AuNPs/Ti3C2 Signal-Enhanced Surface Plasmon Resonance Imaging Biosensor for Ultrasensitive Detection of miRNA
Abstract
:1. Introduction
2. Experimental Section
2.1. Chemicals and Materials
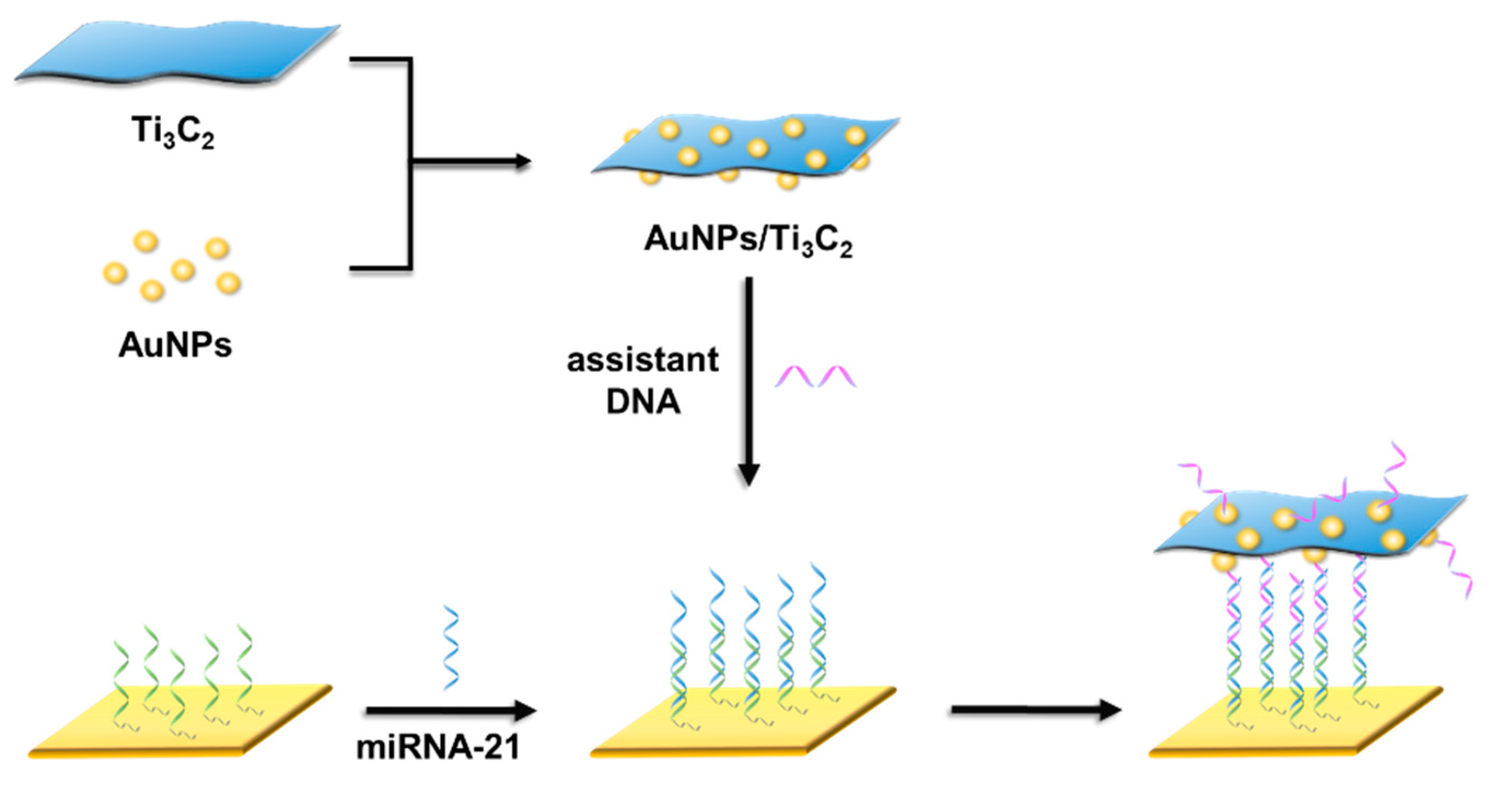
2.2. Preparation of Ti3C2 Nanosheets and AuNPs/Ti3C2
2.3. Preparation of DNA−AuNPs and DNA−AuNPs/Ti3C2
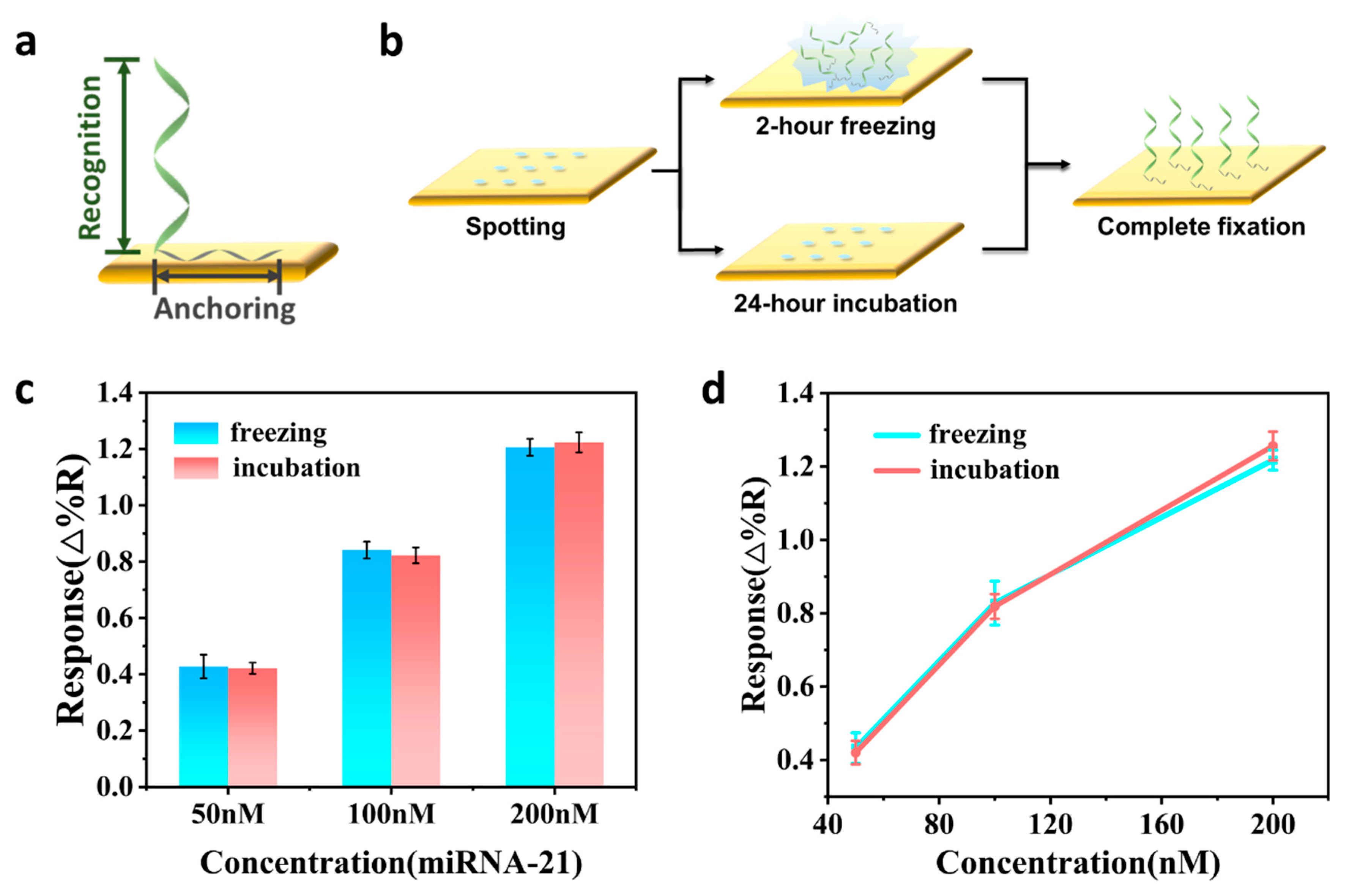
2.4. Au Chip Functionalization
2.5. SPRi Detection
3. Results and Discussion
3.1. SPRi Detection Principle
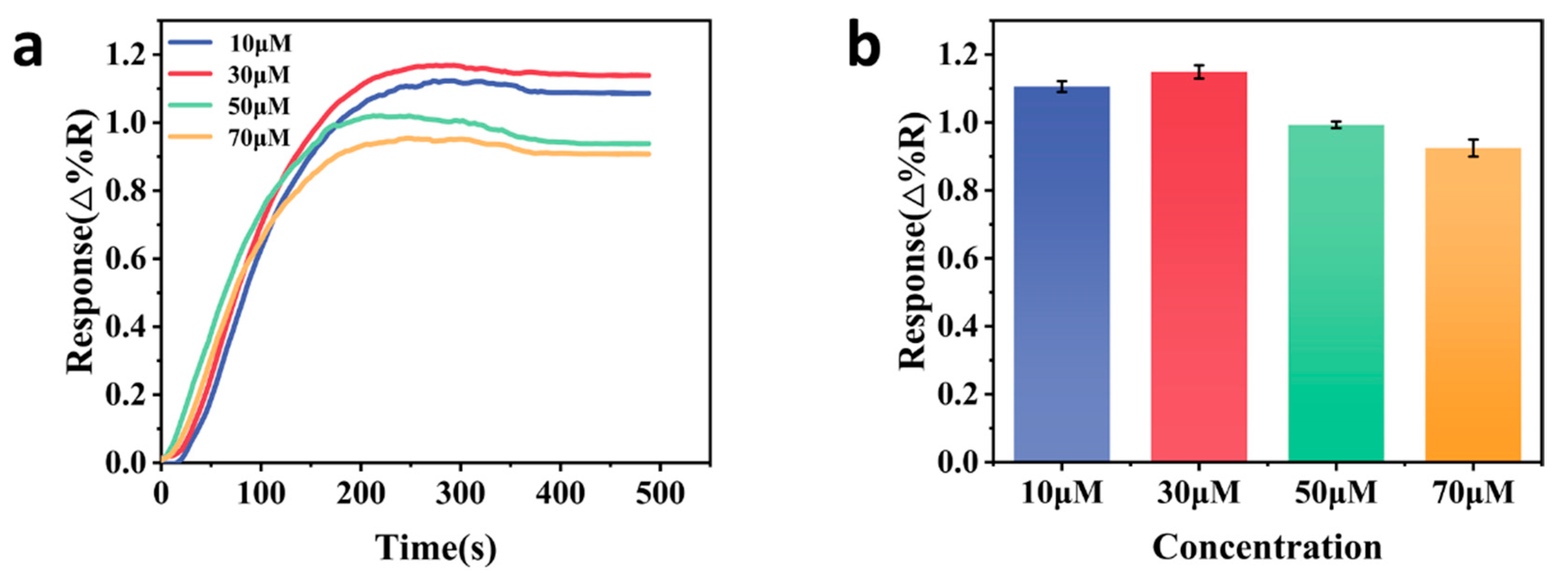
3.2. Optimization of Experimental Conditions
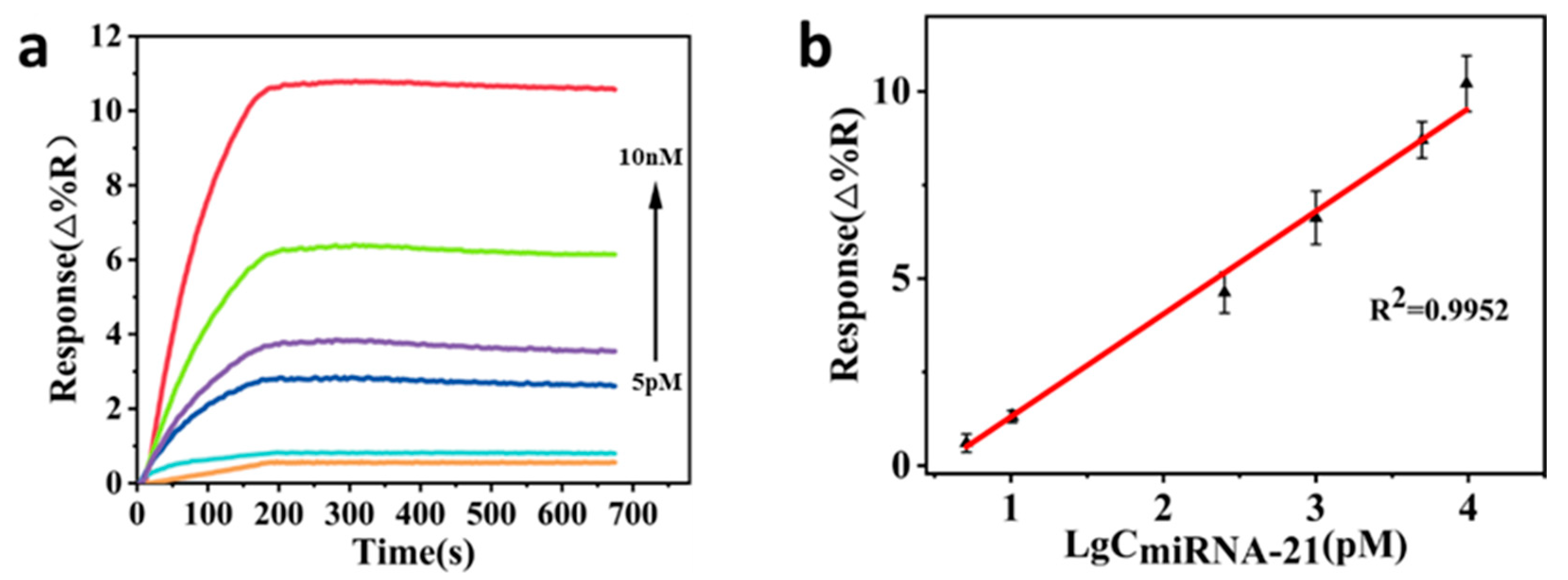
3.3. Direct Detection of Target miRNA-21
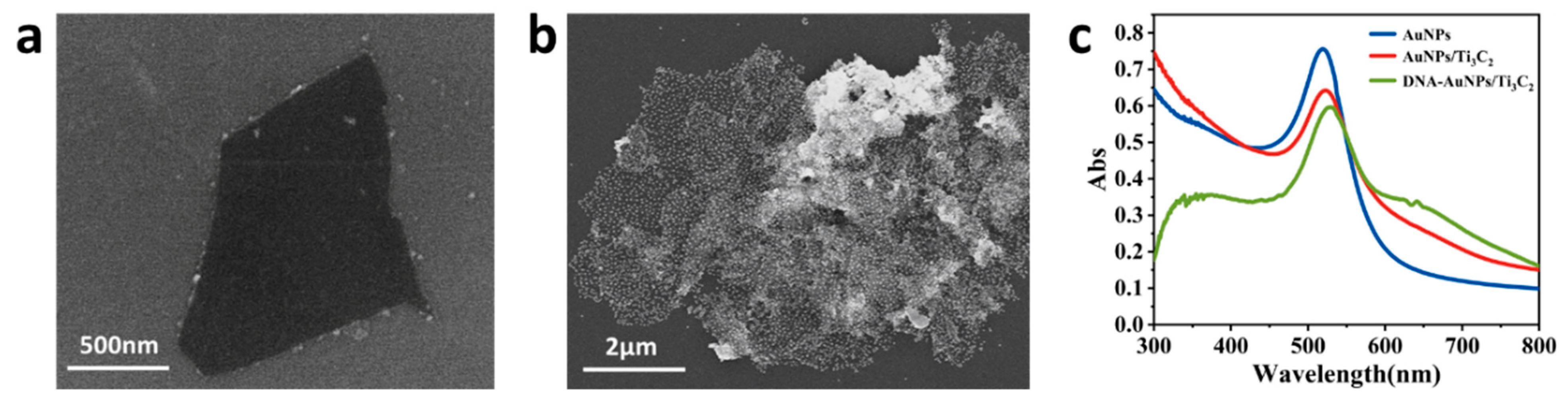
3.4. Characterization of AuNPs/Ti3C2 and DNA−AuNPs/Ti3C2 Composites
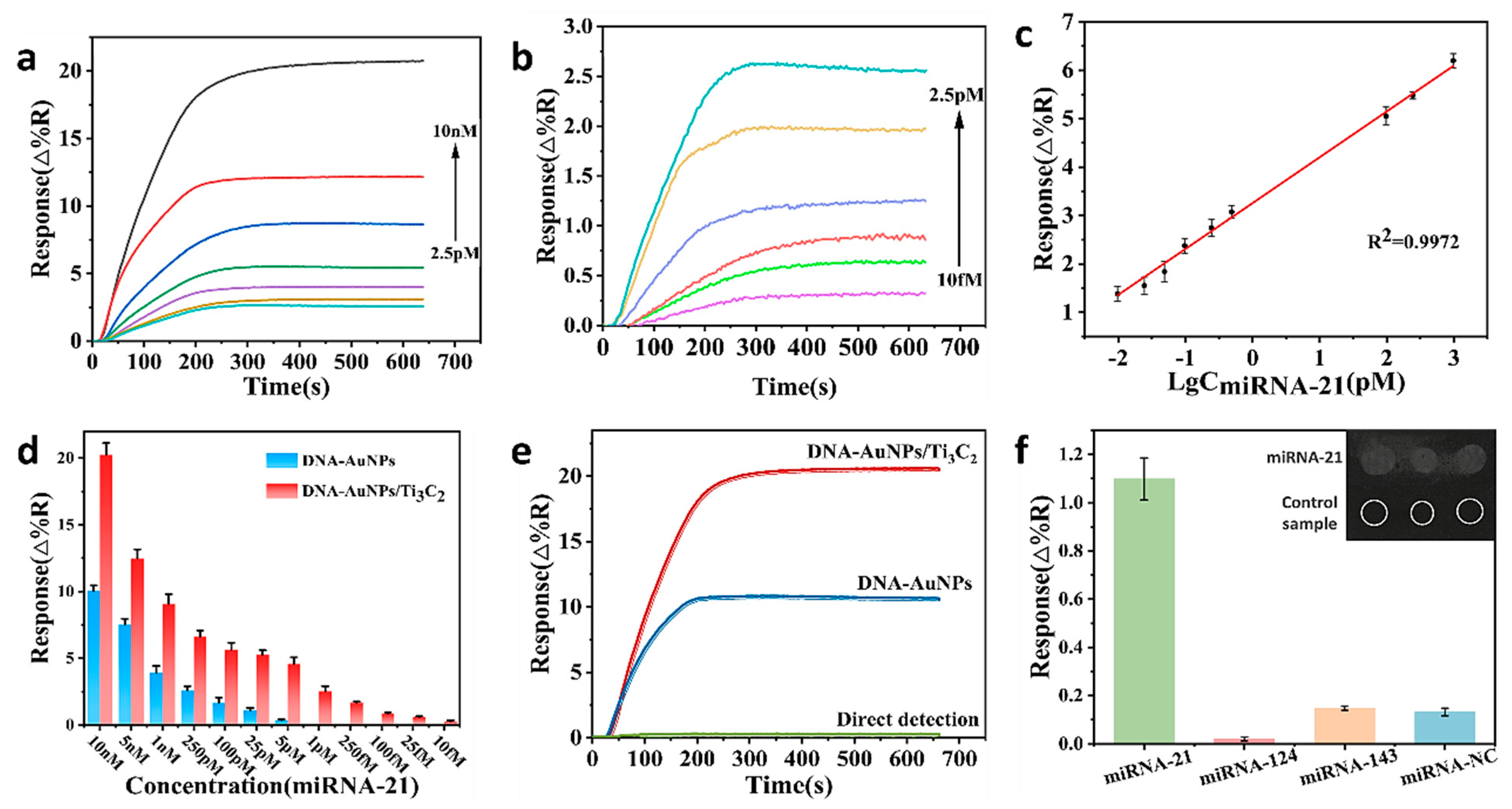
3.5. Enhanced SPRi Signaling by DNA−AuNPs/Ti3C2
3.6. Specificity of SPRi Biosensors
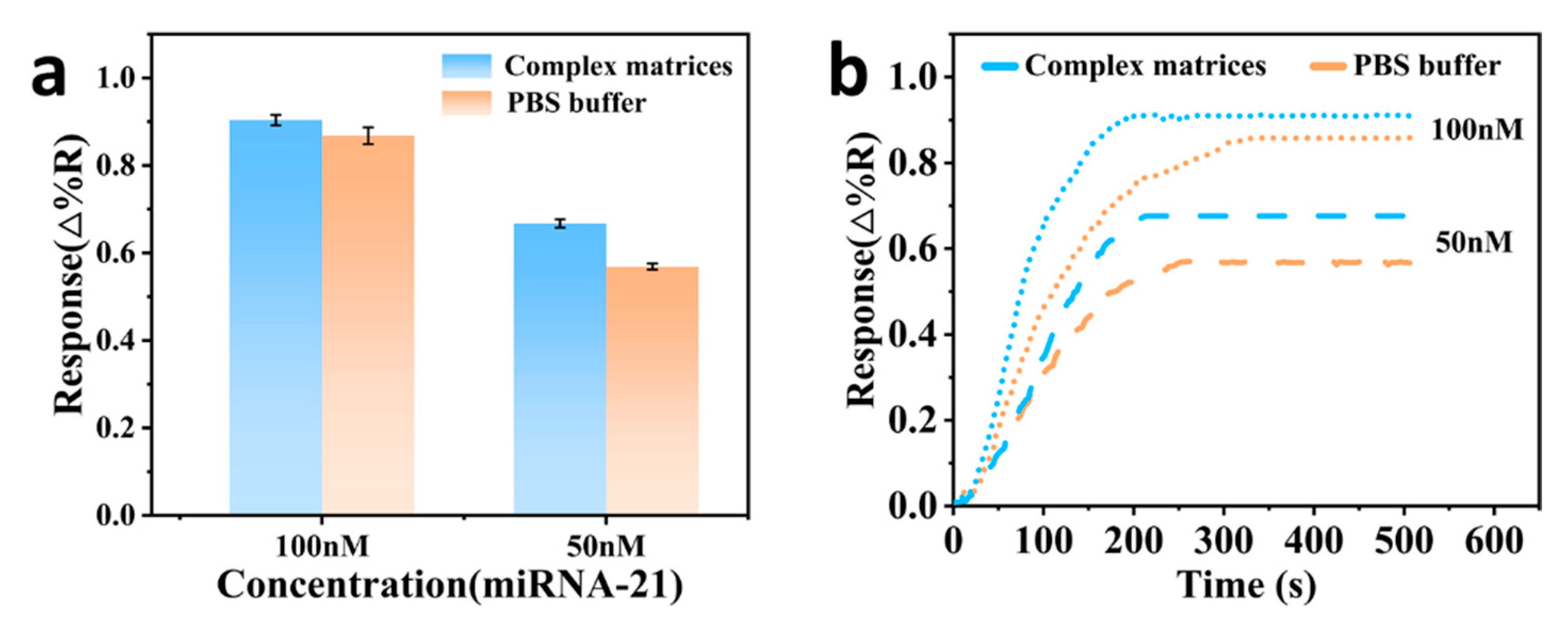
3.7. Detection in Salmon Sperm Sample
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, T.X. Rothenberg M E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA: miRNA Interactions: A Novel Mode of miRNA Regulation and Its Effect on Disease Systems Biology of MicroRNAs in Cancer; Springer International Publishing: Cham, Switzerland, 2022; pp. 241–257. [Google Scholar]
- Shao, T.; Wang, G.; Chen, H.; Xie, Y.; Jin, X.; Bai, J.; Xu, J.; Li, X.; Huang, J.; Jin, Y.; et al. Survey of miRNA-miRNA cooperative regulation principles across cancer types. Brief. Bioinform. 2019, 20, 1621–1638. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, Y.B.; Zhang, X.H.; Yu, X.L.; Bin Wang, Z.; Cheng, X.C. MicroRNA-21 gene and cancer. Med. Oncol. 2013, 30, 376. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell. Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Shen, L.; Wan, Z.; Ma, Y.; Wu, L.; Liu, F.; Zang, H.; Xin, S. The clinical utility of microRNA-21 as novel biomarker for diagnosing human cancers. Tumor Biol. 2015, 36, 1993–2005. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The role of miR-21 in cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef]
- Sara, M.; Tammaro, C.; Misso, G.; Falco, M.; Scrima, M.; Bocchetti, M.; Rea, I.; De Stefano, L.; Caraglia, M. microRNA detection via nanostructured biochips for early cancer diagnostics. Int. J. Mol. Sci. 2023, 24, 7762. [Google Scholar] [CrossRef]
- Liu, Q.; Fan, J.; Zhou, C.; Wang, L.; Zhao, B.; Zhang, H.; Liu, B.; Tong, C. Quantitative detection of miRNA-21 expression in tumor cells and tissues based on molecular beacon. Int. J. Anal. Chem. 2018, 2018, 3625823. [Google Scholar] [CrossRef]
- Kong, L.; Li, H.; Zhang, X.; Zhuo, Y.; Chai, Y.; Yuan, R. A novel ratiometric electrochemical biosensor using only one signal tag for highly reliable and ultrasensitive detection of miRNA-21. Anal. Chem. 2022, 94, 5167–5172. [Google Scholar] [CrossRef]
- Weng, S.; Lin, D.; Lai, S.; Tao, H.; Chen, T.; Peng, M.; Qiu, S.; Feng, S. Highly sensitive and reliable detection of microRNA for clinically disease surveillance using SERS biosensor integrated with catalytic hairpin assembly amplification technology. Biosens. Bioelectron. 2022, 208, 114236. [Google Scholar] [CrossRef] [PubMed]
- Puiu, M.; Bala, C. SPR and SPR imaging: Recent trends in development nanodevices for detection and real-time monitoring of biomolecular events. Sensors 2016, 16, 870. [Google Scholar] [CrossRef] [PubMed]
- Fathi, F.; Rashidi, M.R.; Omidi, Y. Ultra-sensitive detection by metal nanoparticles-mediated enhanced SPR biosensors. Talanta 2019, 192, 118–127. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Q.; Li, Q.; Yang, X.; Wang, K.; Nie, W. Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy. Biosens. Bioelectron. 2017, 87, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, R.; Yang, X.; Wang, K.; Zhu, J.; He, L.; Li, Q. Surface plasmon resonance biosensor for enzyme-free amplified microRNA detection based on gold nanoparticles and DNA supersandwich. Sens. Actuators B Chem. 2016, 223, 613–620. [Google Scholar] [CrossRef]
- Hong, L.; Lu, M.; Dinel, M.P.; Blain, P.; Peng, W.; Gu, H.; Masson, J.-F. Hybridization conditions of oligonucleotide-capped gold nanoparticles for SPR sensing of microRNA. Biosens. Bioelectron. 2018, 109, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.S.; Raghuwanshi, S.K.; Kumar, S. Recent advances in two-dimensional materials-based Kretschmann configuration for SPR sensors: A review. IEEE Sens. J. 2021, 22, 1069–1080. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Q.; Yang, X.; Wang, K.; Du, S.; Zhang, H.; Nie, Y. Graphene oxide—Gold nanoparticles hybrids-based surface plasmon resonance for sensitive detection of microRNA. Biosens. Bioelectron. 2016, 77, 1001–1007. [Google Scholar] [CrossRef]
- Li, Q.; Wang, Q.; Yang, X.; Wang, K.; Zhang, H.; Nie, W. High sensitivity surface plasmon resonance biosensor for detection of microRNA and small molecule based on graphene oxide-gold nanoparticles composites. Talanta 2017, 174, 521–526. [Google Scholar] [CrossRef]
- Nie, W.; Wang, Q.; Yang, X.; Zhang, H.; Li, Z.; Gao, L.; Zheng, Y. High sensitivity surface plasmon resonance biosensor for detection of microRNA based on gold nanoparticles-decorated molybdenum sulfide. Anal. Chim Acta 2017, 993, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.A.; Prakash, P.; Krithiga, T.; Amarnath, D.J.; Premkumar, J.; Rajamohan, N.; Vasseghian, Y.; Saravanan, P.; Rajasimman, M. Methods of synthesis, characteristics, and environmental applications of MXene: A comprehensive review. Chemosphere 2022, 286, 131607. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jiang, L.; Lu, C.; Yu, Z.; Li, F.; Jing, X.; Xu, R.; Zhou, W.; Jin, S. Large-scale two-dimensional titanium carbide MXene as SERS-active substrate for reliable and sensitive detection of organic pollutants. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 236, 118336. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, C.; Cao, Y.; Wang, G. Functional MXene materials: Progress of their applications. Chem. Asian J. 2018, 13, 2742–2757. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, N.; Wang, Y.; Liu, Y.; Xu, Y.; Wei, S.; Wu, J.; Jia, G.; Fang, X.; Chen, F.; et al. A 2D transition metal carbide MXene-based SPR biosensor for ultrasensitive carcinoembryonic antigen detection. Biosens. Bioelectron. 2019, 144, 111697. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, N.; Wang, Y.; Xu, Y.; Wu, J.; Jia, G.; Ji, F.; Fang, X.; Chen, F.; Cui, X. Ultrasensitive and selective determination of carcinoembryonic antigen using multifunctional ultrathin amino-functionalized Ti3C2-MXene nanosheets. Anal. Chem. 2020, 92, 3354–3360. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Y.; Xie, R.; Zeng, Q.; Hong, Y.; Chen, X.; Zhang, P.; Zeng, L.; Zhang, Y.; Zeng, S.; et al. Ultrasensitive label-free miRNA-21 detection based on MXene-enhanced plasmonic lateral displacement measurement. Nanophotonics 2023, 12, 4055–4062. [Google Scholar] [CrossRef]
- Liu, R.; Jiang, L.; Yu, Z.; Jing, X.; Liang, X.; Wang, D.; Yang, B.; Lu, C.; Zhou, W.; Jin, S. MXene (Ti3C2Tx)-Ag nanocomplex as efficient and quantitative SERS biosensor platform by in-situ PDDA electrostatic self-assembly synthesis strategy. Sens. Actuators B Chem. 2021, 333, 129581. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, L.; Liu, R.; Zhao, W.; Yang, Z.; Zhang, J.; Jin, S. Versatile self-assembled MXene-Au nanocomposites for SERS detection of bacteria, antibacterial and photothermal sterilization. Chem. Eng. J. 2021, 426, 131914. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J. Freezing directed construction of bio/nano interfaces: Reagentless conjugation, denser spherical nucleic acids, and better nanoflares. J. Am. Chem. Soc. 2017, 139, 9471–9474. [Google Scholar] [CrossRef]
- Fang, Y.; Jiang, L.; Jin, S.; Li, Y.; Jiang, C.; Zhang, X.; Peng, Y. AuNPs beacons-enhanced surface plasmon resonance imaging sensor for rapid, high-throughput and ultra-sensitive detection of three fusion genes related to acute promyelocytic leukemia. Sens. Actuators B Chem. 2022, 361, 131728. [Google Scholar] [CrossRef]
- Huertas, C.S.; Soler, M.; Estevez, M.C.; Lechuga, L.M. One-step immobilization of antibodies and DNA on gold sensor surfaces via a poly-adenine oligonucleotide approach. Anal. Chem. 2020, 92, 12596–12604. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zou, Y.; Tan, H.; Jiang, L.; Fang, Y.; Jin, S. Simultaneous and sensitive detection of two pathogenic genes of thrombotic diseases using SPRi sensor with one-step fixation probe by a poly-adenine oligonucleotide approach. Colloids Surf. B Biointerfaces 2022, 209, 112184. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Guo, B.; Zhao, M.; Han, D.; Cheng, W.; Song, F.; Ding, S. An enzyme-free “ON-OFF” electrochemiluminescence biosensor for ultrasensitive detection of PML/RARα based on target-switched DNA nanotweezer. ACS Appl. Mater. Interfaces 2019, 11, 3715–3721. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Y.; Jiang, L.; Sun, R.; Fang, Y.; Shi, B.; Jin, S. AuNPs/Ti3C2 Signal-Enhanced Surface Plasmon Resonance Imaging Biosensor for Ultrasensitive Detection of miRNA. Chemosensors 2024, 12, 66. https://doi.org/10.3390/chemosensors12040066
Qin Y, Jiang L, Sun R, Fang Y, Shi B, Jin S. AuNPs/Ti3C2 Signal-Enhanced Surface Plasmon Resonance Imaging Biosensor for Ultrasensitive Detection of miRNA. Chemosensors. 2024; 12(4):66. https://doi.org/10.3390/chemosensors12040066
Chicago/Turabian StyleQin, Yirui, Li Jiang, Rengang Sun, Yunzhu Fang, Boya Shi, and Shangzhong Jin. 2024. "AuNPs/Ti3C2 Signal-Enhanced Surface Plasmon Resonance Imaging Biosensor for Ultrasensitive Detection of miRNA" Chemosensors 12, no. 4: 66. https://doi.org/10.3390/chemosensors12040066
APA StyleQin, Y., Jiang, L., Sun, R., Fang, Y., Shi, B., & Jin, S. (2024). AuNPs/Ti3C2 Signal-Enhanced Surface Plasmon Resonance Imaging Biosensor for Ultrasensitive Detection of miRNA. Chemosensors, 12(4), 66. https://doi.org/10.3390/chemosensors12040066



