SERS-Driven Ceftriaxone Detection in Blood Plasma: A Protein Precipitation Approach
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patsalos, P.N.; Berry, D.J.; Bourgeois, B.F.D.; Cloyd, J.C.; Glauser, T.A.; Johannessen, S.I.; Leppik, I.E.; Tomson, T.; Perucca, E. Antiepileptic Drugs—Best Practice Guidelines for Therapeutic Drug Monitoring: A Position Paper by the Subcommission on Therapeutic Drug Monitoring, ILAE Commission on Therapeutic Strategies. Epilepsia 2008, 49, 1239–1276. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Lee, M.H. Overview of Therapeutic Drug Monitoring. Korean J. Intern. Med. 2009, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.L.; Parés, L.; Ajuria, I.; Bandres, F.; Castanyer, B.; Campos, F.; Farré, C.; Pou, L.; Queraltó, J.M.; To-Figueras, J. State of the Art in Therapeutic Drug Monitoring. Clin. Chem. Lab. Med. 2010, 48, 437–446. [Google Scholar] [CrossRef]
- Dasgupta, A. Usefulness of Monitoring Free (Unbound) Concentrations of Therapeutic Drugs in Patient Management. Clin. Chim. Acta 2007, 377, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Lu, Y.; Li, S.; Zhang, M.; Duan, X.; Liu, C.C.; Jia, J.; Liu, G. Direct Analysis in Real Time-Mass Spectrometry for Rapid Quantification of Five Anti-Arrhythmic Drugs in Human Serum: Application to Therapeutic Drug Monitoring. Sci. Rep. 2020, 10, 15550. [Google Scholar] [CrossRef]
- Losoya-Leal, A.; Estevez, M.C.; Martínez-Chapa, S.O.; Lechuga, L.M. Design of a Surface Plasmon Resonance Immunoassay for Therapeutic Drug Monitoring of Amikacin. Talanta 2015, 141, 253–258. [Google Scholar] [CrossRef]
- Antunes, M.V.; Charão, M.F.; Linden, R. Dried Blood Spots Analysis with Mass Spectrometry: Potentials and Pitfalls in Therapeutic Drug Monitoring. Clin. Biochem. 2016, 49, 1035–1046. [Google Scholar] [CrossRef]
- Decosterd, L.A.; Widmer, N.; André, P.; Aouri, M.; Buclin, T. The Emerging Role of Multiplex Tandem Mass Spectrometry Analysis for Therapeutic Drug Monitoring and Personalized Medicine. TrAC Trends Anal. Chem. 2016, 84, 5–13. [Google Scholar] [CrossRef]
- Ates, H.C.; Roberts, J.A.; Lipman, J.; Cass, A.E.G.; Urban, G.A.; Dincer, C. On-Site Therapeutic Drug Monitoring. Trends Biotechnol. 2020, 38, 1262–1277. [Google Scholar] [CrossRef]
- Panikar, S.S.; Cialla-May, D.; De la Rosa, E.; Salas, P.; Popp, J. Towards Translation of Surface-Enhanced Raman Spectroscopy (SERS) to Clinical Practice: Progress and Trends. TrAC Trends Anal. Chem. 2021, 134, 116122. [Google Scholar] [CrossRef]
- Dina, N.E.; Tahir, M.A.; Bajwa, S.Z.; Amin, I.; Valev, V.K.; Zhang, L. SERS-Based Antibiotic Susceptibility Testing: Towards Point-of-Care Clinical Diagnosis. Biosens. Bioelectron. 2023, 219, 956–5663. [Google Scholar] [CrossRef]
- Fornasaro, S.; Cialla-May, D.; Sergo, V.; Bonifacio, A. The Role of Surface Enhanced Raman Scattering for Therapeutic Drug Monitoring of Antimicrobial Agents. Chemosensors 2022, 10, 128. [Google Scholar] [CrossRef]
- Jaworska, A.; Fornasaro, S.; Sergo, V.; Bonifacio, A. Potential of Surface Enhanced Raman Spectroscopy (SERS) in Therapeutic Drug Monitoring (TDM). A Critical Review. Biosensors 2016, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Weber, S.; Peng, R.; Wu, L.; Zhang, W.-S.; Luppa, P.B.; Popp, J.; Cialla-May, D. Toward SERS-Based Therapeutic Drug Monitoring in Clinical Settings: Recent Developments and Trends. TrAC Trends Anal. Chem. 2023, 164, 117094. [Google Scholar] [CrossRef]
- Fornasaro, S.; Dalla Marta, S.; Rabusin, M.; Bonifacio, A.; Sergo, V. Toward SERS-Based Point-of-Care Approaches for Therapeutic Drug Monitoring: The Case of Methotrexate. Faraday Discuss. 2016, 187, 485–499. [Google Scholar] [CrossRef] [PubMed]
- Surendra Panikar, S.; Banu, N.; Haramati, J.; Yareli Gutierrez-Silerio, G.; Estela Bastidas-Ramirez, B.; Cecilia Tellez-Ba, M.; Camacho-Villegas, T.A.; del Toro-Arreola, S.; De la Rosa, E. Anti-Fouling SERS-Based Immunosensor for Point-of-Care Detection of the B7eH6 Tumor Biomarker in Cervical Cancer Patient Serum. Anal. Chim. Acta 2020, 1138, 110–122. [Google Scholar] [CrossRef]
- Chircov, C.; Grumezescu, A.M.; Holban, A.M. Materials Magnetic Particles for Advanced Molecular Diagnosis. Materials 2019, 12, 2158. [Google Scholar] [CrossRef]
- Markina, N.E.; Goryacheva, I.Y.; Markin, A.V. Sample Pretreatment and SERS-Based Detection of Ceftriaxone in Urine. Anal. Bioanal. Chem. 2018, 410, 2221–2227. [Google Scholar] [CrossRef]
- Giovannini, G.; De Angelis, F. Novel Electro-Magnetophoretic Separation Method for the Highly Sensitive Detection of Analytes. Nanoscale Horiz. 2020, 5, 95–101. [Google Scholar] [CrossRef]
- Pezzi, H.M.; Guckenberger, D.J.; Schehr, J.L.; Rothbauer, J.; Stahlfeld, C.; Singh, A.; Horn, S.; Schultz, Z.D.; Bade, R.M.; Sperger, J.M.; et al. Versatile Exclusion-Based Sample Preparation Platform for Integrated Rare Cell Isolation and Analyte Extraction. Lab Chip 2018, 18, 3446–3458. [Google Scholar] [CrossRef]
- Guselnikova, O.; Lim, H.; Na, J.; Eguchi, M.; Kim, H.-J.; Elashnikov, R.; Postnikov, P.; Svorcik, V.; Semyonov, O.; Miliutina, E.; et al. Enantioselective SERS Sensing of Pseudoephedrine in Blood Plasma Biomatrix by Hierarchical Mesoporous Au Films Coated with a Homochiral MOF. Biosens. Bioelectron. 2021, 180, 113109. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, A.; Dalla Marta, S.; Spizzo, R.; Cervo, S.; Steffan, A.; Colombatti, A.; Sergo, V. Surface-Enhanced Raman Spectroscopy of Blood Plasma and Serum Using Ag and Au Nanoparticles: A Systematic Study. Anal. Bioanal. Chem. 2014, 406, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Ehsan, U.; Nawaz, H.; Irfan Majeed, M.; Rashid, N.; Ali, Z.; Zulfiqar, A.; Tariq, A.; Shahbaz, M.; Meraj, L.; Naheed, I.; et al. Surface-Enhanced Raman Spectroscopy of Centrifuged Blood Serum Samples of Diabetic Type II Patients by Using 50KDa Filter Devices. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 293, 122457. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Hung, H.C.; Sinclair, A.; Zhang, P.; Bai, T.; Galvan, D.D.; Jain, P.; Li, B.; Jiang, S.; Yu, Q. Hierarchical Zwitterionic Modification of a SERS Substrate Enables Real-Time Drug Monitoring in Blood Plasma. Nat. Commun. 2016, 7, 13437. [Google Scholar] [CrossRef]
- Dams, R.; Huestis, M.A.; Lambert, W.E.; Murphy, C.M. Matrix Effect in Bio-Analysis of Illicit Drugs with LC-MS/MS: Influence of Ionization Type, Sample Preparation, and Biofluid. J. Am. Soc. Mass. Spectrom. 2003, 14, 1290–1294. [Google Scholar] [CrossRef]
- Ma, J.; Shi, J.; Le, H.; Cho, R.; Huang, J.C.-j.; Miao, S.; Wong, B.K. A Fully Automated Plasma Protein Precipitation Sample Preparation Method for LC–MS/MS Bioanalysis. J. Chromatogr. B 2008, 862, 219–226. [Google Scholar] [CrossRef]
- Grigoriev, A.; Borisova, I.; Yaroshenko, I.; Sidorova, A. In Vitro and in Vivo Stability of Oseltamivir within a Bioequivalence Trial. Anal. Bioanal. Chem. 2016, 408, 3891–3897. [Google Scholar] [CrossRef]
- Dies, H.; Raveendran, J.; Escobedo, C.; Docoslis, A. Rapid Identification and Quantification of Illicit Drugs on Nanodendritic Surface-Enhanced Raman Scattering Substrates. Sens. Actuators B 2018, 257, 382–388. [Google Scholar] [CrossRef]
- Courlet, P.; Spaggiari, D.; Cavassini, M.; Du Pasquier, R.; Alves Saldanha, S.; Buclin, T.; Marzolini, C.; Csajka, C.; Decosterd, L. Determination of Nucleosidic/Tidic Reverse Transcriptase Inhibitors in Plasma and Cerebrospinal Fluid by Ultra-High-Pressure Liquid Chromatography Coupled with Tandem Mass Spectrometry. Clin. Mass Spectrom. 2018, 8, 8–20. [Google Scholar] [CrossRef]
- Stefancu, A.; Badarinza, M.; Moisoiu, V.; Iancu, S.D.; Serban, O.; Leopold, N.; Fodor, D. SERS-Based Liquid Biopsy of Saliva and Serum from Patients with Sjögren’s Syndrome. Anal. Bioanal. Chem. 2019, 411, 5877–5883. [Google Scholar] [CrossRef]
- Li, P.; Ge, M.; Cao, C.; Lin, D.; Yang, L. High-Affinity Fe3O4/Au Probe with Synergetic Effect of Surface Plasmon Resonance and Charge Transfer Enabling Improved SERS Sensing of Dopamine in Biofluids. Analyst 2019, 144, 4526–4533. [Google Scholar] [CrossRef] [PubMed]
- Meenks, S.; Le Noble, J.; Foudraine, N.; de Vries, F.; Janssen, P. Liquid Chromatography-Tandem Mass Spectrometry to Monitor Unbound and Total Ceftriaxone in Serum of Critically Ill Patients. Curr. Rev. Clin. Exp. Pharmacol. 2021, 16, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Markin, A.V. Application of Aluminum Hydroxide for Improvement of Label-Free SERS Detection of Some Cephalosporin Antibiotics in Urine. Biosensors 2019, 9, 91. [Google Scholar] [CrossRef]
- Norrby, S.R. Side Effects of Cepbalosporins. Drugs 1987, 34 (Suppl. 2), 105–120. [Google Scholar] [CrossRef]
- Markina, N.E.; Ustinov, S.N.; Zakharevich, A.M.; Markin, A.V. Copper Nanoparticles for SERS-Based Determination of Some Cephalosporin Antibiotics in Spiked Human Urine. Anal. Chim. Acta 2020, 1138, 9–17. [Google Scholar] [CrossRef]
- Markina, N.E.; Volkova, E.K.; Zakharevich, A.M.; Goryacheva, I.Y.; Markin, A.V. SERS Detection of Ceftriaxone and Sulfadimethoxine Using Copper Nanoparticles Temporally Protected by Porous Calcium Carbonate. Microchim. Acta 2018, 185, 481. [Google Scholar] [CrossRef]
- Guo, S.; Popp, J.; Bocklitz, T. Chemometric Analysis in Raman Spectroscopy from Experimental Design to Machine Learning–Based Modeling. Nat. Protoc. 2021, 16, 5426–5459. [Google Scholar] [CrossRef]
- Villa, J.E.L.; Quiñones, N.R.; Fantinatti-Garboggini, F.; Poppi, R.J. Fast Discrimination of Bacteria Using a Filter Paper–Based SERS Platform and PLS-DA with Uncertainty Estimation. Anal. Bioanal. Chem. 2019, 411, 705–713. [Google Scholar] [CrossRef]
- Guo, S.; Bocklitz, T.; Neugebauer, U.; Urgen Popp, J. Common Mistakes in Cross-Validating Classification Models. Anal. Methods 2017, 9, 4410–4417. [Google Scholar] [CrossRef]
- Storozhuk, D.; Ryabchykov, O.; Popp, J.; Bocklitz, T. RAMANMETRIX: A Delightful Way to Analyze Raman Spectra. arXiv 2022, arXiv:2201.07586. [Google Scholar]
- Ryabchykov, O.; Bocklitz, T.; Ramoji, A.; Neugebauer, U.; Foerster, M.; Kroegel, C.; Bauer, M.; Kiehntopf, M.; Popp, J. Automatization of Spike Correction in Raman Spectra of Biological Samples. Chemom. Intell. Lab. Syst. 2016, 155, 1–6. [Google Scholar] [CrossRef]
- Erb, D. Pybaselines: A Python Library of Algorithms for the Baseline Correction of Experimental Data; Zenodo: Geneva, Switzerland, 2022. [Google Scholar] [CrossRef]
- Koleva, B.B.; Kolev, T.M.; Spiteller, M. Determination of Cephalosporins in Solid Binary Mixtures by Polarized IR- and Raman Spectroscopy. J. Pharm. Biomed. Anal. 2008, 48, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Ingraham, H.M.; Premasiri, W.R.; Ziegler, L.D. NIR Surface Enhanced Raman Spectra of Biological Hemes: Solvation and Plasmonic Metal Effects. J. Raman Spectrosc. 2021, 52, 2500–2515. [Google Scholar] [CrossRef]
- Feng, S.; Chen, R.; Lin, J.; Pan, J.; Chen, G.; Li, Y.; Cheng, M.; Huang, Z.; Chen, J.; Zeng, H. Nasopharyngeal Cancer Detection Based on Blood Plasma Surface-Enhanced Raman Spectroscopy and Multivariate Analysis. Biosens. Bioelectron. 2010, 25, 2414–2419. [Google Scholar] [CrossRef] [PubMed]
- Premasiri, W.R.; Lee, J.C.; Ziegler, L.D. Surface-Enhanced Raman Scattering of Whole Human Blood, Blood Plasma, and Red Blood Cells: Cellular Processes and Bioanalytical Sensing. J. Phys. Chem. B 2012, 116, 9376–9386. [Google Scholar] [CrossRef]
- Bruce, S.J.; Tavazzi, I.; Parisod, V.; Rezzi, S.; Kochhar, S.; Guy, P.A. Investigation of Human Blood Plasma Sample Preparation for Performing Metabolomics Using Ultrahigh Performance Liquid Chromatography/Mass Spectrometry. Anal. Chem. 2009, 81, 3285–3296. [Google Scholar] [CrossRef]
- Roberts, J.A.; Paul, S.K.; Akova, M.; Bassetti, M.; De Waele, J.J.; Dimopoulos, G.; Kaukonen, K.-M.; Koulenti, D.; Martin, C.; Montravers, P.; et al. DALI: Defining Antibiotic Levels in Intensive Care Unit Patients: Are Current β-Lactam Antibiotic Doses Sufficient for Critically Ill Patients? Clin. Infect. Dis. 2014, 58, 1072–1083. [Google Scholar] [CrossRef]
- Pollock, A.A.; Tee, P.E.; Patel, I.H.; Spicehandler, J.; Simberkoff, M.S.; Rahal, J.J., Jr. Pharmacokinetic Characteristics of Intravenous Ceftriaxone in Normal Adults. Antimicrob. Agents Chemother. 1982, 22, 816–823. [Google Scholar] [CrossRef]
- Liu, C.; Franceschini, C.; Weber, S.; Dib, T.; Liu, P.; Wu, L.; Farnesi, E.; Zhang, W.-S.; Sivakov, V.; Luppa, P.B.; et al. SERS-Based Detection of the Antibiotic Ceftriaxone in Spiked Fresh Plasma and Microdialysate Matrix by Using Silver-Functionalized Silicon Nanowire Substrates. Talanta 2024, 271, 125697. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Kong, D.; Yuan, L.; Yang, J.; Zhang, J.; Luo, M.; Teng, P.; Gao, D.; Yang, X.; et al. Optofluidic In-Fiber Integrated Surface-Enhanced Raman Spectroscopy Detection Based on a Hollow Optical Fiber with a Suspended Core. Opt. Lett. 2019, 44, 5173–5176. [Google Scholar] [CrossRef]
- Avci, E.; Yilmaz, H.; Sahiner, N.; Tuna, B.G.; Cicekdal, M.B.; Eser, M.; Basak, K.; Altıntoprak, F.; Zengin, I.; Dogan, S.; et al. Label-Free Surface Enhanced Raman Spectroscopy for Cancer Detection. Cancers 2022, 14, 5021. [Google Scholar] [CrossRef] [PubMed]
- Bonifacio, A. Label-Free Surface-Enhanced Raman Scattering for Clinical Applications. In Principles and Clinical Diagnostic Applications of Surface-Enhanced Raman Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2021; pp. 125–170. ISBN 9780128211212. [Google Scholar] [CrossRef]
- Bonifacio, A.; Cervo, S.; Sergo, V. Label-Free Surface-Enhanced Raman Spectroscopy of Biofluids: Fundamental Aspects and Diagnostic Applications. Anal. Bioanal. Chem. 2015, 407, 8265–8277. [Google Scholar] [CrossRef] [PubMed]
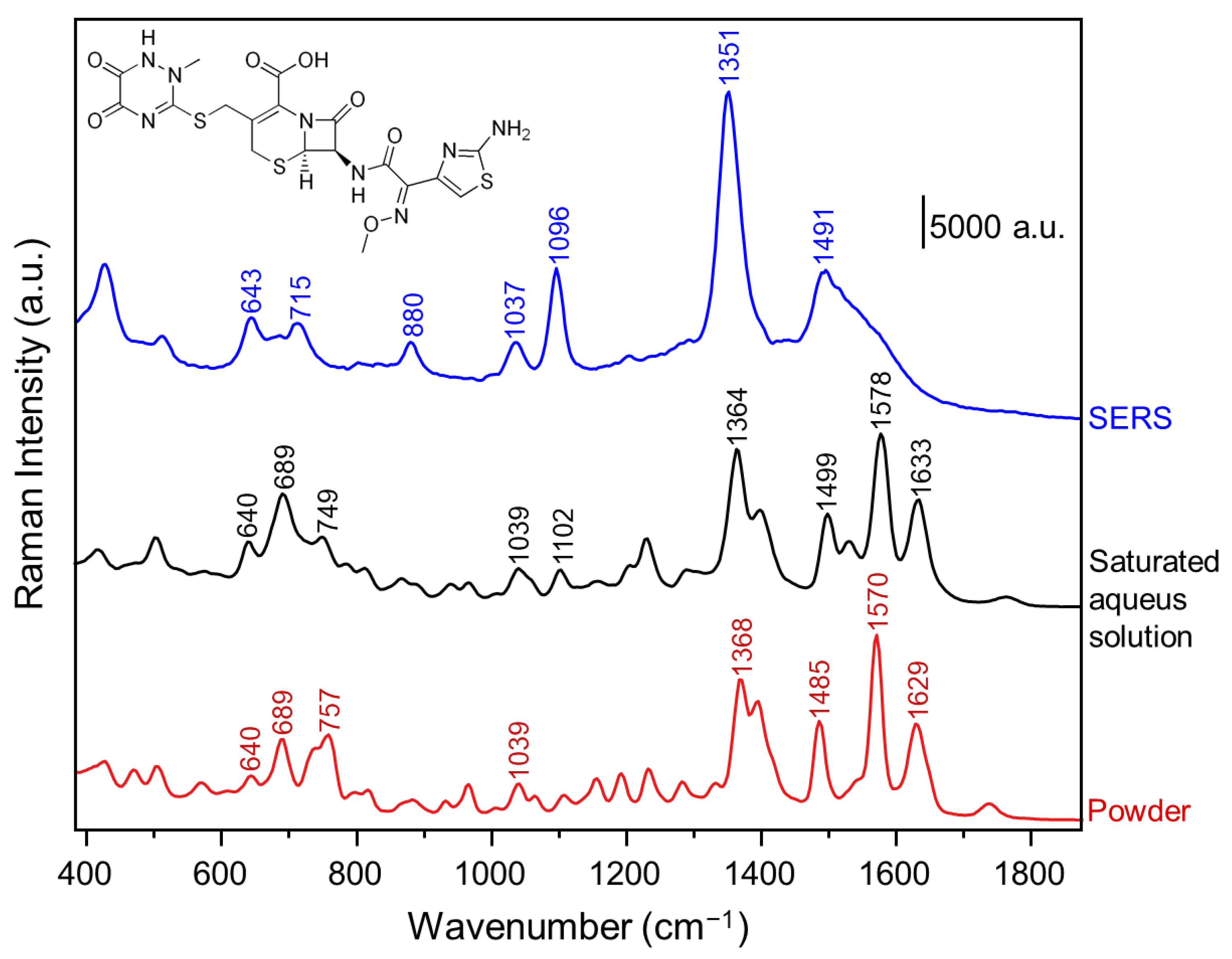


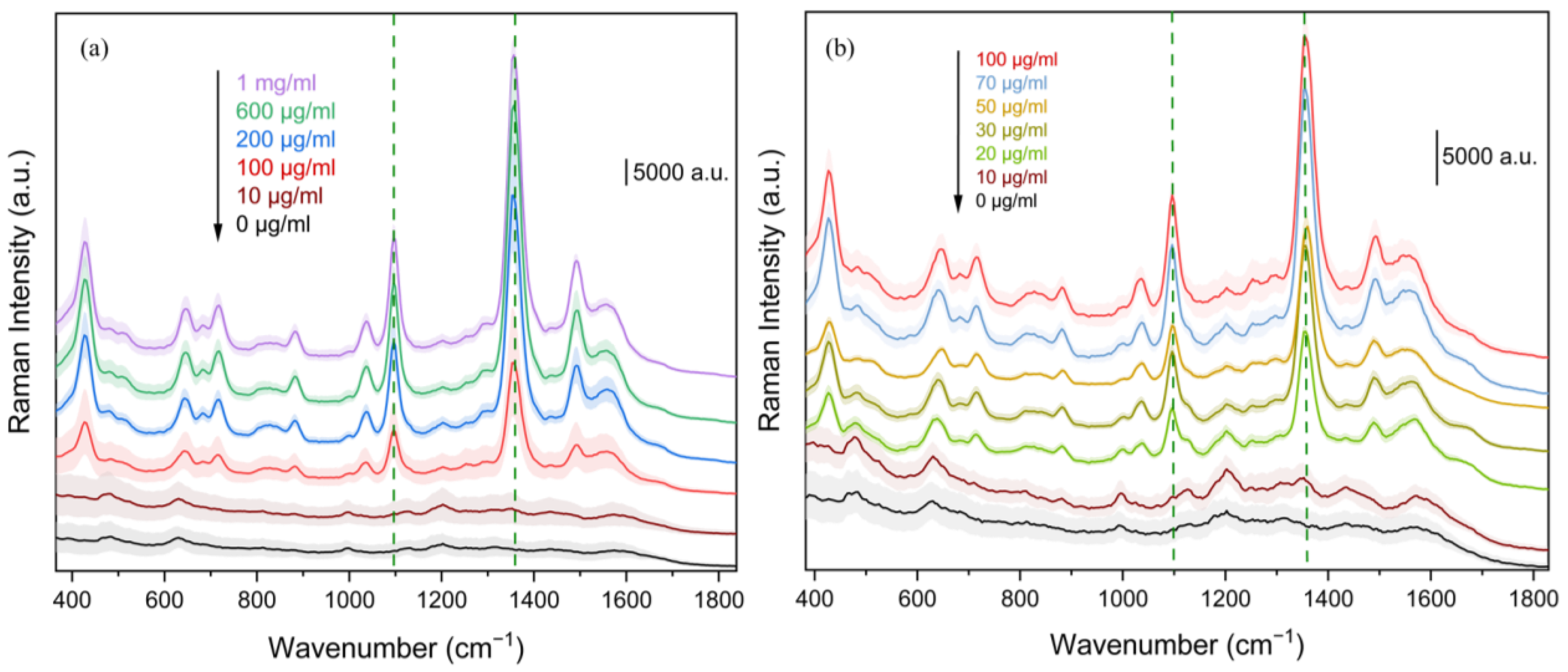

| SERS Substrate | Matrix | Lowest Detected Concentration (LOD) | Detection Range | Reference |
|---|---|---|---|---|
| AgNPs | urine | 0.4 μg mL−1 | 5–500 μg mL−1 | [18] |
| SERS optical fiber | water | 1 μM (0.5 μg mL−1) | 10–10,000 μM | [51] |
| CuNP | urine | 7.5 μg mL−1 | 50–500 μg mL−1 | [35] |
| AgNPs | urine | 92 μg mL−1 | 100–500 μg mL−1 | [33] |
| Ag@SiNWs | Blood plasma | 94 μM (52 μg mL−1) | 1–1000 μM | [50] |
| Ag@SiNWs | Microdialysate | 1.4 μM (0.8 μg mL−1) | 2.5–1000 μM | [50] |
| Commercial substrate (Silmeco) | Blood plasma | 20 μg mL−1 | 10–1000 μg mL−1 | This work |
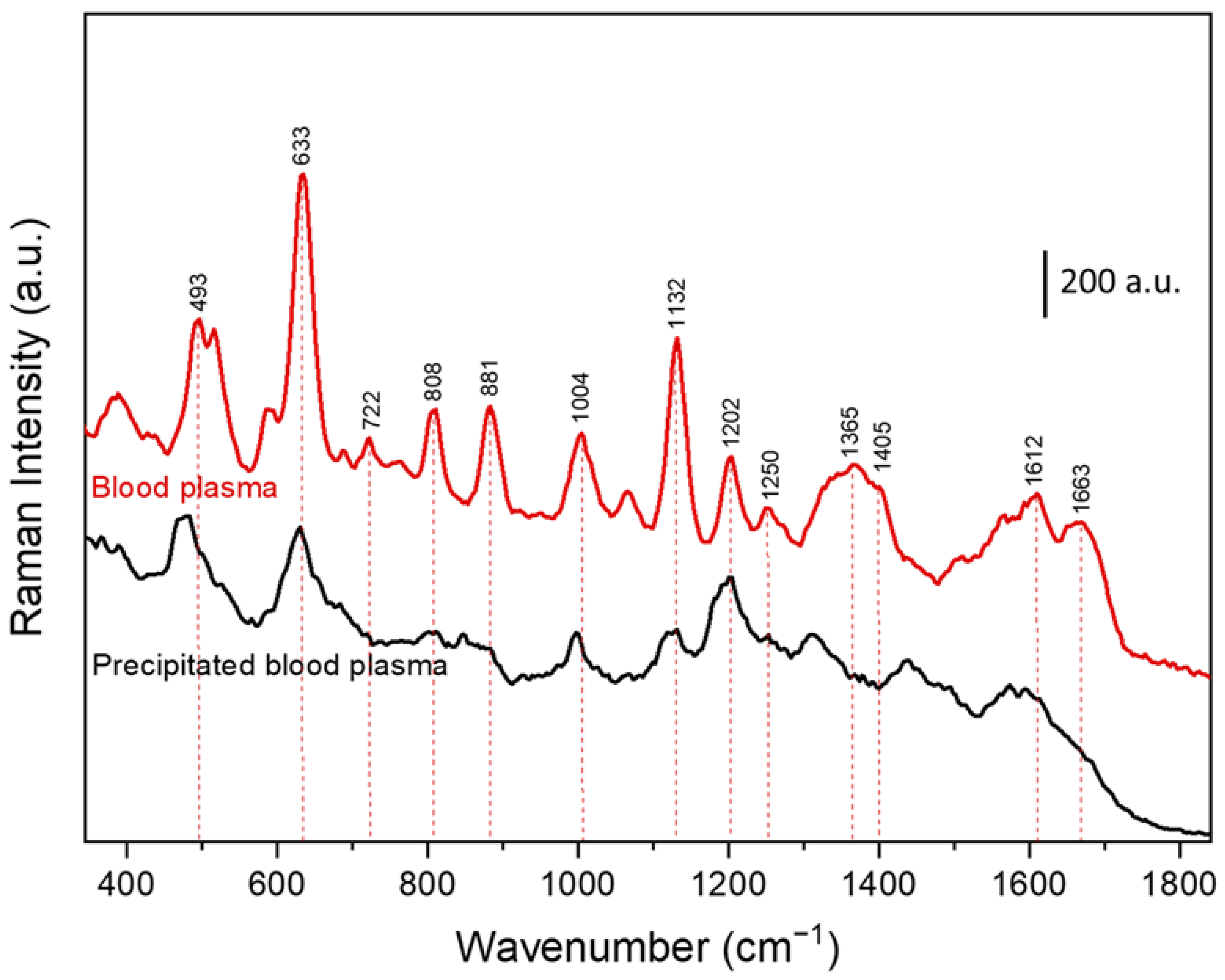
| SERS Band Positions (cm−1) | Assignments |
|---|---|
| 493 | L-Arginine [24,45] |
| 633 | Uric Acid [22,24] |
| 722 | Hypoxanthine or Adenine [24,45] |
| 808 | L-serine, Glutathione [45] |
| 881 | Uric Acid [22,24] |
| 1004 | Phenylalanine, Hypoxanthine [22,44] |
| 1132 | Uric Acid [22,24] |
| 1202 | L-tryptophan, Phenylalanine [45] |
| 1250 | Amide Ⅲ [22] |
| 1365 | Adenine [24] |
| 1405 | CH2 deformation, phospholipids [22,24,45] |
| 1612 | Uric acid [22] |
| 1663 | Amide Ⅰ [22,24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dwivedi, A.; Ryabchykov, O.; Liu, C.; Farnesi, E.; Schmidt, M.S.; Bocklitz, T.; Popp, J.; Cialla-May, D. SERS-Driven Ceftriaxone Detection in Blood Plasma: A Protein Precipitation Approach. Chemosensors 2024, 12, 213. https://doi.org/10.3390/chemosensors12100213
Dwivedi A, Ryabchykov O, Liu C, Farnesi E, Schmidt MS, Bocklitz T, Popp J, Cialla-May D. SERS-Driven Ceftriaxone Detection in Blood Plasma: A Protein Precipitation Approach. Chemosensors. 2024; 12(10):213. https://doi.org/10.3390/chemosensors12100213
Chicago/Turabian StyleDwivedi, Aradhana, Oleg Ryabchykov, Chen Liu, Edoardo Farnesi, Michael Stenbæk Schmidt, Thomas Bocklitz, Jürgen Popp, and Dana Cialla-May. 2024. "SERS-Driven Ceftriaxone Detection in Blood Plasma: A Protein Precipitation Approach" Chemosensors 12, no. 10: 213. https://doi.org/10.3390/chemosensors12100213
APA StyleDwivedi, A., Ryabchykov, O., Liu, C., Farnesi, E., Schmidt, M. S., Bocklitz, T., Popp, J., & Cialla-May, D. (2024). SERS-Driven Ceftriaxone Detection in Blood Plasma: A Protein Precipitation Approach. Chemosensors, 12(10), 213. https://doi.org/10.3390/chemosensors12100213









