Methylene Blue-Modified Biochar from Sugarcane for the Simultaneous Electrochemical Detection of Four DNA Bases
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Instrumentation
2.3. Preparation of Methylene Blue-Modified Sugarcane Biochar (SC-MB)
2.4. Preparation of Graphite Paste Electrode (GPE) with SC-MB
2.5. Human Saliva Samples
3. Discussion
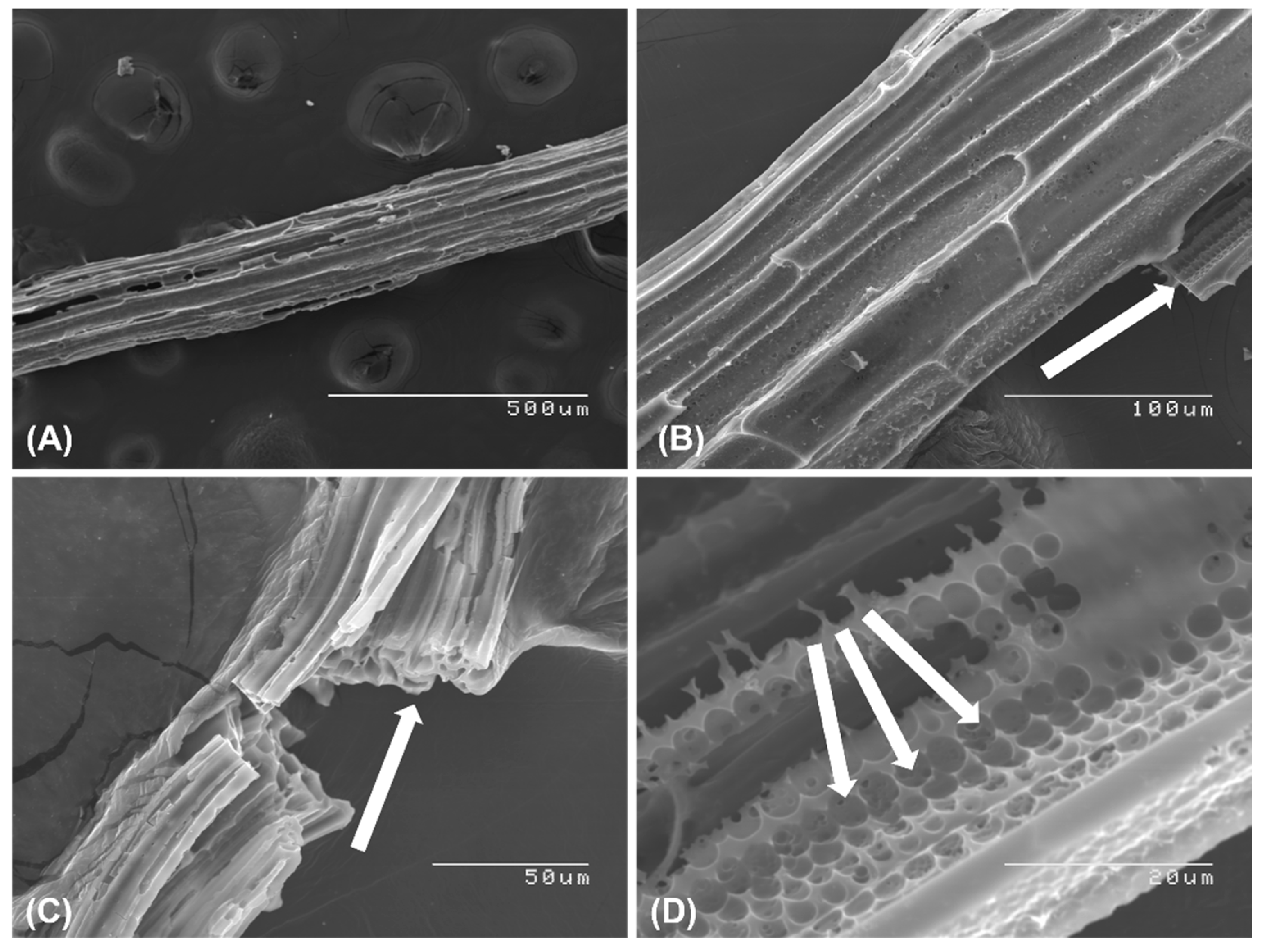
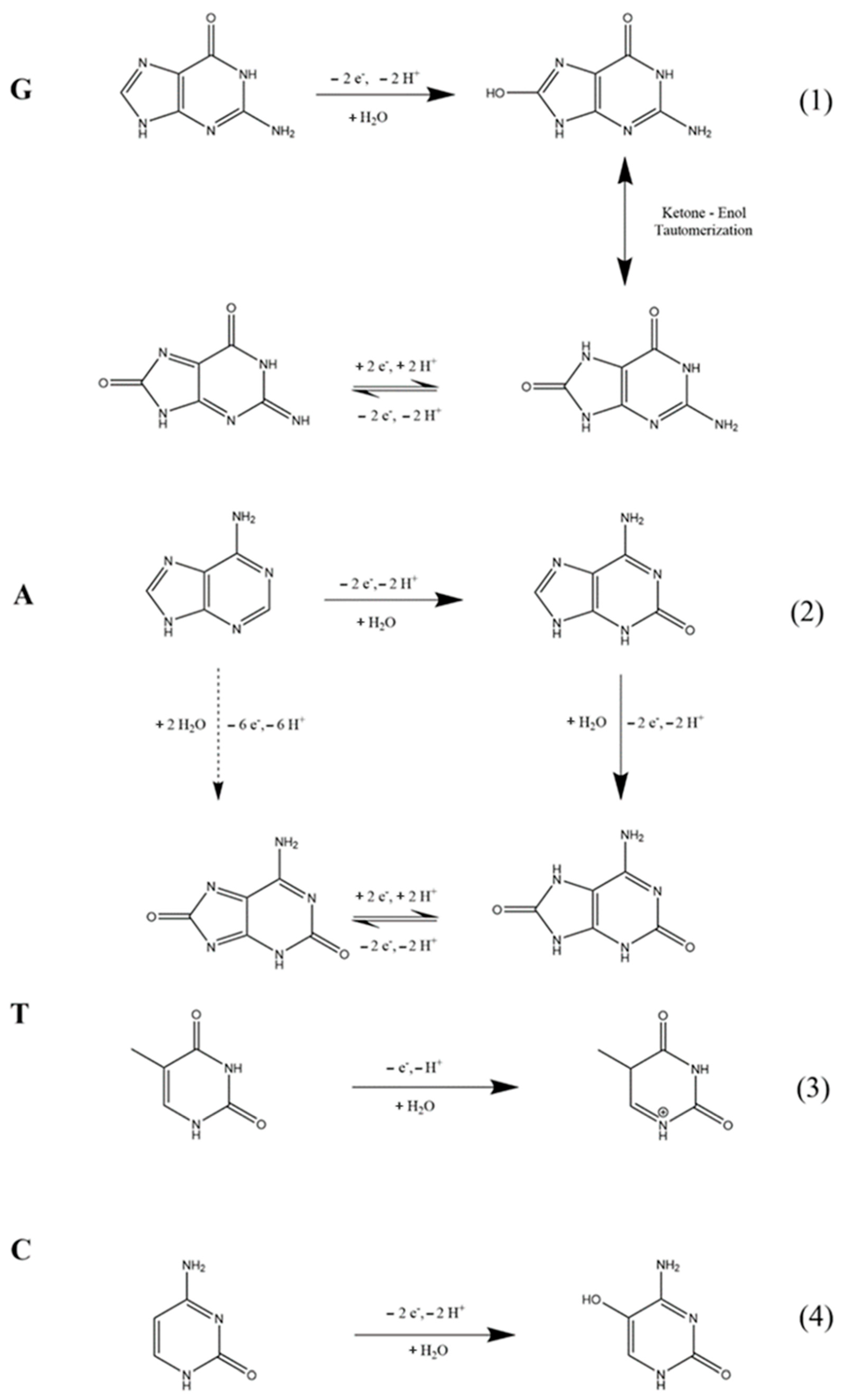
3.1. Characterization of GPE-SC-MB Nanocomposite
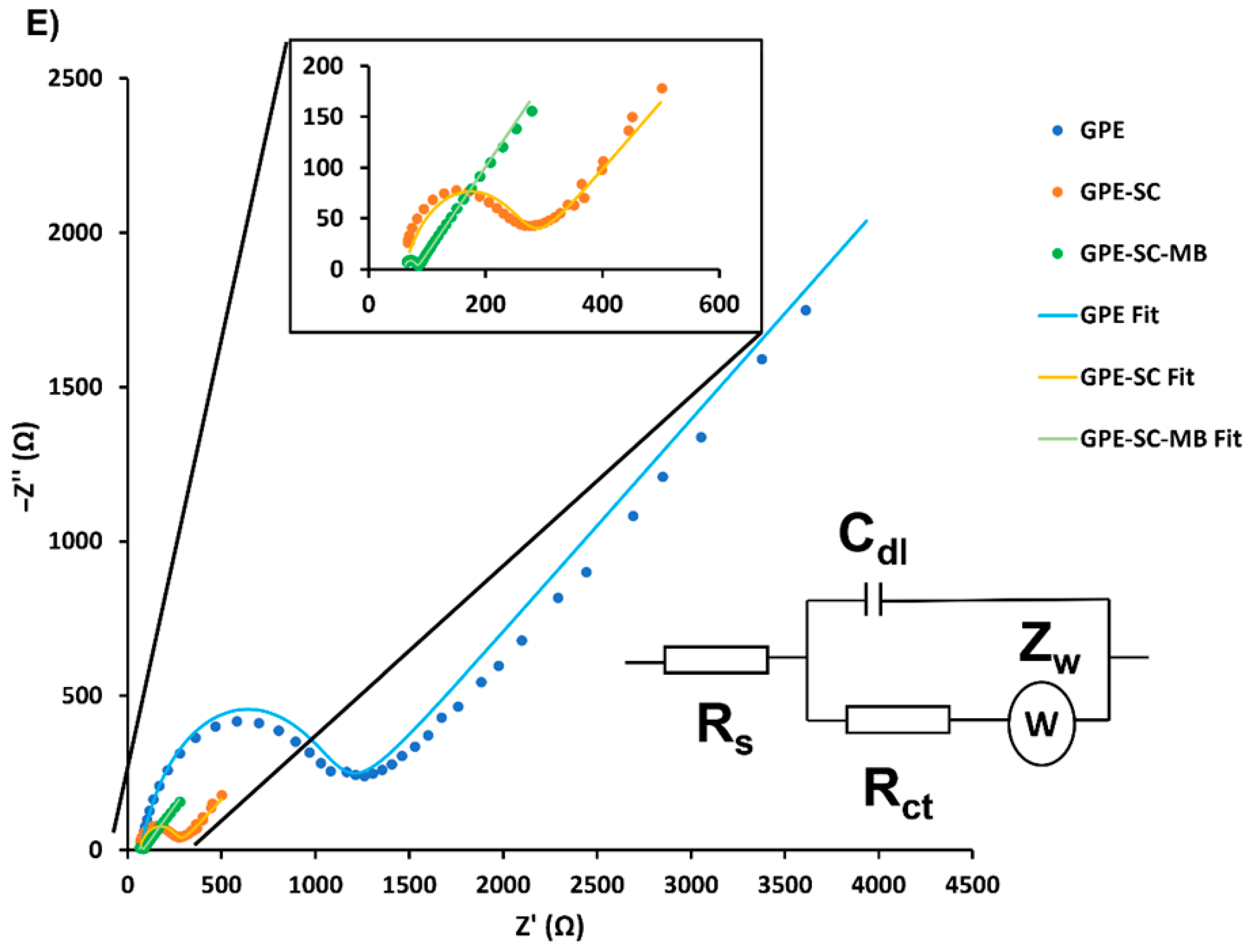
3.2. Comparison of the Modified Electrodes
3.3. pH Effect and Reproducibility
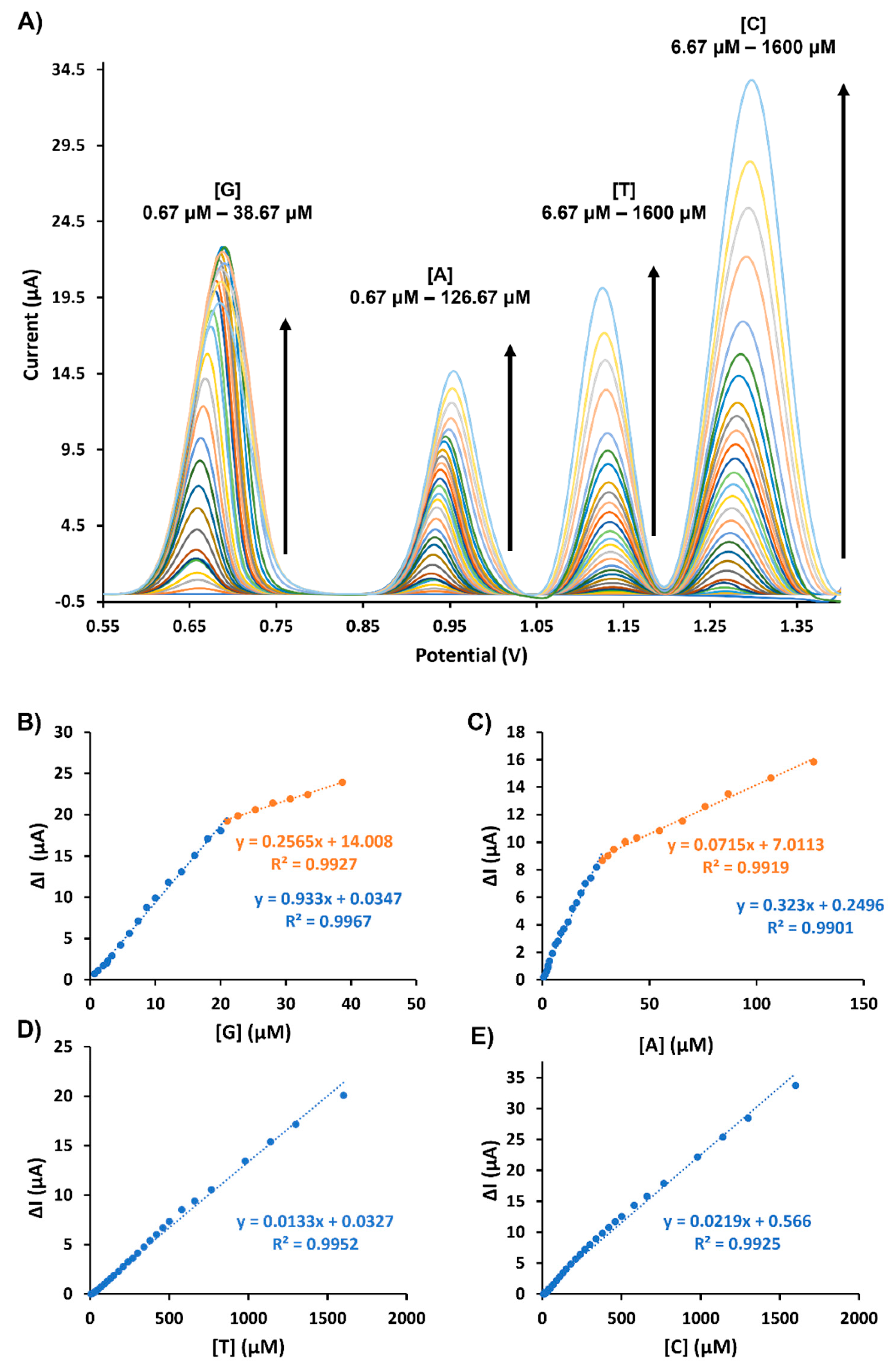
3.4. Calibration Plots for the Simultaneous Electrochemical Detection of DNA Bases
3.5. Scan Rate
3.6. Chronoamperometry
3.7. Real Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cobb, M. 60 Years Ago, Francis Crick Changed the Logic of Biology. PLoS Biol. 2017, 15, e2003243. [Google Scholar] [CrossRef] [PubMed]
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–562. [Google Scholar] [CrossRef]
- Soukup, G.A. Nucleic Acids: General Properties. In Encyclopedia of Life Sciences; Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar] [CrossRef]
- Crick, F.; Watson, J. Molecular Structure of Nucleic Acids. Nature 1953, 171, 737–738. [Google Scholar]
- Idzko, M.; Ferrari, D.; Riegel, A.K.; Eltzschig, H.K. Extracellular Nucleotide and Nucleoside Signaling in Vascular and Blood Disease. Blood 2014, 124, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Dudzinska, W.; Lubkowska, A.; Dolegowska, B.; Safranow, K.; Jakubowska, K. Adenine, Guanine and Pyridine Nucleotides in Blood during Physical Exercise and Restitution in Healthy Subjects. Eur. J. Appl. Physiol. 2010, 110, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, B.A.; Ashihara, H. Purine and Pyrimidine Nucleotide Synthesis and Metabolism. Arab. B. 2002, 1, e0018. [Google Scholar] [CrossRef]
- Peplinska-Miaskowska, J.; Wichowicz, H.; Smolenski, R.T.; Jablonska, P.; Kaska, L. Comparison of Plasma Nucleotide Metabolites and Amino Acids Pattern in Patients with Binge Eating Disorder and Obesity. Nucl. Nucl. Nucleic Acids 2020, 40, 32–42. [Google Scholar] [CrossRef]
- Kelley, R.E.; Andersson, H.C. Disorders of Purines and Pyrimidines, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 120. [Google Scholar]
- Fumagalli, M.; Lecca, D.; Abbracchio, M.P.; Ceruti, S. Pathophysiological Role of Purines and Pyrimidines in Neurodevelopment: Unveiling New Pharmacological Approaches to Congenital Brain Diseases. Front. Pharmacol. 2017, 8, 941. [Google Scholar] [CrossRef]
- Traut, T.W. Physiological Concentrations of Purines and Pyrimidines. Mol. Cell. Biochem. 1994, 140, 1–22. [Google Scholar] [CrossRef]
- Wang, M.; Guo, H.; Xue, R.; Guan, Q.; Zhang, J.; Zhang, T.; Sun, L.; Yang, F.; Yang, W. A Novel Electrochemical Sensor Based on MWCNTs-COOH/Metal-Covalent Organic Frameworks (MCOFs)/Co NPs for Highly Sensitive Determination of DNA Base. Microchem. J. 2021, 167, 106336. [Google Scholar] [CrossRef]
- Lu, N.; Liu, H.; Huang, R.; Gu, Y.; Yan, X.; Zhang, T.; Xu, Z.; Xu, H.; Xing, Y.; Song, Y.; et al. Charge Transfer Platform and Catalytic Amplification of Phenanthroimidazole Derivative: A New Strategy for DNA Bases Recognition. Anal. Chem. 2019, 91, 11938–11945. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ferrag, C.; Noroozifar, M.; Kerman, K. Simultaneous Determination of Four DNA Bases at Graphene Oxide/Multi-Walled Carbon Nanotube Nanocomposite-Modified Electrode. Micromachines 2020, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Kaiser, K.; Benner, R. A Simple High Performance Liquid Chromatography Method for the Measurement of Nucleobases and the RNA and DNA Content of Cellular Material. Limnol. Oceanogr. Methods 2012, 10, 608–616. [Google Scholar] [CrossRef]
- Muguruma, Y.; Tsutsui, H.; Akatsu, H.; Inoue, K. Comprehensive Quantification of Purine and Pyrimidine Metabolism in Alzheimer’s Disease Postmortem Cerebrospinal Fluid by LC–MS/MS with Metal-Free Column. Biomed. Chromatogr. 2020, 34. [Google Scholar] [CrossRef] [PubMed]
- Stentoft, C.; Vestergaard, M.; Løvendahl, P.; Kristensen, N.B.; Moorby, J.M.; Jensen, S.K. Simultaneous Quantification of Purine and Pyrimidine Bases, Nucleosides and Their Degradation Products in Bovine Blood Plasma by High Performance Liquid Chromatography Tandem Mass Spectrometry. J. Chromatogr. A 2014, 1356, 197–210. [Google Scholar] [CrossRef]
- Laourdakis, C.D.; Merino, E.F.; Neilson, A.P.; Cassera, M.B. Comprehensive Quantitative Analysis of Purines and Pyrimidines in the Human Malaria Parasite Using Ion-Pairing Ultra-Performance Liquid Chromatography-Mass Spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 967, 127–133. [Google Scholar] [CrossRef]
- Liu, C.; Gu, C.; Huang, W.; Sheng, X.; Du, J.; Li, Y. Targeted UPLC-MS/MS High-Throughput Metabolomics Approach to Assess the Purine and Pyrimidine Metabolism. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1113, 98–106. [Google Scholar] [CrossRef]
- Brohi, R.O.Z.; Khuhawar, M.Y.; Channa, A.; Laghari, A.J.; Abbasi, K. Gas Chromatographic Determination of Purines and Pyrimidines from DNA Using Ethyl Chloroformate as Derivatizing Reagent. Pak. J. Anal. Environ. Chem. 2016, 17, 50–57. [Google Scholar] [CrossRef]
- Brohi, R.O.Z.Z.; Khuhawar, M.Y.; Khuhawar, T.M.J. GC-FID Determination of Nucleobases Guanine, Adenine, Cytosine, and Thymine from DNA by Precolumn Derivatization with Isobutyl Chloroformate. J. Anal. Sci. Technol. 2016, 7, 5. [Google Scholar] [CrossRef]
- Wang, P.; Ren, J. Separation of Purine and Pyrimidine Bases by Capillary Electrophoresis Using β-Cyclodextrin as an Additive. J. Pharm. Biomed. Anal. 2004, 34, 277–283. [Google Scholar] [CrossRef]
- Friedeck, D.; Adam, T.; Bartk, P. Capillary Electrophoresis for Detection of Inherited Disorders of Purine and Pyrimidine Metabolism: A Selective Approach. Electrophoresis 2002, 23, 565–571. [Google Scholar] [CrossRef]
- Carrillo-Carrión, C.; Armenta, S.; Simonet, B.M.; Valcárcel, M.; Lendl, B. Determination of Pyrimidine and Purine Bases by Reversed-Phase Capillary Liquid Chromatography with at-Line Surface-Enhanced Raman Spectroscopic Detection Employing a Novel SERS Substrate Based on ZnS/CdSe Silver-Quantum Dots. Anal. Chem. 2011, 83, 9391–9398. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, L.F.; Bendich, A.; Tinker, J.F.; Brown, G.B. Ultraviolet Absorption Spectra of Purines, Pyrimidines, and Triazolopyrimidines. J. Am. Chem. Soc. 1948, 70, 3875–3880. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Brett, A.M.; Diculescu, V.; Piedade, J.A.P. Electrochemical Oxidation Mechanism of Guanine and Adenine Using a Glassy Carbon Microelectrode. Bioelectrochemistry 2002, 55, 61–62. [Google Scholar] [CrossRef]
- Oliveira-Brett, A.M.; Piedade, J.A.P.; Silva, L.A.; Diculescu, V.C. Voltammetric Determination of All DNA Nucleotides. Anal. Biochem. 2004, 332, 321–329. [Google Scholar] [CrossRef]
- Ng, K.L.; Khor, S.M. Graphite-Based Nanocomposite Electrochemical Sensor for Multiplex Detection of Adenine, Guanine, Thymine, and Cytosine: A Biomedical Prospect for Studying DNA Damage. Anal. Chem. 2017, 89, 10004–10012. [Google Scholar] [CrossRef]
- Anu Prathap, M.U.; Srivastava, R.; Satpati, B. Simultaneous Detection of Guanine, Adenine, Thymine, and Cytosine at Polyaniline/MnO2 Modified Electrode. Electrochim. Acta 2013, 114, 285–295. [Google Scholar] [CrossRef]
- Geng, X.; Bao, J.; Huang, T.; Wang, X.; Hou, C.; Hou, J.; Samalo, M.; Yang, M.; Huo, D. Electrochemical Sensor for the Simultaneous Detection of Guanine and Adenine Based on a PPyox/MWNTs-MoS 2 Modified Electrode. J. Electrochem. Soc. 2019, 166, B498–B504. [Google Scholar] [CrossRef]
- Wang, P.; Wu, H.; Dai, Z.; Zou, X. Simultaneous Detection of Guanine, Adenine, Thymine and Cytosine at Choline Monolayer Supported Multiwalled Carbon Nanotubes Film. Biosens. Bioelectron. 2011, 26, 3339–3345. [Google Scholar] [CrossRef]
- Sfragano, P.S.; Laschi, S.; Palchetti, I. Sustainable Printed Electrochemical Platforms for Greener Analytics. Front. Chem. 2020, 8, 644. [Google Scholar] [CrossRef]
- Geetha Bai, R.; Muthoosamy, K.; Zhou, M.; Ashokkumar, M.; Huang, N.M.; Manickam, S. Sonochemical and Sustainable Synthesis of Graphene-Gold (G-Au) Nanocomposites for Enzymeless and Selective Electrochemical Detection of Nitric Oxide. Biosens. Bioelectron. 2017, 87, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Kalambate, P.K.; Rao, Z.; Dhanjai, Z.; Wu, J.; Shen, Y.; Boddula, R.; Huang, Y. Electrochemical (Bio) Sensors Go Green. Biosens. Bioelectron. 2020, 163, 112270. [Google Scholar] [CrossRef] [PubMed]
- Baby, J.N.; Sriram, B.; Wang, S.F.; George, M. Effect of Various Deep Eutectic Solvents on the Sustainable Synthesis of MgFe2O4 Nanoparticles for Simultaneous Electrochemical Determination of Nitrofurantoin and 4-Nitrophenol. ACS Sustain. Chem. Eng. 2020, 8, 1479–1486. [Google Scholar] [CrossRef]
- Zaidi, S.A. Utilization of an Environmentally-Friendly Monomer for an Efficient and Sustainable Adrenaline Imprinted Electrochemical Sensor Using Graphene. Electrochim. Acta 2018, 274, 370–377. [Google Scholar] [CrossRef]
- Chaithra, C.; Bhat, V.S.; Akshaya, A.K.; Maiyalagan, T.; Hegde, G.; Varghese, A.; George, L. Unique Host Matrix to Disperse Pd Nanoparticles for Electrochemical Sensing of Morin: Sustainable Engineering Approach. ACS Biomater. Sci. Eng. 2020, 6, 5264–5273. [Google Scholar] [CrossRef]
- He, L.; Yang, Y.; Kim, J.; Yao, L.; Dong, X.; Li, T.; Piao, Y. Multi-Layered Enzyme Coating on Highly Conductive Magnetic Biochar Nanoparticles for Bisphenol A Sensing in Water. Chem. Eng. J. 2020, 384, 123276. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, L.; He, L.; Liu, N.; Piao, Y. Electrochemical Enzyme Biosensor Bearing Biochar Nanoparticle as Signal Enhancer for Bisphenol a Detection in Water. Sens. (Switz.) 2019, 19, 1619. [Google Scholar] [CrossRef]
- Al-Mokhalelati, K.; Al-Bakri, I.; Al Shibeh Al Wattar, N. Adsorption of Methylene Blue onto Sugarcane Bagasse-Based Adsorbent Materials. J. Phys. Org. Chem. 2021, 34, e4193. [Google Scholar] [CrossRef]
- Zhou, F.; Li, K.; Hang, F.; Zhang, Z.; Chen, P.; Wei, L.; Xie, C. Efficient Removal of Methylene Blue by Activated Hydrochar Prepared by Hydrothermal Carbonization and NaOH Activation of Sugarcane Bagasse and Phosphoric Acid. RSC Adv. 2022, 12, 1885–1896. [Google Scholar] [CrossRef]
- Siqueira, T.C.A.; da Silva, I.Z.; Rubio, A.J.; Bergamasco, R.; Gasparotto, F.; Paccola, E.A.d.S.; Yamaguchi, N.U. Sugarcane Bagasse as an Efficient Biosorbent for Methylene Blue Removal: Kinetics, Isotherms and Thermodynamics. Int. J. Environ. Res. Public Health 2020, 17, 526. [Google Scholar] [CrossRef]
- Erdem, A.; Kerman, K.; Meric, B.; Akarca, U.S.; Ozsoz, M. Novel Hybridization Indicator Methylene Blue for the Electrochemical Detection of Short DNA Sequences Related to the Hepatitis B Virus. Anal. Chim. Acta 2000, 422, 139–149. [Google Scholar] [CrossRef]
- Kara, P.; Kerman, K.; Ozkan, D.; Meric, B.; Erdem, A.; Ozkan, Z.; Ozsoz, M. Electrochemical Genosensor for the Detection of Interaction between Methylene Blue and DNA. Electrochem. Commun. 2002, 4, 705–709. [Google Scholar] [CrossRef]
- Yang, W.; Ozsoz, M.; Hibbert, D.B.; Gooding, J.J. Evidence for the Direct Interaction between Methylene Blue and Guanine Bases Using DNA-Modified Carbon Paste Electrodes. Electroanalysis 2002, 14, 1299–1302. [Google Scholar] [CrossRef]
- Zhou, J.; Li, S.; Noroozifar, M.; Kerman, K. Graphene Oxide Nanoribbons in Chitosan for Simultaneous Electrochemical Detection of Guanine, Adenine, Thymine and Cytosine. Biosensors 2020, 10, 30. [Google Scholar] [CrossRef]
- Chandel, A.K.; Antunes, F.A.F.; Anjos, V.; Bell, M.J.V.; Rodrigues, L.N.; Polikarpov, I.; De Azevedo, E.R.; Bernardinelli, O.D.; Rosa, C.A.; Pagnocca, F.C.; et al. Multi-Scale Structural and Chemical Analysis of Sugarcane Bagasse in the Process of Sequential Acid-Base Pretreatment and Ethanol Production by Scheffersomyces Shehatae and Saccharomyces Cerevisiae. Biotechnol. Biofuels 2014, 7, 63. [Google Scholar] [CrossRef]
- Khan, S.; Alam, F.; Azam, A.; Khan, A.U. Gold Nanoparticles Enhance Methylene Blue-Induced Photodynamic Therapy: A Novel Therapeutic Approach to Inhibit Candida Albicans Biofilm. Int. J. Nanomed. 2012, 7, 3245–3257. [Google Scholar] [CrossRef]
- Li, S.; Noroozifar, M.; Kerman, K. Nanocomposite of Ferricyanide-Doped Chitosan with Multi-Walled Carbon Nanotubes for Simultaneous Senary Detection of Redox-Active Biomolecules. J. Electroanal. Chem. 2019, 849, 113376. [Google Scholar] [CrossRef]
- Ganesh, H.; Noroozifar, M.; Kerman, K. Epigallocatechin Gallate-Modified Graphite Paste Electrode for Simultaneous Detection of Redox-Active Biomolecules. Sensors 2017, 18, 23. [Google Scholar] [CrossRef]
- Zhang, S.; Li, B.Q.; Zheng, J. Bin an Electrochemical Sensor for the Sensitive Determination of Nitrites Based on Pt-PANI-Graphene Nanocomposites. Anal. Methods 2015, 7, 8366–8372. [Google Scholar] [CrossRef]
- Dai, Z.; Chen, J.; Yan, F.; Ju, H. Electrochemical Sensor for Immunoassay of Carcinoembryonic Antigen Based on Thionine Monolayer Modified Gold Electrode. Cancer Detect. Prev. 2005, 29, 233–240. [Google Scholar] [CrossRef]
- Li, B.Q.; Nie, F.; Sheng, Q.L.; Zheng, J. Bin An Electrochemical Sensor for Sensitive Determination of Nitrites Based on Ag-Fe3O4-Graphene Oxide Magnetic Nanocomposites. Chem. Pap. 2015, 69, 911–920. [Google Scholar] [CrossRef]
- Thangaraj, R.; Senthil Kumar, A. Simultaneous Detection of Guanine and Adenine in DNA and Meat Samples Using Graphitized Mesoporous Carbon Modified Electrode. J. Solid State Electrochem. 2013, 17, 583–590. [Google Scholar] [CrossRef]
- Lei, P.; Zhou, Y.; Zhu, R.; Liu, Y.; Dong, C.; Shuang, S. Novel Strategy of Electrochemical Analysis of DNA Bases with Enhanced Performance Based on Copper−nickel Nanosphere Decorated N,B−doped Reduced Graphene Oxide. Biosens. Bioelectron. 2020, 147, 111735. [Google Scholar] [CrossRef] [PubMed]
- Yari, A.; Derki, S. New MWCNT-Fe3O4@PDA-Ag Nanocomposite as a Novel Sensing Element of an Electrochemical Sensor for Determination of Guanine and Adenine Contents of DNA. Sens. Actuators B Chem. 2016, 227, 456–466. [Google Scholar] [CrossRef]
- Qin, J.; Gao, S.; Li, H.; Li, C.; Li, M. Growth of Monolayer and Multilayer Graphene on Glassy Carbon Electrode for Simultaneous Determination of Guanine, Adenine, Thymine, and Cytosine. J. Electroanal. Chem. 2021, 895, 115403. [Google Scholar] [CrossRef]
- Yin, H.; Zhou, Y.; Ma, Q.; Ai, S.; Ju, P.; Zhu, L.; Lu, L. Electrochemical Oxidation Behavior of Guanine and Adenine on Graphene-Nafion Composite Film Modified Glassy Carbon Electrode and the Simultaneous Determination. Process Biochem. 2010, 45, 1707–1712. [Google Scholar] [CrossRef]
- Emran, M.Y.; El-Safty, S.A.; Selim, M.M.; Shenashen, M.A. Selective Monitoring of Ultra-Trace Guanine and Adenine from Hydrolyzed DNA Using Boron-Doped Carbon Electrode Surfaces. Sens. Actuators B Chem. 2021, 329, 129192. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Wei, Y.; Xing, T.; Cao, T.; Wu, S.; Zhu, F. A Copper-Based Metal-Organic Framework/Graphene Nanocomposite for the Sensitive and Stable Electrochemical Detection of DNA Bases. Analyst 2020, 145, 1933–1942. [Google Scholar] [CrossRef]
- Rajabi, H.; Noroozifar, M.; Khorasani-Motlagh, M. Graphite Paste Electrode Modified with Lewatit® FO36 Nano-Resin for Simultaneous Determination of Ascorbic Acid, Acetaminophen and Tryptophan. Anal. Methods 2016, 8, 1924–1934. [Google Scholar] [CrossRef]

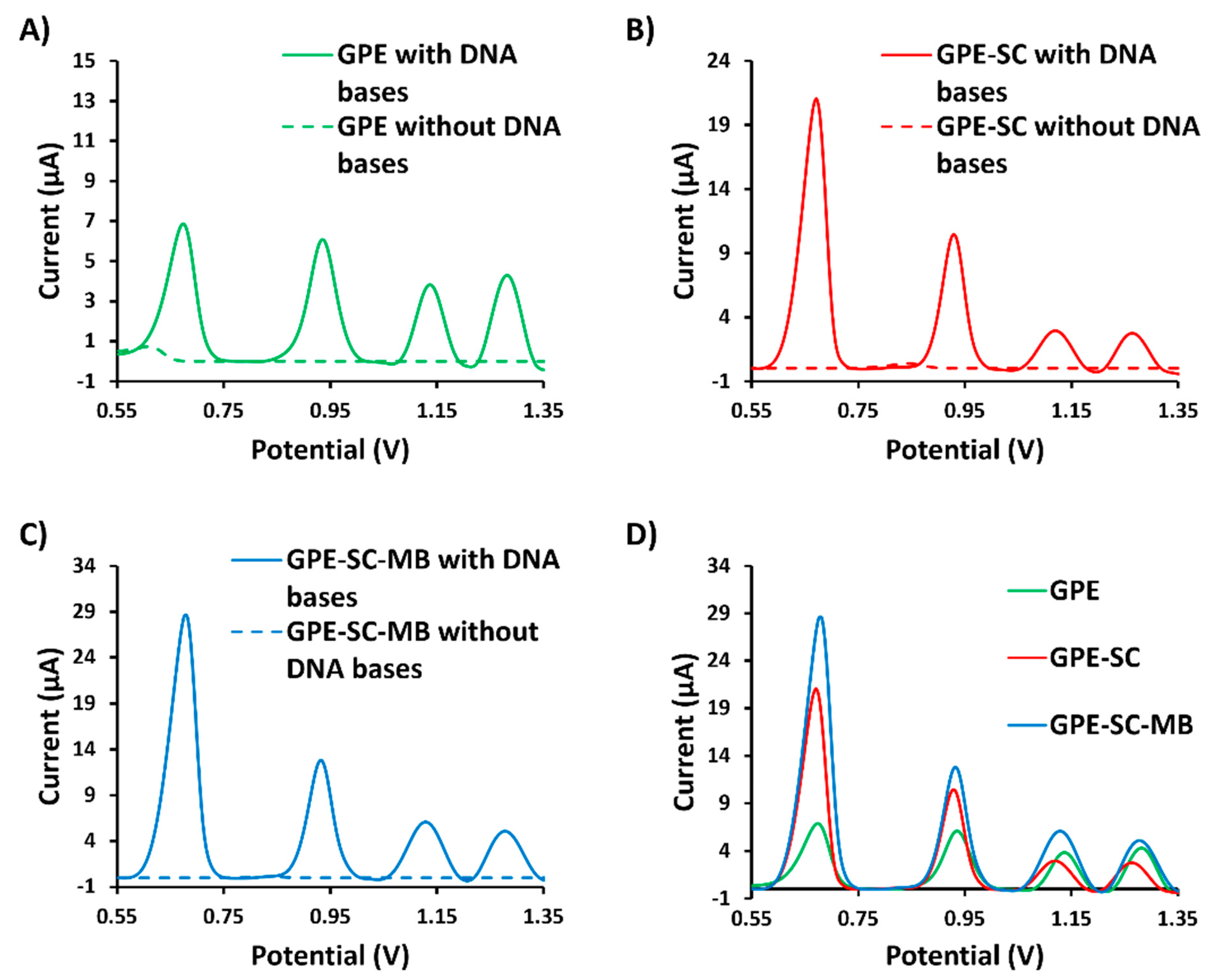

| Ip,a Guanine (μA) | Ip,a Adenine (μA) | Ip,a Thymine (μA) | Ip,a Cytosine (μA) | |
|---|---|---|---|---|
| GPE | 6.83 | 6.04 | 3.80 | 4.28 |
| GPE-SC | 21.04 | 10.45 | 2.94 | 2.74 |
| GPE-SC-MB | 28.55 | 12.83 | 6.08 | 5.07 |
| Analytes | Equations | R2 | LOD (μM) | LOQ (μM) |
|---|---|---|---|---|
| G | ΔIp(G1) = 0.9330 [G] + 0.0347 | 0.9967 | 0.037 | 0.12 |
| ΔIp(G2) = 0.2565[G] + 14.008 | 0.9927 | |||
| A | ΔIp(A1) = 0.3230 [A] + 0.2496 | 0.9901 | 0.042 | 0.14 |
| ΔIp(A2) = 0.0715 [A] + 7.0113 | 0.9919 | |||
| T | ΔIp(T) = 0.0133 [T] + 0.0327 | 0.9952 | 4.25 | 14.17 |
| C | ΔIp(C) = 0.0219 [C] + 0.5660 | 0.9925 | 5.33 | 17.79 |
| Platform | Method | DNA Base | LOD (μM) | Linear Range (μM) | Reference | ||
|---|---|---|---|---|---|---|---|
| Graphene oxide nanoribbon in chitosan modified glassy carbon electrode (GCE) | DPV | G | 0.0018 | 0.013–256 | [46] | ||
| A | 0.023 | 0.11–172 | |||||
| T | 1.330 | 6.0–855 | |||||
| C | 0.640 | 3.5–342 | |||||
| Graphene oxide-MWCNT modified GCE | DPV | G | 0.11 | 1–78 | [14] | ||
| A | 0.43 | 2–119.5 | |||||
| T | 1.71 | 12.5–227.5 | |||||
| C | 0.80 | 5–132.5 | |||||
| Graphitized mesoporous modified carbon paste electrode | DPV | G | 0.76 | 25–200 | [54] | ||
| A | 0.118 | 25–150 | |||||
| Cu-Ni nanosphere decorated N,B-doped reduced graphene oxide-modified glassy carbon electrode (GCE) | DPV | G | 0.118 | 1.0–160 | [55] | ||
| A | 0.134 | 1.0–120 | |||||
| MWCNT-Fe3O4 polydopamine Ag-nanoparticle-modified carbon paste electrode | DPV | G | 1.47 | 8–130 | [56] | ||
| A | 5.66 | 10–120 | |||||
| Graphene multilayer-modified GCE | DPV | G | 0.3 | 2–120 | [57] | ||
| A | 0.4 | 2–110 | |||||
| T | 6.5 | 20–1100 | |||||
| C | 4.5 | 20–1100 | |||||
| MWCNT embedded with Au/reduced graphene oxide GPE | SWV | G | 3.3 | 3–170 | [28] | ||
| A | 3.7 | 3–190 | |||||
| T | 7.9 | 7.5–800 | |||||
| C | 9.0 | 9–900 | |||||
| Graphene-Nafion™-modified GCE | DPV | G | 0.58 | 2–120 | [58] | ||
| A | 0.75 | 5–170 | |||||
| 2-(4-bromophenyl)-1-phenyl-1H-phenanthro-[9,10-d]-imidazole-modified GCE | SWV | G | 0.28 | 3–300 | [13] | ||
| A | 0.24 | 1–300 | |||||
| T | 3.2 | 30–800 | |||||
| C | 6.8 | 20–750 | |||||
| Boron-doped carbon nanospheres modified GCE | SWV | G | 4 × 10−4 | 0.01–0.5 | [59] | ||
| A | 2 × 10−4 | 0.01–0.5 | |||||
| Copper metal-organic framework with reduced graphene oxide-modified GCE | DPV | G | 0.012 | 0.02–100 | [60] | ||
| A | 0.002 | 0.005–200 | |||||
| MWCNT-COOH with a Cu -porphyrin metal covalent organic framework with Co nanoparticles on GCE | DPV | G | 0.0055 | 0.04–130 | [12] | ||
| A | 0.0072 | 0.06–130 | |||||
| GPE-SC-MB | DPV | G | 0.037 | 0.67–38.67 | This work | ||
| A | 0.042 | 0.67–126.67 | |||||
| T | 4.25 | 6.67–1600 | |||||
| C | 5.33 | 6.67–1600 | |||||
| Sample Matrix | DNA Base | Spiked Concentration (µM) | Detected Concentration ± S.D. (µM) | % Recovery ± %RSD |
|---|---|---|---|---|
| Human Saliva | G | 12.0 | 12.3 ± 0.2 | 102.5 ± 1.7 |
| A | 12.0 | 12.1 ± 0.3 | 100.8 + 2.5 | |
| T | 146.7 | 145.7 ± 0.7 | 99.3 ± 0.5 | |
| C | 146.7 | 149.1 ± 0.6 | 101.7 ± 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, Q.; Meng, Z.; Noroozifar, M.; Kerman, K. Methylene Blue-Modified Biochar from Sugarcane for the Simultaneous Electrochemical Detection of Four DNA Bases. Chemosensors 2023, 11, 169. https://doi.org/10.3390/chemosensors11030169
Hassan Q, Meng Z, Noroozifar M, Kerman K. Methylene Blue-Modified Biochar from Sugarcane for the Simultaneous Electrochemical Detection of Four DNA Bases. Chemosensors. 2023; 11(3):169. https://doi.org/10.3390/chemosensors11030169
Chicago/Turabian StyleHassan, Qusai, Zhixin Meng, Meissam Noroozifar, and Kagan Kerman. 2023. "Methylene Blue-Modified Biochar from Sugarcane for the Simultaneous Electrochemical Detection of Four DNA Bases" Chemosensors 11, no. 3: 169. https://doi.org/10.3390/chemosensors11030169
APA StyleHassan, Q., Meng, Z., Noroozifar, M., & Kerman, K. (2023). Methylene Blue-Modified Biochar from Sugarcane for the Simultaneous Electrochemical Detection of Four DNA Bases. Chemosensors, 11(3), 169. https://doi.org/10.3390/chemosensors11030169









