Surface-Enhanced Raman Spectroscopic Analysis of Flavoenzyme Cofactors: Guidance for Flavin-Related Bio- and Chemo- Sensors
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation of Ag nanoparticles
2.2.2. Preparation of Self-Assembly Ag Nanofilm on Glass
2.2.3. Measurement
2.2.4. Calculation Method of Density-Functional Theory
3. Results and Discussion
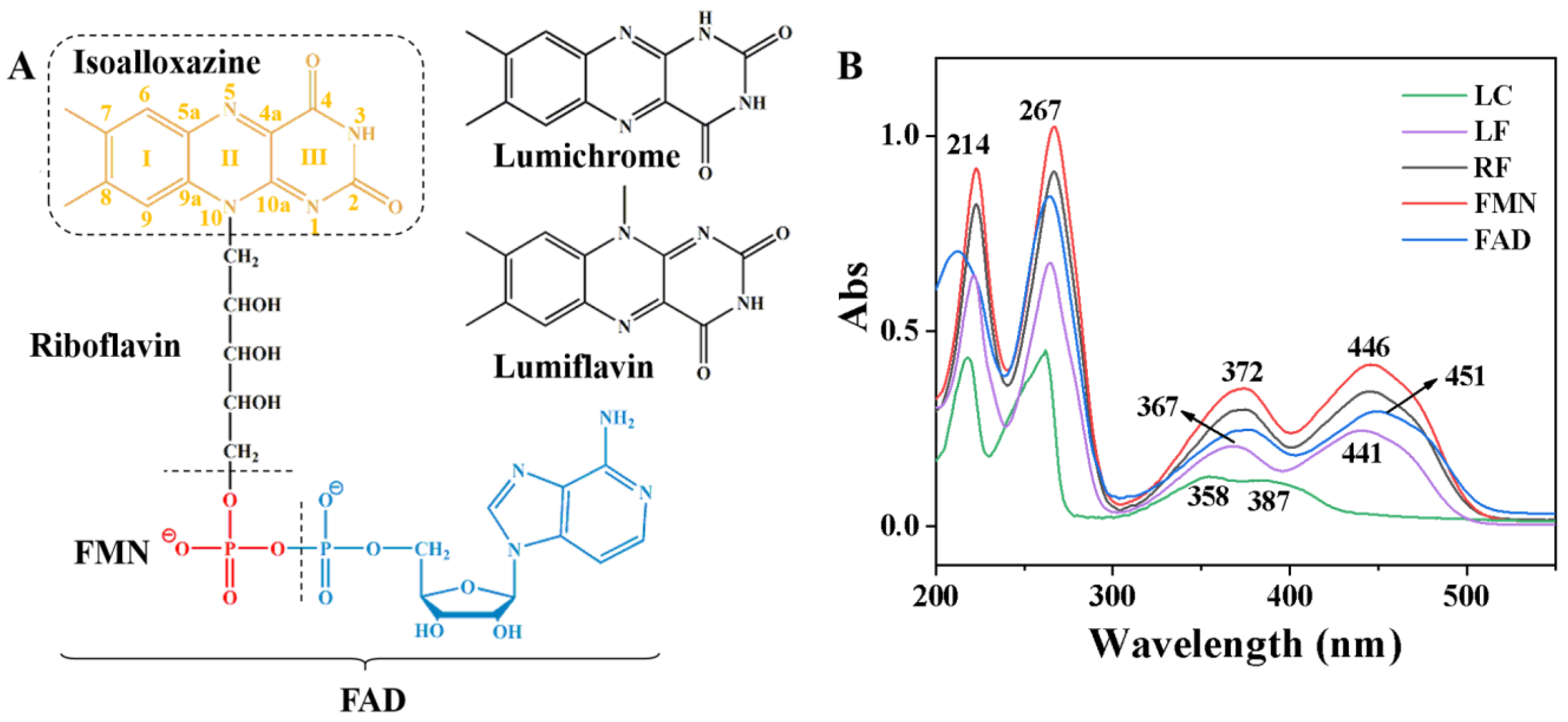
3.1. Characterization of Ultraviolet-Visible Spectroscopic Absorption Spectra of Flavins
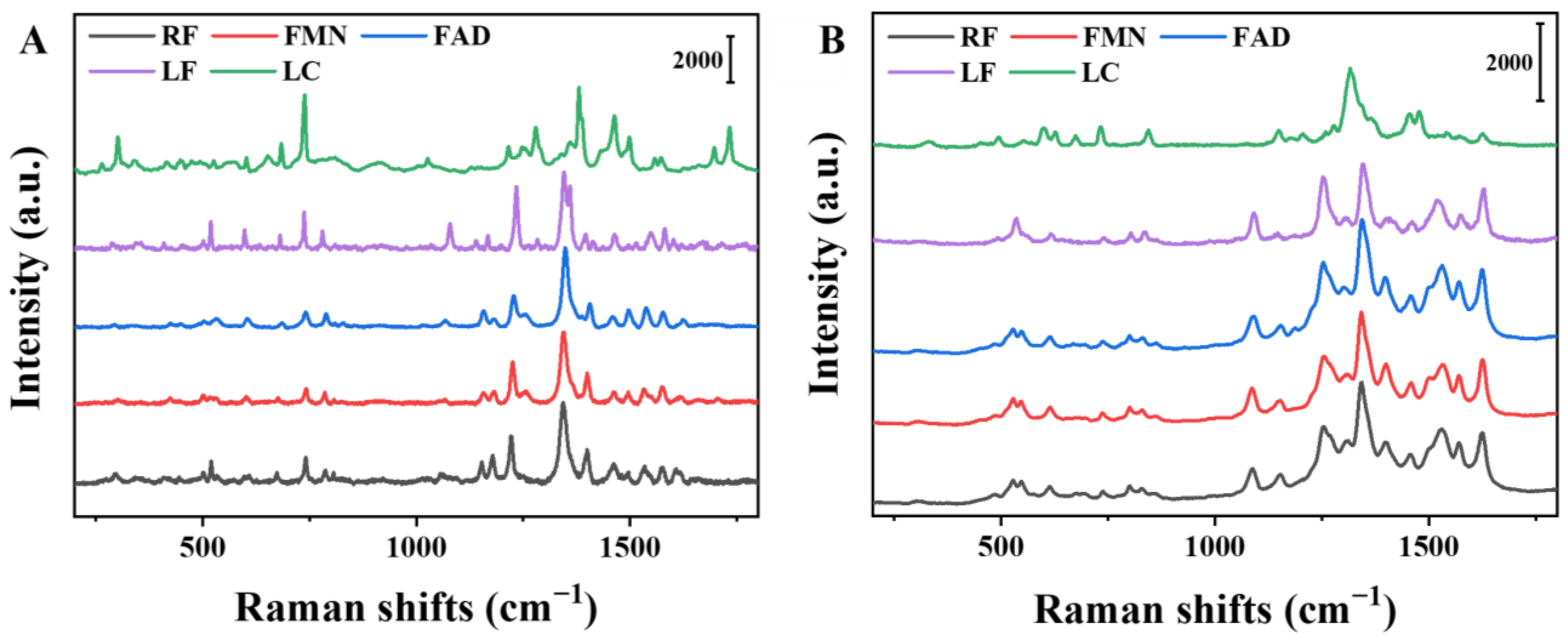
3.2. Solid-State Raman Spectra and Theoretical Analysis of Flavin Molecules
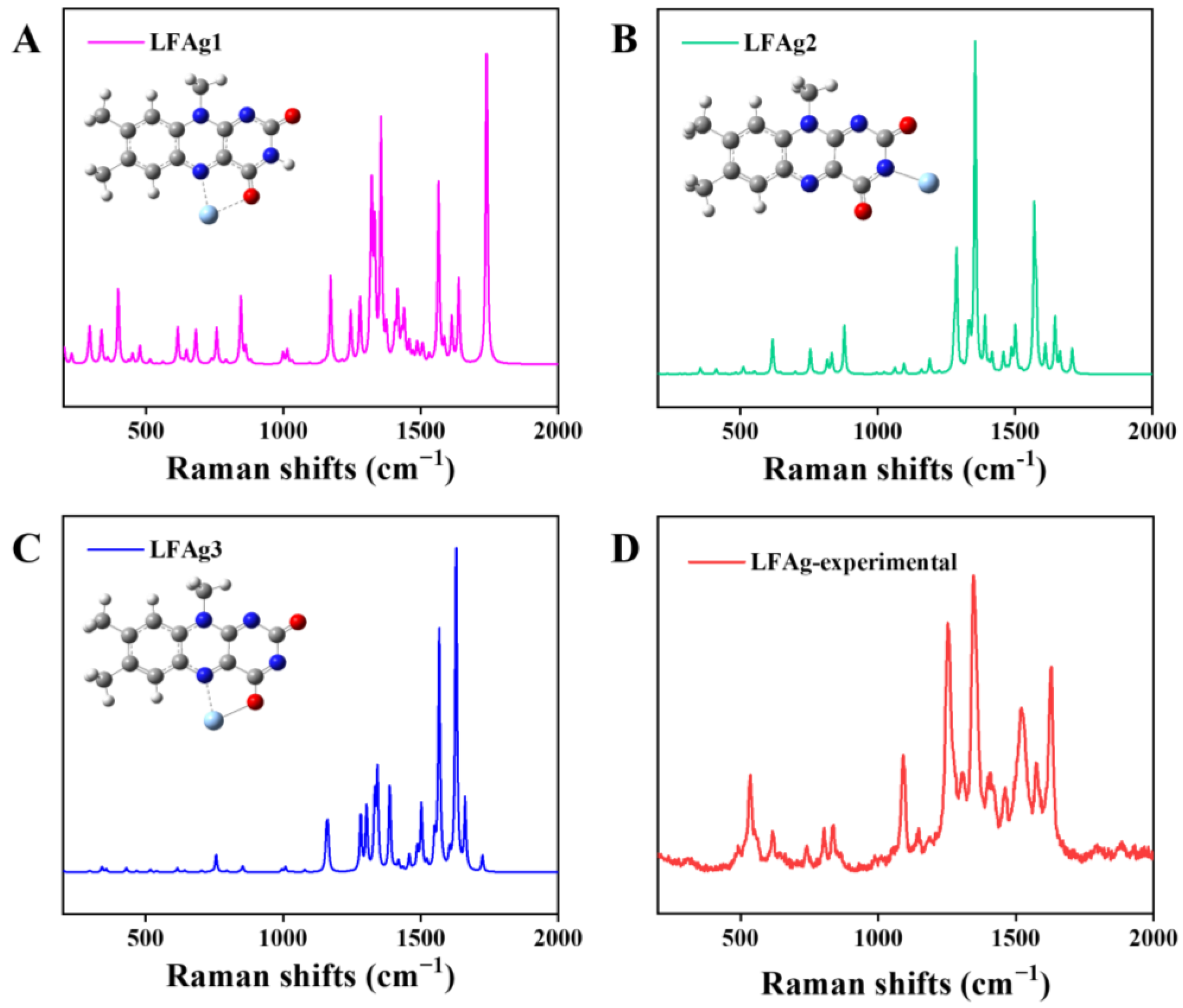
3.2.1. LC and LF
3.2.2. RF, FMN, and FAD
3.3. SERS Spectra of Flavin Molecules
3.3.1. Characterization of Ag nanoparticles
3.3.2. SERS Spectra of Flavins
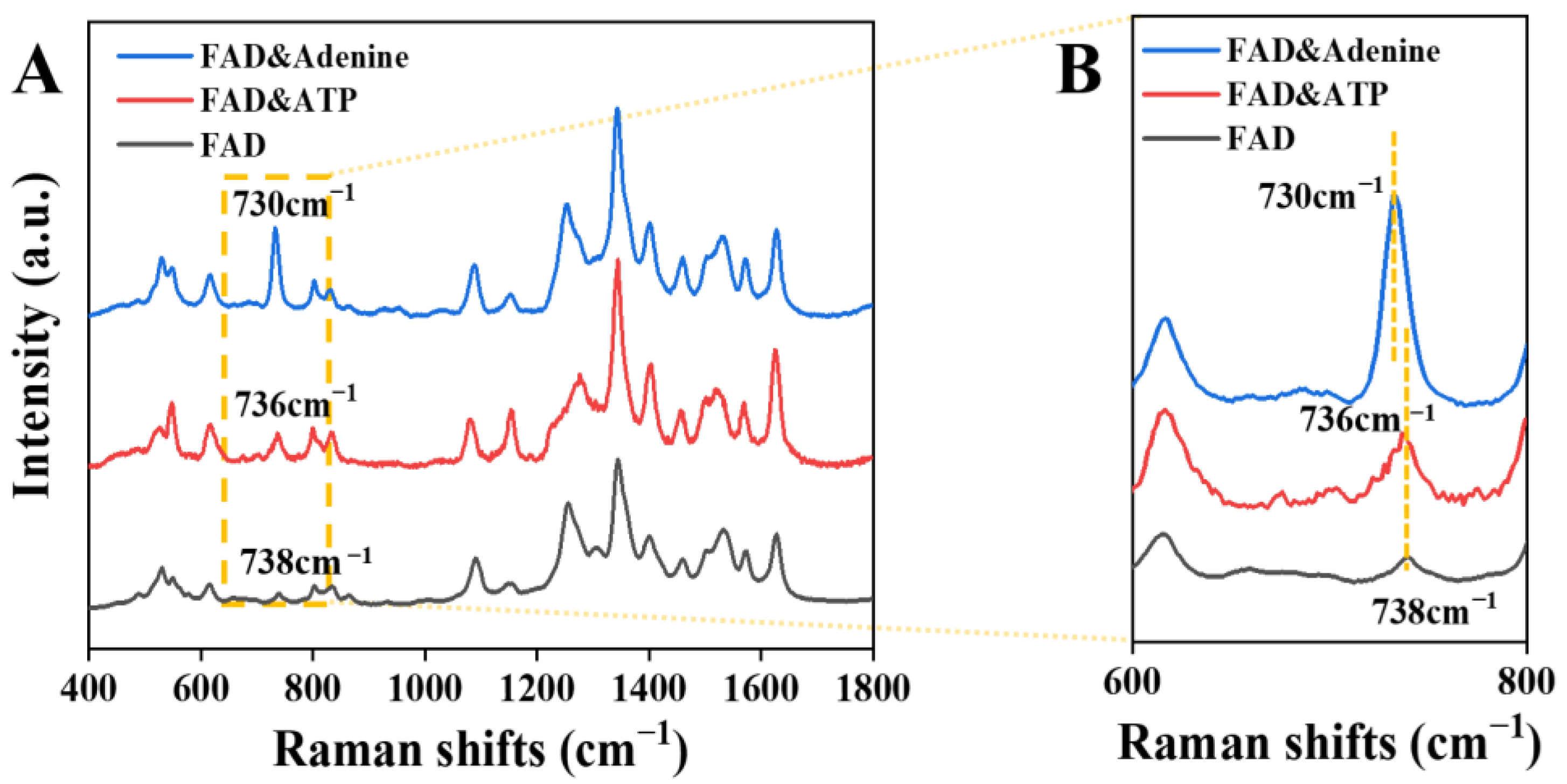
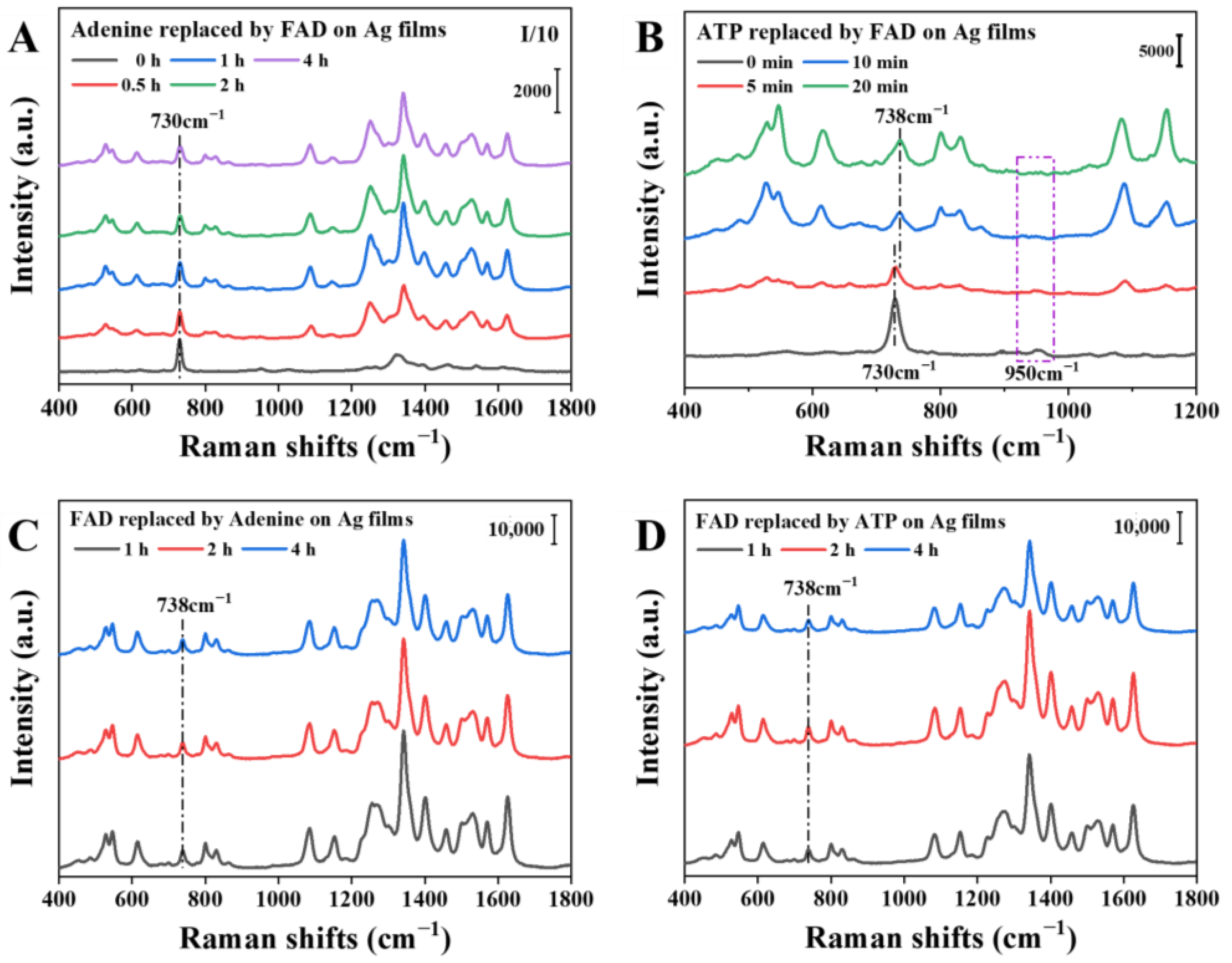
3.4. Analysis of FAD Molecules on Ag-Nanoparticle Substrates
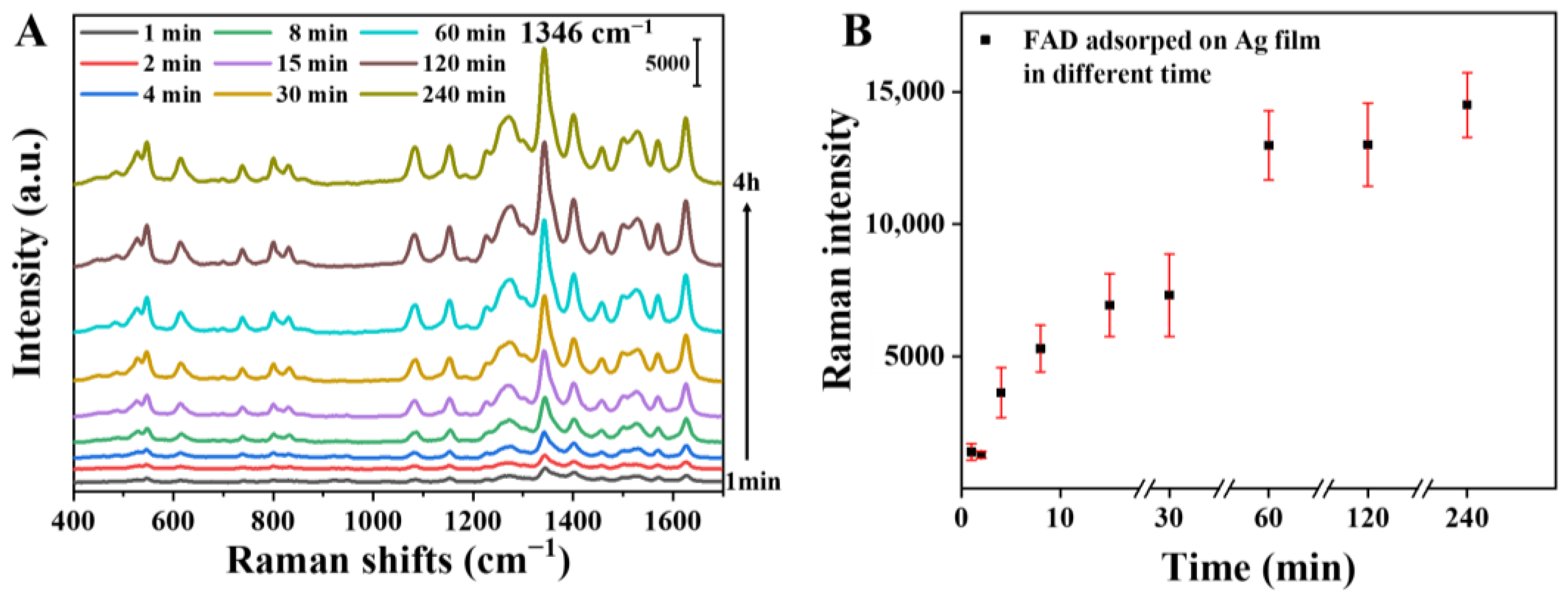
3.4.1. Adsorption Time
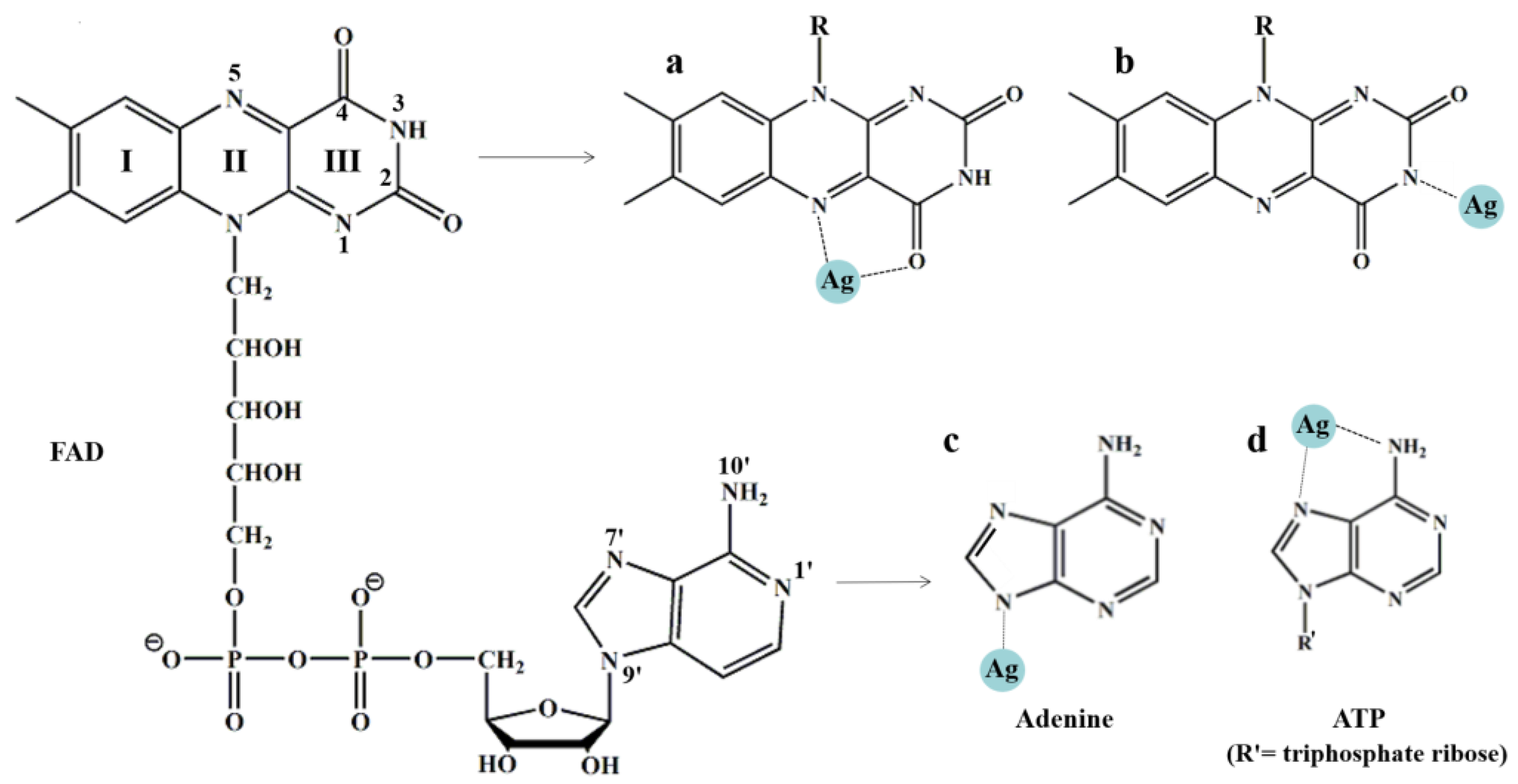
3.4.2. Adsorption Mode
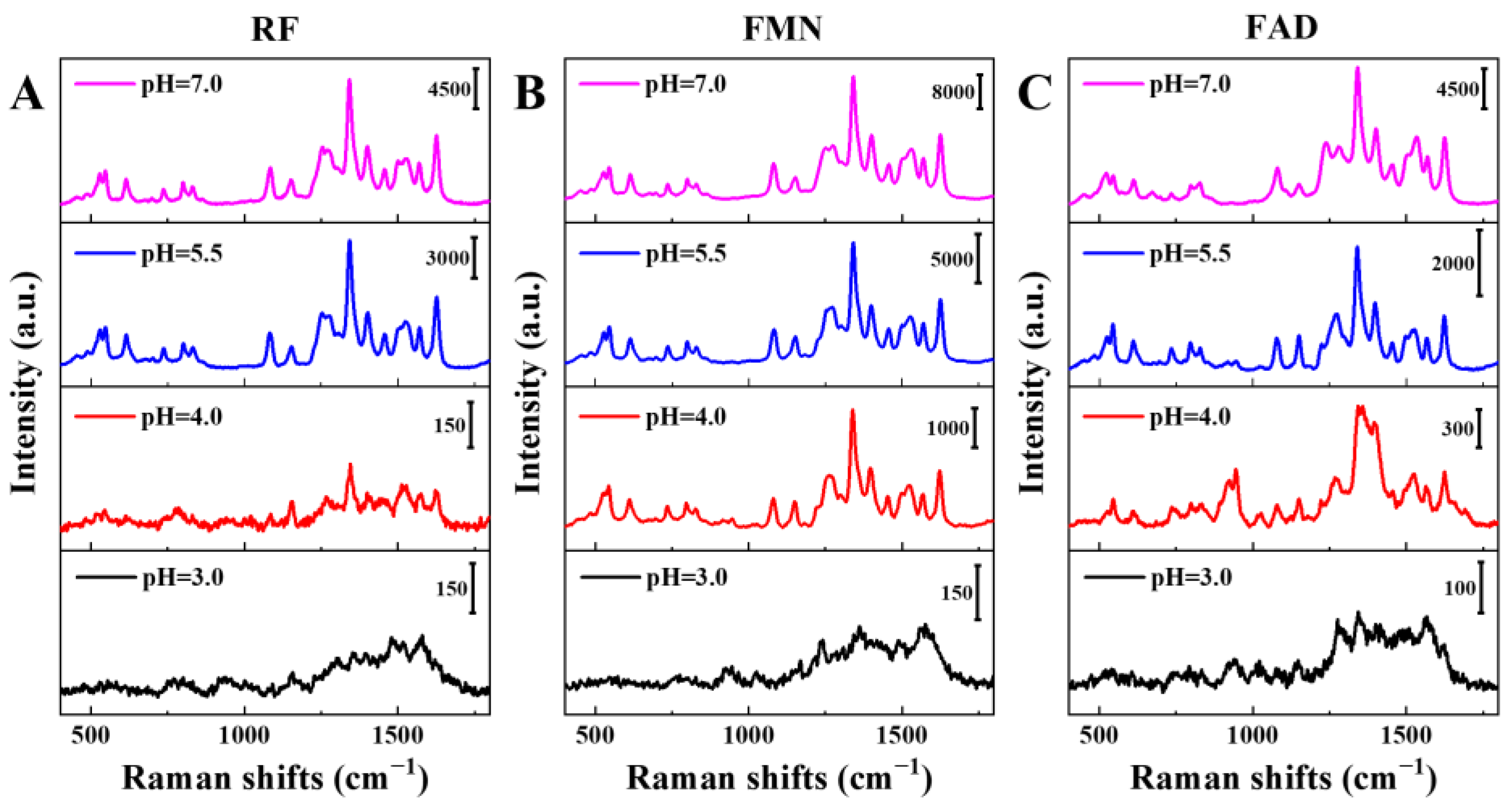
3.5. Analysis of Flavin Molecules under Different pH Conditions
3.6. Concentration Dependence of Flavin Molecules on Ag-Nanoparticle Substrates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pinto, J.T.; Zempleni, J. Riboflavin. Adv. Nutr. 2016, 7, 973–975. [Google Scholar] [CrossRef]
- Powers, H.J. Riboflavin (vitamin B-2) and health. Am. J. Clin. Nutr. 2003, 77, 1352–1360. [Google Scholar] [CrossRef]
- Viñas, P.; Balsalobre, N.; López-Erroz, C.; Hernández-Córdoba, M. Liquid Chromatographic Analysis of Riboflavin Vitamers in Foods Using Fluorescence Detection. J. Agric. Food Chem. 2004, 52, 1789–1794. [Google Scholar] [CrossRef]
- Díez-Pascual, A.M.; García-García, D.; Andrés, M.P.S.; Vera, S. Determination of riboflavin based on fluorescence quenching by graphene dispersions in polyethylene glycol. RSC Adv. 2016, 6, 19686–19699. [Google Scholar] [CrossRef]
- Shumyantseva, V.V.; Bulko, T.V.; Petushkova, N.A.; Samenkova, N.F.; Kuznetsova, G.P.; Archakov, A.I. Fluorescent assay for riboflavin binding to cytochrome P450 2B4. J. Inorg. Biochem. 2003, 98, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Bie, Z.; Wang, F.; Guo, E. Efficient synthesis of riboflavin-imprinted magnetic nanoparticles by boronate affinity-based surface imprinting for the selective recognition of riboflavin. Analyst 2018, 143, 4936–4943. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Muhammad, P.; Guo, Z.; Peng, Q.; Lu, H.; Liu, Z. Controllably prepared molecularly imprinted core-shell plasmonic nanostructure for plasmon-enhanced fluorescence assay. Biosens. Bioelectron. 2019, 146, 111733. [Google Scholar] [CrossRef]
- McNay, G.; Eustace, D.; Smith, W.E.; Faulds, K.; Graham, D. Surface-Enhanced Raman Scattering (SERS) and Surface-Enhanced Resonance Raman Scattering (SERRS): A Review of Applications. Appl. Spectrosc. 2011, 65, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Cialla, D.; März, A.; Böhme, R.; Theil, F.; Weber, K.; Schmitt, M.; Popp, J. Surface-enhanced Raman spectroscopy (SERS): Progress and trends. Anal. Bioanal. Chem. 2011, 403, 27–54. [Google Scholar] [CrossRef]
- Li, J.F.; Huang, Y.F.; Ding, Y.; Yang, Z.L.; Li, S.B.; Zhou, X.S.; Fan, F.R.; Zhang, W.; Zhou, Z.Y.; Wu, D.Y.; et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature 2010, 464, 392–395. [Google Scholar] [CrossRef]
- Wang, H.-L.; You, E.-M.; Panneerselvam, R.; Ding, S.-Y.; Tian, Z.-Q. Advances of surface-enhanced Raman and IR spectroscopies: From nano/microstructures to macro-optical design. Light. Sci. Appl. 2021, 10, 161. [Google Scholar] [CrossRef]
- Li, P.; Wang, X.; Zhang, X.; Zhang, L.; Yang, X.; Zhao, B. Investigation of the Charge-Transfer Between Ga-Doped ZnO Nanoparticles and Molecules Using Surface-Enhanced Raman Scattering: Doping Induced Band-Gap Shrinkage. Front. Chem. 2019, 7, 144. [Google Scholar] [CrossRef]
- Guo, L.; Mao, Z.; Jin, S.; Zhu, L.; Zhao, J.; Zhao, B.; Jung, Y. A SERS Study of Charge Transfer Process in Au Nanorod–MBA@Cu2O Assemblies: Effect of Length to Diameter Ratio of Au Nanorods. Nanomaterials 2021, 11, 867. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Guo, S.; Jin, S.; Park, E.; Chen, L.; Jung, Y.M. Charge transfer study for semiconductor and semiconductor/ metal composites based on surface-enhanced Raman scattering. Bull. Korean Chem. Soc. 2021, 42, 1411–1418. [Google Scholar] [CrossRef]
- Liu, Y.; Ma, H.; Han, X.X.; Zhao, B. Metal–semiconductor heterostructures for surface-enhanced Raman scattering: Synergistic contribution of plasmons and charge transfer. Mater. Horizons 2020, 8, 370–382. [Google Scholar] [CrossRef]
- Shan, Y.; Zheng, Z.; Liu, J.; Yang, Y.; Li, Z.; Huang, Z.; Jiang, D. Niobium pentoxide: A promising surface-enhanced Raman scattering active semiconductor substrate. npj Comput. Mater. 2017, 3, 11. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, E.; Shi, H.; Tao, Y.; Ren, X. Semiconductor-based surface enhanced Raman scattering (SERS): From active materials to performance improvement. Anal. 2022, 147, 1257–1272. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Tang, X.; Liu, Y.; Han, X.X.; He, C.; Lu, H.; Zhao, B. Surface-Enhanced Raman Scattering for Direct Protein Function Investigation: Controlled Immobilization and Orientation. Anal. Chem. 2019, 91, 8767–8771. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Wang, J.; Zhou, H.; Tang, X.; Yang, Y.; Wang, Z.; Chen, D.; Zhou, X.; Guo, J.; et al. Click-Reaction-Triggered SERS Signals for Specific Detection of Monoamine Oxidase B Activity. Anal. Chem. 2020, 92, 15050–15058. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Y.; Zhao, X.; Ji, Y.; Mei, R.; Fu, L.; Man, M.; Ma, J.; Wang, X.; Chen, L. Surface-enhanced Raman scattering labeled nanoplastic models for reliable bio-nano interaction investigations. J. Hazard. Mater. 2021, 425, 127959. [Google Scholar] [CrossRef]
- Plou, J.; Valera, P.S.; García, I.; de Albuquerque, C.D.L.; Carracedo, A.; Liz-Marzán, L.M. Prospects of Surface-Enhanced Raman Spectroscopy for Biomarker Monitoring toward Precision Medicine. ACS Photonics 2022, 9, 333–350. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Goryacheva, I.Y.; Markin, A.V. Surface-Enhanced Raman Spectroscopy for the Determination of Medical and Narcotic Drugs in Human Biofluids. J. Anal. Chem. 2022, 77, 930–947. [Google Scholar] [CrossRef]
- Samal, A.K.; Polavarapu, L.; Rodal-Cedeira, S.; Liz-Marzán, L.M.; Pérez-Juste, J.; Pastoriza-Santos, I. Size Tunable Au@Ag Core–Shell Nanoparticles: Synthesis and Surface-Enhanced Raman Scattering Properties. Langmuir 2013, 29, 15076–15082. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, Z.; Peng, B.; Cao, C.; Zhang, C.; You, H.; Xiong, Q.; Li, Z.; Fang, J. Highly sensitive, uniform, and reproducible surface-enhanced Raman spectroscopy from hollow Au-Ag alloy nanourchins. Adv. Mater. 2014, 26, 2431–2439. [Google Scholar] [CrossRef]
- Bonifacio, A.; Marta, S.D.; Spizzo, R.; Cervo, S.; Steffan, A.; Colombatti, A.; Sergo, V. Surface-enhanced Raman spectroscopy of blood plasma and serum using Ag and Au nanoparticles: A systematic study. Anal. Bioanal. Chem. 2014, 406, 2355–2365. [Google Scholar] [CrossRef]
- Wang, X.; Li, P.; Han, X.X.; Kitahama, Y.; Zhao, B.; Ozaki, Y. An enhanced degree of charge transfer in dye-sensitized solar cells with a ZnO-TiO 2/N3/Ag structure as revealed by surface-enhanced Raman scattering. Nanoscale 2017, 9, 15303–15313. [Google Scholar] [CrossRef]
- Guo, L.; Mao, Z.; Ma, C.; Wu, J.; Zhu, L.; Zhao, B.; Jung, Y.M. Charge Transfer in 4-Mercaptobenzoic Acid-Stabilized Au Nanorod@Cu2O Nanostructures: Implications for Photocatalysis and Photoelectric Devices. ACS Appl. Nano Mater. 2021, 4, 381–388. [Google Scholar] [CrossRef]
- Schmidt, J.; Coudron, P.; Thompson, A.W.; Watters, K.L.; McFarland, J.T. Hydrogen bonding between flavin and protein: A resonance Raman study. Biochemistry 1983, 22, 76–84. [Google Scholar] [CrossRef]
- Murgida, D.H.; Schleicher, E.; Bacher, A.; Richter, G.; Hildebrandt, P. Resonance Raman spectroscopic study of the neutral flavin radical complex of DNA photolyase fromEscherichia coli. J. Raman Spectrosc. 2001, 32, 551–556. [Google Scholar] [CrossRef]
- Schelvis, J.P.M.; Pun, D.; Goyal, N.; Sokolova, O. Resonance Raman spectra of the neutral and anionic radical semiquinones of flavin adenine dinucleotide in glucose oxidase revisited. J. Raman Spectrosc. 2006, 37, 822–829. [Google Scholar] [CrossRef]
- Green, D.; Roy, P.; Hall, C.R.; Iuliano, J.N.; Jones, G.A.; Lukacs, A.; Tonge, P.J.; Meech, S.R. Excited State Resonance Raman of Flavin Mononucleotide: Comparison of Theory and Experiment. J. Phys. Chem. A 2021, 125, 6171–6179. [Google Scholar] [CrossRef] [PubMed]
- Zeiri, L.; Efrima, S. Surface-enhanced Raman spectroscopy of bacteria: The effect of excitation wavelength and chemical modification of the colloidal milieu. J. Raman Spectrosc. 2005, 36, 667–675. [Google Scholar] [CrossRef]
- Xu, B.-B.; Ma, Z.-C.; Wang, L.; Zhang, R.; Niu, L.-G.; Yang, Z.; Zhang, Y.-L.; Zheng, W.-H.; Zhao, B.; Xu, Y.; et al. Localized flexible integration of high-efficiency surface enhanced Raman scattering (SERS) monitors into microfluidic channels. Lab Chip 2011, 11, 3347–3351. [Google Scholar] [CrossRef]
- Qi, G.; Wang, Y.; Zhang, B.; Sun, D.; Fu, C.; Xu, W.; Xu, S. Glucose oxidase probe as a surface-enhanced Raman scattering sensor for glucose. Anal. Bioanal. Chem. 2016, 408, 7513–7520. [Google Scholar] [CrossRef]
- Hernández-Sánchez, D.; Villabona-Leal, G.; Saucedo-Orozco, I.; Bracamonte, V.; Pérez, E.; Bittencourt, C.; Quintana, M. Stable graphene oxide–gold nanoparticle platforms for biosensing applications. Phys. Chem. Chem. Phys. 2017, 20, 1685–1692. [Google Scholar] [CrossRef]
- Maskevich, S.; Strekal, N.; Artsukevich, I.; Kivach, L.; Chernikevich, I. Surface enhanced Raman scattering investigation of protein-bound flavin adenine dinucleotide structure. J. Mol. Struct. 1995, 349, 5–8. [Google Scholar] [CrossRef]
- Abdelsalam, M.; Bartlett, P.N.; Russell, A.; Baumberg, J.J.; Calvo, E.J.; Tognalli, N.G.; Fainstein, A. Quantitative Electrochemical SERS of Flavin at a Structured Silver Surface. Langmuir 2008, 24, 7018–7023. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Meisel, D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J. Phys. Chem. 1982, 86, 3391–3395. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef] [PubMed]
- Becke, A.D. Density-functional thermochemistry III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Jamróz, M.H. Vbrational energy distribution analysis (VEDA): Scopes and limitations Spectrochim. Acta. A 2013, 114, 220–230. [Google Scholar] [CrossRef]
- Scott, A.P.; Radom, L. Harmonic vibrational frequencies: An evaluation of Hartree− Fock, Møller− Plesset, quadratic configuration interaction, density functional theory, and semiempirical scale factors. J. Phys. Chem. 1996, 100, 16502–16513. [Google Scholar] [CrossRef]
- Merrick, J.P.; Moran, D.; Radom, L. An Evaluation of Harmonic Vibrational Frequency Scale Factors. J. Phys. Chem. A 2007, 111, 11683–11700. [Google Scholar] [CrossRef]
- Alecu, I.M.; Zheng, J.; Zhao, Y.; Truhlar, D.G. Computational Thermochemistry: Scale Factor Databases and Scale Factors for Vibrational Frequencies Obtained from Electronic Model Chemistries. J. Chem. Theory Comput. 2010, 6, 2872–2887. [Google Scholar] [CrossRef]
- Laury, M.L.; Carlson, M.J.; Wilson, A.K. Vibrational frequency scale factors for density functional theory and the polarization consistent basis sets. J. Comput. Chem. 2012, 33, 2380–2387. [Google Scholar] [CrossRef]
- Kashinski, D.O.; Chase, G.M.; Nelson, R.G.; Di Nallo, O.E.; Scales, A.N.; VanderLey, D.L.; Byrd, E.F.C. Harmonic Vibrational Frequencies: Approximate Global Scaling Factors for TPSS, M06, and M11 Functional Families Using Several Common Basis Sets. J. Phys. Chem. A 2017, 121, 2265–2273. [Google Scholar] [CrossRef]
- Ouyang, L.; Zhu, L.; Jiang, J.; Tang, H. A surface-enhanced Raman scattering method for detection of trace glutathione on the basis of immobilized silver nanoparticles and crystal violet probe. Anal. Chim. Acta 2014, 816, 41–49. [Google Scholar] [CrossRef]
- Hussain, S.; Pang, Y. Surface geometry of tryptophan adsorbed on gold colloidal nanoparticles. J. Mol. Struct. 2015, 1096, 121–128. [Google Scholar] [CrossRef]
- Maruyama, Y.; Ishikawa, M.; Futamata, M. Surface-Enhanced Raman Scattering of Single Adenine Molecules on Silver Colloidal Particles. Chem. Lett. 2001, 30, 834–835. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Bell, S.E.J. Structure of Adenine on Metal Nanoparticles: pH Equilibria and Formation of Ag+ Complexes Detected by Surface-Enhanced Raman Spectroscopy. J. Phys. Chem. C 2010, 114, 22644–22651. [Google Scholar] [CrossRef]
- Holt, R.E.; Cotton, T.M. Surface-enhanced resonance Raman and electrochemical investigation of glucose oxidase catalysis at a silver electrode. J. Am. Chem. Soc. 1989, 111, 2815–2821. [Google Scholar] [CrossRef]
- Choudhury, S.D.; Vir, P.; Mohanty, J.; Bhasikuttan, A.C.; Pal, H. Selective prototropism of lumichrome in cationic micelles and reverse micelles: A photophysical perspective. RSC Adv. 2016, 6, 6111–6124. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Ma, H.; Zhao, J.; Wang, J.; Han, X.; Zhao, B. Surface-Enhanced Raman Spectroscopic Analysis of Flavoenzyme Cofactors: Guidance for Flavin-Related Bio- and Chemo- Sensors. Chemosensors 2023, 11, 190. https://doi.org/10.3390/chemosensors11030190
Liu Y, Ma H, Zhao J, Wang J, Han X, Zhao B. Surface-Enhanced Raman Spectroscopic Analysis of Flavoenzyme Cofactors: Guidance for Flavin-Related Bio- and Chemo- Sensors. Chemosensors. 2023; 11(3):190. https://doi.org/10.3390/chemosensors11030190
Chicago/Turabian StyleLiu, Yawen, Hao Ma, Junqi Zhao, Jihong Wang, Xiaoxia Han, and Bing Zhao. 2023. "Surface-Enhanced Raman Spectroscopic Analysis of Flavoenzyme Cofactors: Guidance for Flavin-Related Bio- and Chemo- Sensors" Chemosensors 11, no. 3: 190. https://doi.org/10.3390/chemosensors11030190
APA StyleLiu, Y., Ma, H., Zhao, J., Wang, J., Han, X., & Zhao, B. (2023). Surface-Enhanced Raman Spectroscopic Analysis of Flavoenzyme Cofactors: Guidance for Flavin-Related Bio- and Chemo- Sensors. Chemosensors, 11(3), 190. https://doi.org/10.3390/chemosensors11030190





