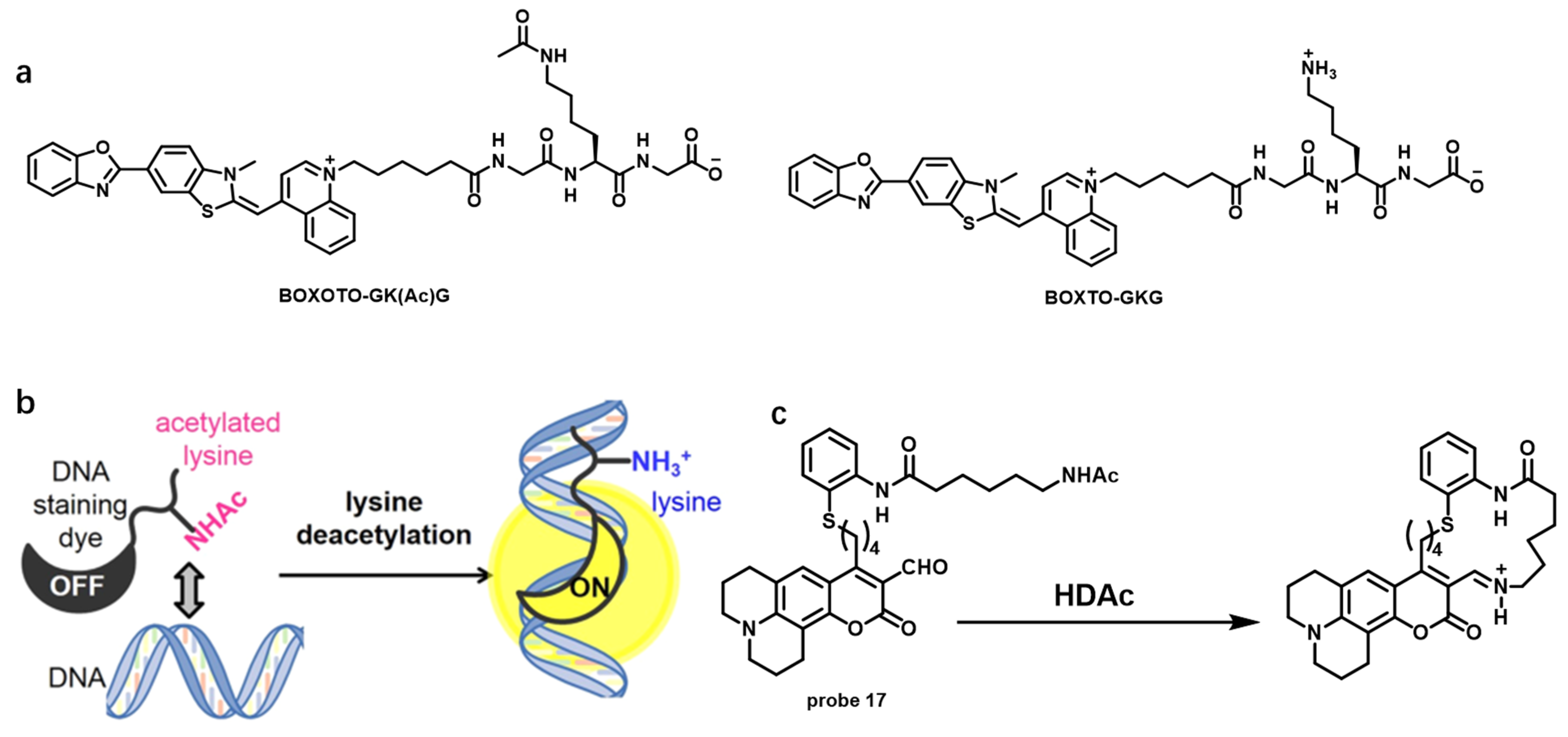





| Name [Ref.] | λex (nm) | λem (nm) | Stokes Shift (nm) | Binding Constant | Fluorescent Quantum Yield | Species of Detection | Extinction Coefficient |
|---|---|---|---|---|---|---|---|
| probe 8 [45] | 451 | 548 | 97 | Kb = 2 × 106 M−1 (DNA d[A]10:d[T]10) | 0.13 | DNA | — |
| probe 9 [46] | 430 | 585 | 155 | — | 0.024 | AT-rich DNA | 26,700 M−1 cm−1 |
| probe 10 [47] | 475 | 636 | 161 | — | 0.090 | DNA | 28,500 M−1 cm−1 |
| probe 11 [47] | 491 | 656 | 165 | — | 0.11 | DNA | 37,400 M−1 cm−1 |
| probe 12 [47] | 491 | 665 | 174 | — | 0.12 | DNA | 66,000 M−1 cm−1 |
| PA1 (probe 13) [48] | 473 | 522 | 49 | 7.3 × 102 M−1 | 0.008 | DNA | 41,200 M−1 cm−1 |
| PA2 (probe 13) | 480 | 560 | 80 | 5.9 × 102 M−1 | 0.009 | DNA | 38,700 M−1 cm−1 |
| PA3 (probe 13) | 454 | 490 | 36 | 1.2 × 103 M−1 | 0.010 | DNA | 38,600 M−1 cm−1 |
| PA4 (probe 13) | 481 | 520 | 39 | 2.4 × 103 M−1 | 0.030 | DNA | 49,200 M−1 cm−1 |
| PA5 (probe 13) | 504 | 534 | 30 | 7.4 × 103 M−1 | 0.070 | DNA | 29,600 M−1 cm−1 |
| probe 14 [49] | 340 | 406; 490 | 66; 150 | 2 × 105 M−1 (p(dA·dT)2) 3 × 104 M−1 (p(dG·dC)2) | — | AT-rich DNA | — |
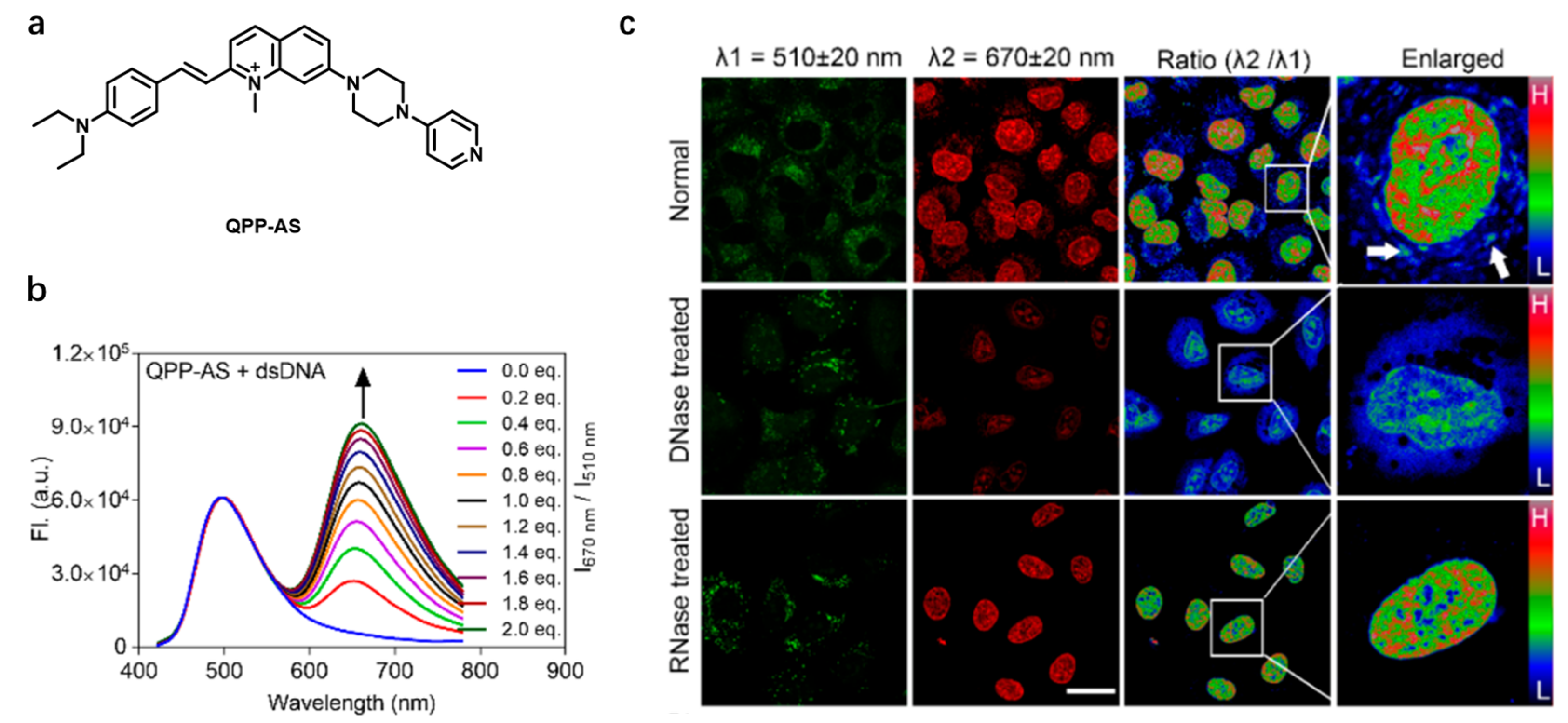
| QPP-AS [50] | 405; 488 (in DCM) | 500 and 650; 650 (in DCM) | — | — | — | DNA | 22,900 M−1 cm−1; 54,300 M−1 cm−1 (in DCM) |
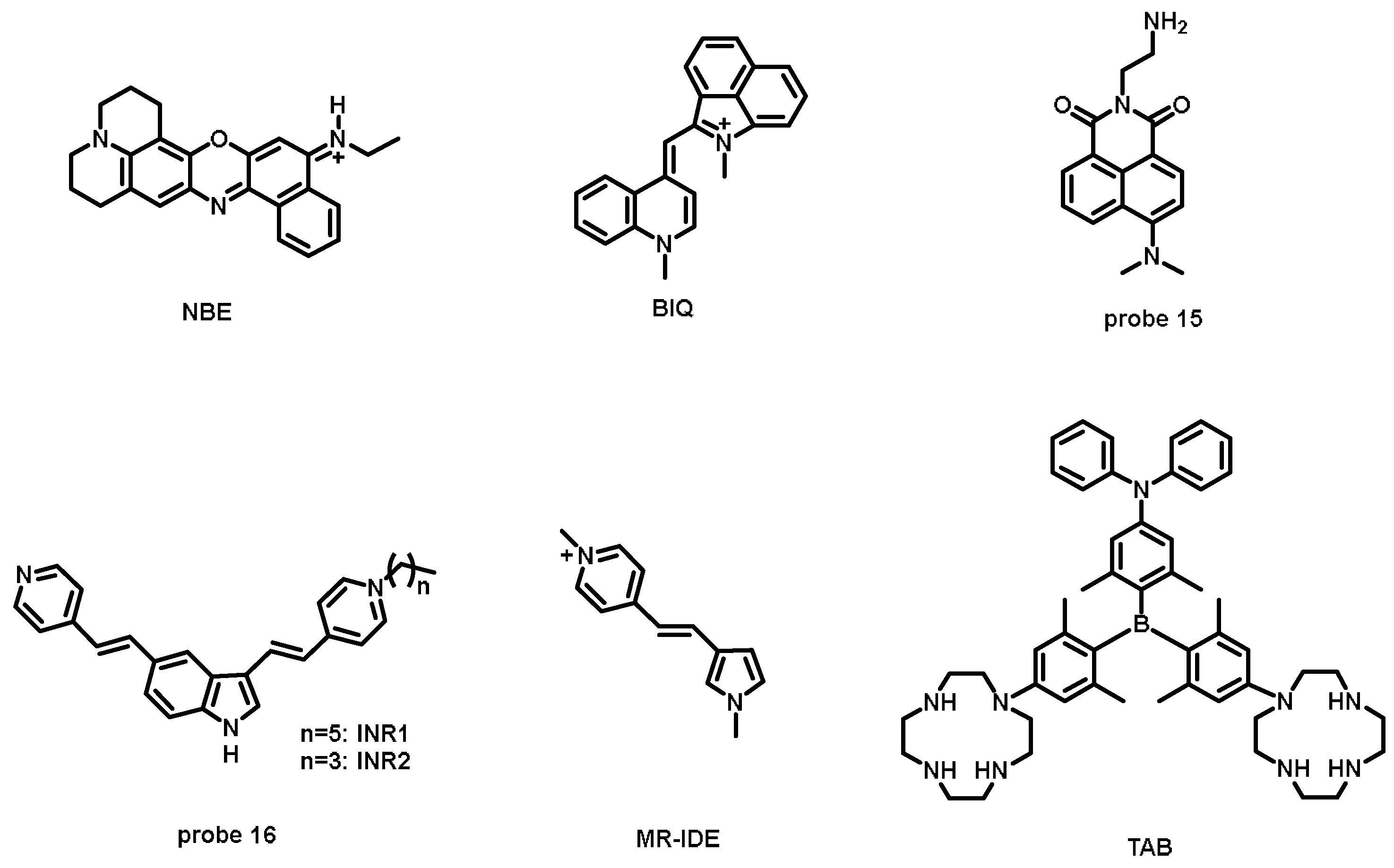
| NBE [52] | 618 | 680 | 62 | — | 0.040 | RNA | — |
| BIQ [53] | 611 | 657 | 46 | 1.2 × 108 M−1 | 0.010 | RNA | — |
| probe 15 [54] | 444 | 545 | 101 | — | 0.0378 | rRNA | 11,250 M−1·cm−1 |
| INR1 (probe 16) [56] | 467 | 533 (single-fluorescent peak); 540 (two-photo fluorescent peak) | 66; 73 | — | 0.028 | RNA | 25,000 M−1 cm−1 |
| INR2 (probe 16) [56] | 465 | 532 (single-fluorescent peak); 540 (two-photo fluorescent peak) | 67; 75 | — | 0.027 | RNA | 25,000 M−1 cm−1 |
| MR-IDE [55] | 438 (in DMF) | ~531 (in DMF) | ~93 | — | 0.0157 | mtRNA | — |
| TAB [57] | 405 | 560 | 155 | — | 0.500 (in DMSO) | RNA | — |
| 4a [58] | 475 | 630 | 155 | 9.65 × 105 M−1 | 0.170 | GQ | — |
| SQgI [59] | 660 | 744 | 84 | ~105 M−1 | 0.610 | Parallel GQ | — |
| CAS-C1 [60] | 698 | 720 | 22 | 9.7 × 106 M−1 | ~0.700 | Parallel GQ | ~90,000 M−1 cm−1 |
| K21 [65] | 426 | 480 | 54 | 6.32 × 106 M−1 (10AT dsDNA) | — | AT-rich DNA | — |
| Rh-Gly [66] | 365 | 585 | 220 | — | — | H2B | — |
| BOXTO-GK(Ac)G [68] | 520 | 545 | 25 | ~1.41 × 106 M−1 | — | HADCs | — |
| probe 17 [69] | 499 | 523 | 24 | — | — | HADCs | — |
| CZtpyZn [70] | 400 | 515 | 115 | — | — | PPi | — |
| NucPE1 [73] | 505 | 530 | 25 | — | 0.626 | H2O2 | 19,100 M−1 cm−1 |
| NP1 [74] | 446 | 555 | 109 | — | 0.087 | H2O2 | 10,820 M−1 cm−1 |
| pep-NP1 [74] | 403 | 551 | 148 | — | — | H2O2 | ~5900 M−1 cm−1 |
| STDBT [76] | 506 | 600 | 94 | 1.32 × 103 M−1 | — | Ca2+ | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.; Xu, S.; Song, Z.; Li, H.; Liu, H. Recent Advance in Nucleus-Targeted Fluorescent Probes for Bioimaging, Detection and Therapy. Chemosensors 2023, 11, 125. https://doi.org/10.3390/chemosensors11020125
Hu C, Xu S, Song Z, Li H, Liu H. Recent Advance in Nucleus-Targeted Fluorescent Probes for Bioimaging, Detection and Therapy. Chemosensors. 2023; 11(2):125. https://doi.org/10.3390/chemosensors11020125
Chicago/Turabian StyleHu, Cong, Shuai Xu, Zhiling Song, Haixia Li, and Hongwen Liu. 2023. "Recent Advance in Nucleus-Targeted Fluorescent Probes for Bioimaging, Detection and Therapy" Chemosensors 11, no. 2: 125. https://doi.org/10.3390/chemosensors11020125





