Quick Plant Sample Preparation Methods Using a Micro-Homogenizer for the Detection of Multiple Citrus Pathogens
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Buffers
2.2. Pathogens and Plant Materials
2.3. Primers and Probes
2.4. Citrus Tissue Processing
2.4.1. High-Throughput Benchtop Protocol for Lysis of Positive and Negative Controls
2.4.2. Semi-Automated Protocol for Lysis of Test Samples
2.4.3. Manual Extraction Protocol for Preparation of Test Samples
2.5. High-Throughput Protocol for Extraction of Nucleic Acids from Lysates
2.6. Quick Sample Preparation Protocols for qPCR/RT–qPCR Assays
2.6.1. Serial Dilution Method
2.6.2. Paper Disk Method
2.7. Singleplex and Multiplex qPCR/RT–qPCR Assays
3. Results and Discussion
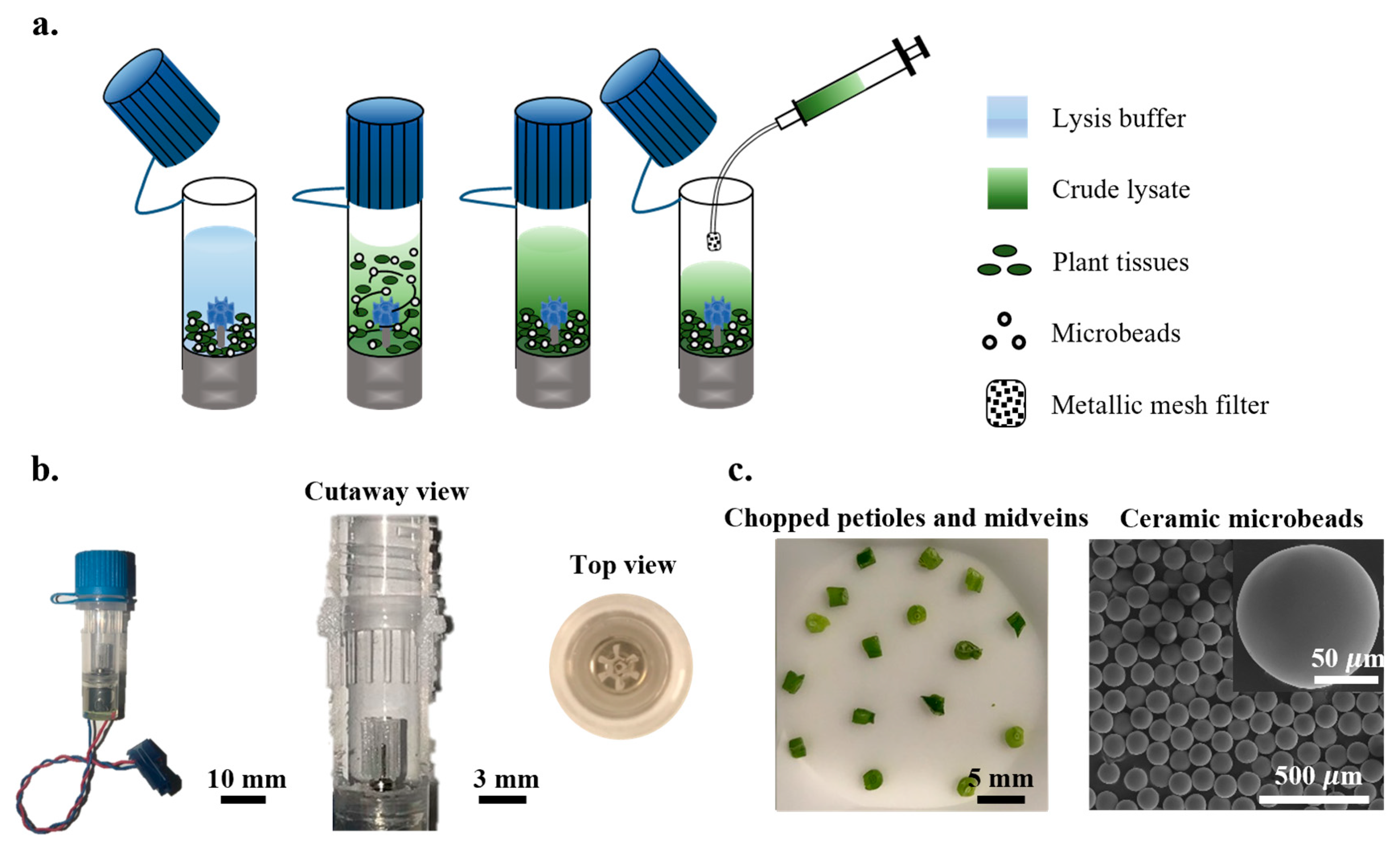
3.1. Characterization of OmniLyse Micro-Homogenizer
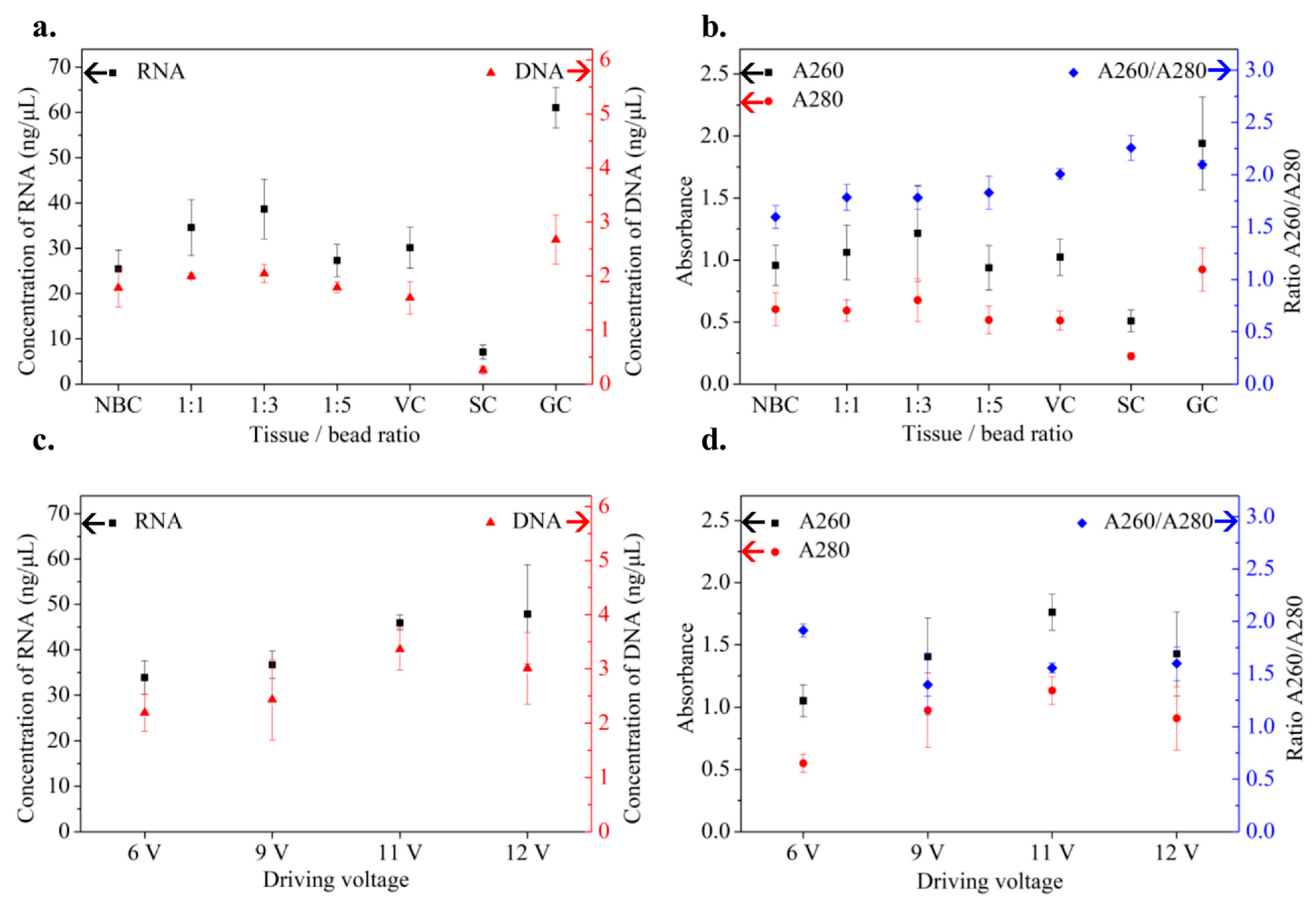
3.1.1. Quality and Quantity of Extracts
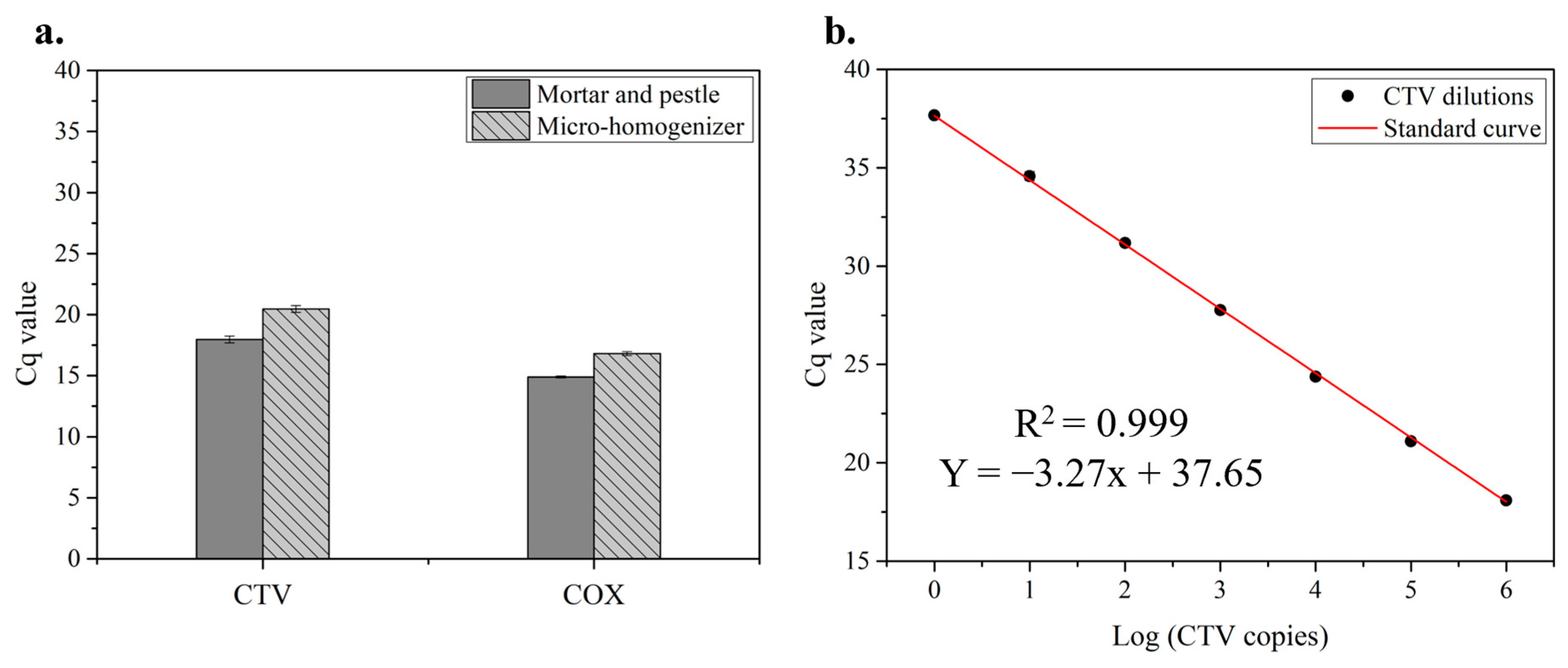
3.1.2. Verification with RT–qPCR Assays
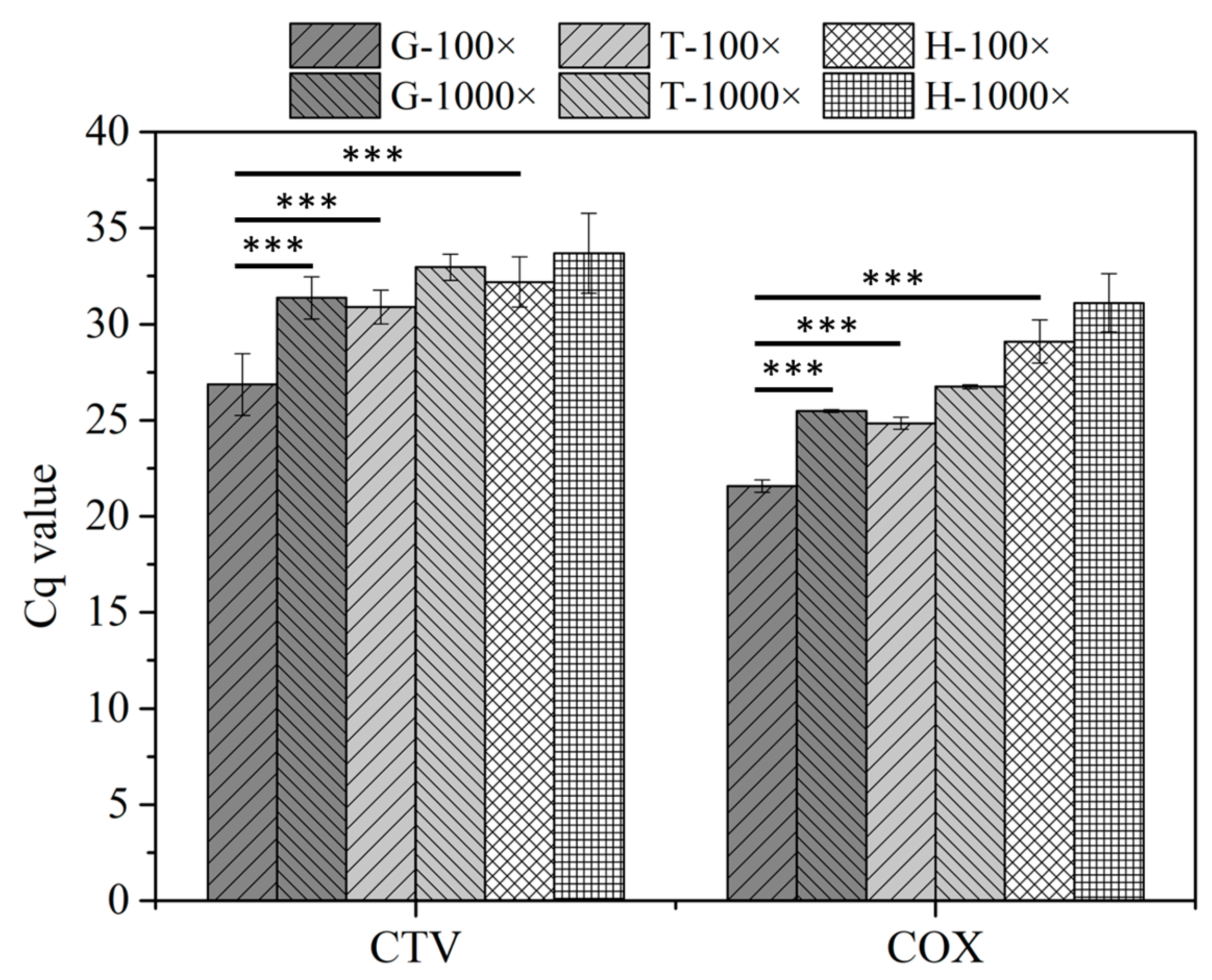
3.2. RT–qPCR Assays Combined with Serial Dilution
3.3. qPCR and RT–qPCR Assays Combined with a Paper Disk
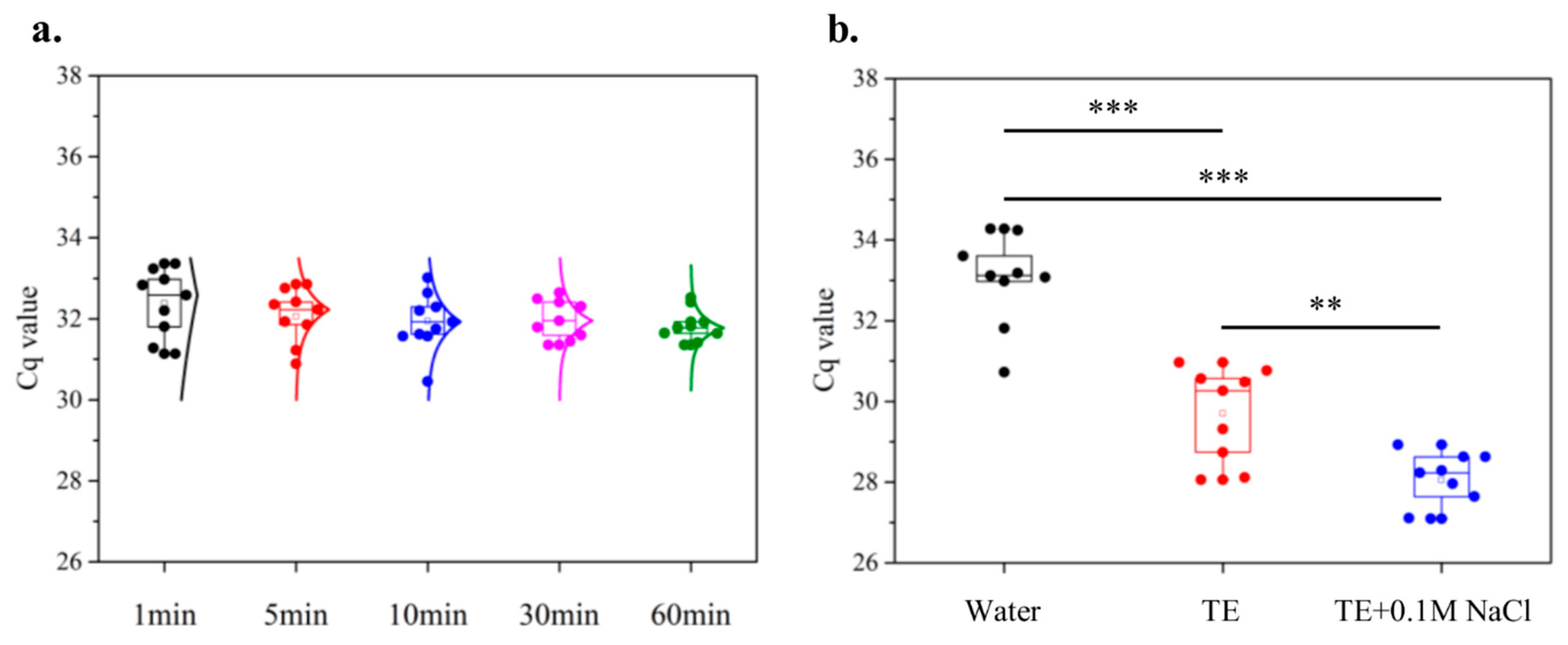
3.3.1. Characterization of Paper Disk Method
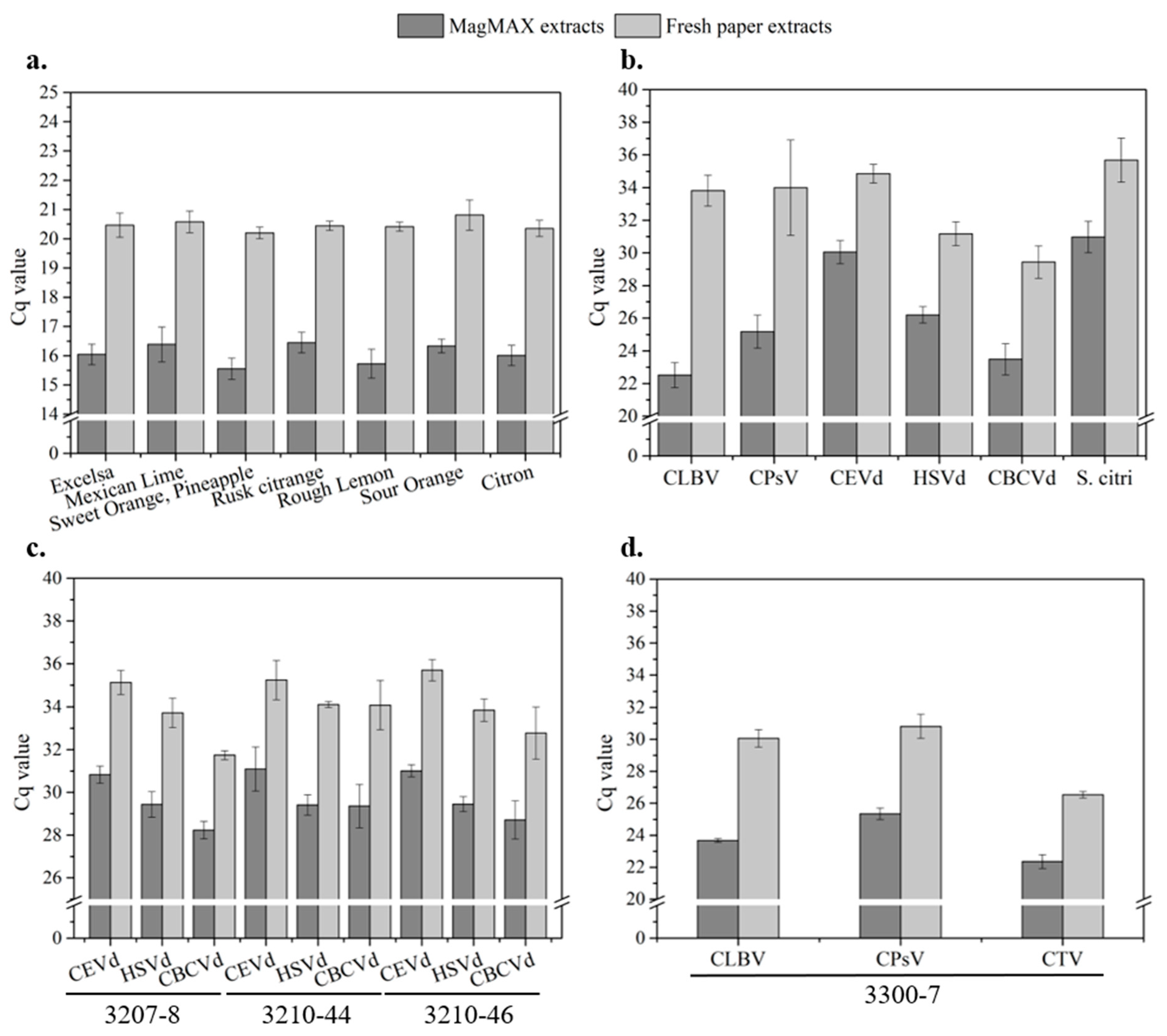
3.3.2. Validations with Healthy, Single-Infected, and Multi-Infected Sources
Healthy Samples
Single-Infected Samples
Multi-Infected Samples
3.3.3. Long-Term Storage Tests
Healthy Samples
Single-Infected Samples
Multi-Infected Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benítez-Galeano, M.J.; Hernández-Rodríguez, L.; Dalmao, F.; Bertoni, E.; Bertalmío, A.; Rubio, L.; Rivas, F.; Maeso, D.; Colina, R. First comprehensive sanitary report of citrus-infecting viruses and viroids in Uruguay. J. Citrus Pathol. 2021, 8, c481049181. [Google Scholar] [CrossRef]
- Folimonova, S.Y. Citrus tristeza virus: A large RNA virus with complex biology turned into a valuable tool for crop protection. PLoS Pathog. 2020, 16, e1008416. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, F.; Duan, Y.; Singerman, A.; Guan, Z. Citrus greening: Management strategies and their economic impact. HortScience 2020, 55, 604–612. [Google Scholar] [CrossRef]
- Wang, J.; Boubourakas, I.; Voloudakis, A.; Agorastou, T.; Magripis, G.; Rucker, T.; Kyriakopoulou, P.; Vidalakis, G. Identification and characterization of known and novel viroid variants in the Greek national citrus germplasm collection: Threats to the industry. Eur. J. Plant Pathol. 2013, 137, 17–27. [Google Scholar] [CrossRef]
- Davis, T.J.; Gómez, M.I.; Harper, S.J.; Twomey, M. The Economic Impact of Hop Stunt Viroid and Certified Clean Planting Materials. HortScience 2021, 56, 1471–1475. [Google Scholar] [CrossRef]
- Osman, F.; Dang, T.; Bodaghi, S.; Vidalakis, G. One-step multiplex RT-qPCR detects three citrus viroids from different genera in a wide range of hosts. J. Virol. Methods 2017, 245, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Osman, F.; Hodzic, E.; Kwon, S.-J.; Wang, J.; Vidalakis, G. Development and validation of a multiplex reverse transcription quantitative PCR (RT-qPCR) assay for the rapid detection of Citrus tristeza virus, Citrus psorosis virus, and Citrus leaf blotch virus. J. Virol. Methods 2015, 220, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Stelinski, L.L. Ecological aspects of the vector-borne bacterial disease, citrus greening (Huanglongbing): Dispersal and host use by Asian citrus psyllid, Diaphorina citri Kuwayama. Insects 2019, 10, 208. [Google Scholar] [CrossRef] [PubMed]
- Yokomi, R.; Rattner, R.; Osman, F.; Maheshwari, Y.; Selvaraj, V.; Pagliaccia, D.; Chen, J.; Vidalakis, G. Whole genome sequence of five strains of Spiroplasma citri isolated from different host plants and its leafhopper vector. BMC Res. Notes 2020, 13, 320. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, D.; Tan, Y.; Zong, X.; Wei, H.; Liu, Q. First Report of Citrus leaf blotch virus in Sweet Cherry. Plant Dis. 2016, 100, 1027. [Google Scholar] [CrossRef]
- Jakse, J.; Radisek, S.; Pokorn, T.; Matousek, J.; Javornik, B. Deep-sequencing revealed Citrus bark cracking viroid (CBCVd) as a highly aggressive pathogen on hop. Plant Pathol. 2015, 64, 831–842. [Google Scholar] [CrossRef]
- Eiras, M.; Targon, M.L.P.N.; Fajardo, T.V.M.; Flores, R.; Kitajima, E.W. Citrus exocortis viroid and Hop Stunt viroid Doubly infecting grapevines in Brazil. Fitopatol. Bras. 2006, 31, 440–446. [Google Scholar] [CrossRef]
- Atta, S.; Cao, M.; Umar, U.u.d.; Zhou, Y.; Yang, F.; Zhou, C. Biological indexing and genetic analysis of Citrus tristeza virus in Pakistan. J. Gen. Plant Pathol. 2017, 83, 382–389. [Google Scholar] [CrossRef]
- Lee, R.F.; Keremane, M.L.; Ramadugu, C. Use of young plants for biological indexing of graft transmissible pathogens of citrus. Crop Prot. 2021, 143, 105524. [Google Scholar] [CrossRef]
- Chalupowicz, L.; Dombrovsky, A.; Gaba, V.; Luria, N.; Reuven, M.; Beerman, A.; Lachman, O.; Dror, O.; Nissan, G.; Manulis-Sasson, S. Diagnosis of plant diseases using the Nanopore sequencing platform. Plant Pathol. 2019, 68, 229–238. [Google Scholar] [CrossRef]
- Kishore, K.; Rahman, H.; Kalita, H.; Pandey, B.; Monika, N. Prevalence of Citrus tristeza virus in mandarin of Sikkim Himalayan Region. Indian J. Virol. 2010, 21, 140–143. [Google Scholar] [CrossRef][Green Version]
- Tarafdar, A.; Godara, S.; Dwivedi, S.; Jayakumar, B.; Biswas, K.K. Characterization of Citrus tristeza virus and determination of genetic variability in North-east and South India. Indian Phytopathol. 2013, 66, 302–307. [Google Scholar]
- Warghane, A.; Kokane, A.; Kokane, S.; Motghare, M.; Surwase, D.; Chodhury, S.P.; Biswas, K.K.; Ghosh, D.K. Molecular detection and coat protein gene based characterization of Citrus tristeza virus prevalent in Sikkim state of India. Indian Phytopathol. 2020, 73, 135–143. [Google Scholar] [CrossRef]
- Ghosh, A.; Das, A.; Lepcha, R.; Majumdar, K.; Baranwal, V. Identification and distribution of aphid vectors spreading Citrus tristeza virus in Darjeeling hills and Dooars of India. J. Asia-Pac. Entomol. 2015, 18, 601–605. [Google Scholar] [CrossRef]
- Abubaker, M.Y.A.; Elhassan, S.M.; Irabi, A.I. First report of Citrus tristeza virus (CTV) disease in commercial citrus Orchards in Sudan. Asian Res. J. Agric. 2017, 3, 1–11. [Google Scholar] [CrossRef]
- Ding, F.; Duan, Y.; Paul, C.; Brlansky, R.H.; Hartung, J.S. Localization and distribution of ’Candidatus Liberibacter asiaticus’ in citrus and periwinkle by direct tissue blot immuno assay with an anti-OmpA polyclonal antibody. PLoS ONE 2015, 10, e0123939. [Google Scholar] [CrossRef] [PubMed]
- Duran-Vila, N. Detection of Viroids by sPAGE Gel Electrophoresis. In Viroids: Methods and Protocols; Humana: New York, NY, USA, 2022; pp. 71–77. [Google Scholar]
- Murcia, N.; Serra, P.; Olmos, A.; Durán-Vila, N. A novel hybridization approach for detection of citrus viroids. Mol. Cell. Probes 2009, 23, 95–102. [Google Scholar] [CrossRef]
- Pallás, V.; Sánchez-Navarro, J.A.; Kinard, G.R.; Di Serio, F. Molecular hybridization techniques for detecting and studying viroids. In Viroids and Satellites; Elsevier: Amsterdam, The Netherlands, 2017; pp. 369–379. [Google Scholar]
- Simeone, M.; Gómez, C.; Bertalmío, A.; Ruiz, E.; Hauteville, C.; Godoy Suarez, L.; Tito, B.; García, M.L. Detection of citrus psorosis virus by RT-qPCR validated by diagnostic parameters. Plant Pathol. 2021, 70, 980–986. [Google Scholar] [CrossRef]
- Chambers, G.A.; Geering, A.D.; Holford, P.; Vidalakis, G.; Donovan, N.J. Development of a one-step RT-qPCR detection assay for the newly described citrus viroid VII. J. Virol. Methods 2022, 299, 114330. [Google Scholar] [CrossRef] [PubMed]
- Osman, F.; Vidalakis, G. Real-Time Detection of Viroids Using Singleplex and Multiplex Quantitative Polymerase Chain Reaction. In Viroids: Methods and Protocols; Humana: New York, NY, USA, 2022; pp. 181–194. [Google Scholar]
- Huatang, W.; Xinnian, Z.; Peipei, X.; Liu, Y. Rapid DNA Extraction Methods for Direct-PCR Detection Citrus Huanglongbing. Plant Dis. Pests 2015, 6, 1. [Google Scholar]
- Rezaian, M.; Krake, L. Nucleic acid extraction and virus detection in grapevine. J. Virol. Methods 1987, 17, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Mock, R.; Huang, Q.; Abad, J.; Hartung, J.; Kinard, G. A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J. Virol. Methods 2008, 154, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Barbier, F.F.; Chabikwa, T.G.; Ahsan, M.U.; Cook, S.E.; Powell, R.; Tanurdzic, M.; Beveridge, C.A. A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods 2019, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Osman, F.; Wang, J.; Rucker, T.; Bodaghi, S.; Tan, S.-h.; Pagliaccia, D.; Lavagi-Craddock, I.; Vidalakis, G. High-Throughput RNA Extraction from Citrus Tissues for the Detection of Viroids. In Viroids: Methods and Protocols; Humana: New York, NY, USA, 2022; pp. 57–64. [Google Scholar]
- Liu, C.-W.; Tsutsui, H. Sample–to-answer sensing technologies for nucleic acid preparation and detection in the field. SLAS Technol. 2023, 28, 302–323. [Google Scholar] [CrossRef]
- Carpinetti, P.d.A.; Fioresi, V.S.; Ignez da Cruz, T.; de Almeida, F.A.N.; Canal, D.; Ferreira, A.; Ferreira, M.F.d.S. Efficient method for isolation of high-quality RNA from Psidium guajava L. tissues. PLoS ONE 2021, 16, e0255245. [Google Scholar] [CrossRef]
- Fujiwara, K.; Inoue, H.; Sonoda, R.; Iwamoto, Y.; Kusaba, M.; Tashiro, N.; Miyasaka, A. Real-time PCR detection of the onion downy mildew pathogen Peronospora destructor from symptomless onion seedlings and soils. Plant Dis. 2021, 105, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Qiagen. RNeasy Plant Mini Kit. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/rna-purification/total-rna/rneasy-plant-mini-kit (accessed on 26 March 2024).
- Qiagen. DNeasy Plant Pro and Plant Kits. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/dna-rna-purification/dna-purification/genomic-dna/dneasy-plant-pro-and-plant-kits (accessed on 26 March 2024).
- Jia, Z.; Ding, M.; Nakano, M.; Hong, K.; Huang, R.; Becker, D.; Glazebrook, J.; Katagiri, F.; Han, X.; Tsuda, K. DNA purification-free PCR from plant tissues. Plant Cell Physiol. 2021, 62, 1503–1505. [Google Scholar] [CrossRef] [PubMed]
- Qiagen. QIAcard FTA PlantSaver. Available online: https://www.qiagen.com/us/products/discovery-and-translational-research/sample-collection-stabilization/qiacard-fta/qiacard-fta-plantsaver (accessed on 26 March 2024).
- Paul, R.; Saville, A.C.; Hansel, J.C.; Ye, Y.; Ball, C.; Williams, A.; Chang, X.; Chen, G.; Gu, Z.; Ristaino, J.B. Extraction of plant DNA by microneedle patch for rapid detection of plant diseases. ACS Nano 2019, 13, 6540–6549. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Ostermann, E.; Gu, Z.; Ristaino, J.B.; Wei, Q. DNA extraction from plant leaves using a microneedle patch. Curr. Protoc. Plant Biol. 2020, 5, e20104. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Mason, M.G.; Wang, Y.; Wee, E.; Turni, C.; Blackall, P.J.; Trau, M.; Botella, J.R. Nucleic acid purification from plants, animals and microbes in under 30 seconds. PLoS Biol. 2017, 15, e2003916. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.M.; Bodaghi, S.; Heick, J.A.; Dodds, J.A. Detection of Avocado Sunblotch and Other Viroids Using RNA Filter Paper Capture and RT-PCR. In Viroids: Methods and Protocols; Humana: New York, NY, USA, 2023; pp. 219–233. [Google Scholar]
- Pretorius, L.-S.; Chandra, K.A.; Jooste, A.E.; Motaung, L.C.; Parkinson, L.E.; Geering, A.D. Adaptation of a filter paper method for RNA template preparation for the detection of avocado sunblotch viroid by reverse transcription qPCR. J. Virol. Methods 2022, 301, 114455. [Google Scholar] [CrossRef] [PubMed]
- Haveman, N.J.; Schuerger, A.C.; Yu, P.-L.; Brown, M.; Doebler, R.; Paul, A.-L.; Ferl, R.J. Advancing the automation of plant nucleic acid extraction for rapid diagnosis of plant diseases in space. Front. Plant Sci. 2023, 14, 1194753. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; da Graça, J.V.; Freitas-Astua, J.; Vidalakis, G.; Durán-Vila, N.; Lavagi, I. Citrus viruses and viroids. In The Genus Citrus; Elsevier: Amsterdam, The Netherlands, 2020; pp. 391–410. [Google Scholar]
- Lavagi, I.; Matoušek, J.; Vidalakis, G. Other cocadviroids. In Viroids and Satellites; Elsevier: Amsterdam, The Netherlands, 2017; pp. 275–287. [Google Scholar]
- Shi, J.; Pagliaccia, D.; Morgan, R.; Qiao, Y.; Pan, S.; Vidalakis, G.; Ma, W. Novel diagnosis for citrus stubborn disease by detection of a Spiroplasma citri-secreted protein. Phytopathology 2014, 104, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Dotti, I.; Bonin, S. Integrity assessment of nucleic acids. In Guidel. Mol. Anal. Arch. Tissues: Springer: Berlin, Heidelberg, 2011, pp. 81–85.
- Gallagher, S.R.; Desjardins, P.R. Quantitation of DNA and RNA with absorption and fluorescence spectroscopy. Curr. Protoc. Mol. Biol. 2006, 76, A.3D.1–A.3D.21. [Google Scholar] [CrossRef]
- Schultheiss, O.C.; Stanton, S.J. Assessment of salivary hormones. In Methods Soc. Neurosci.; Guilford Press: New York, NY, USA, 2009; pp. 17–44. [Google Scholar]
- Tan, S.-h.; Osman, F.; Bodaghi, S.; Dang, T.; Greer, G.; Huang, A.; Hammado, S.; Abu-Hajar, S.; Campos, R.; Vidalakis, G. Full genome characterization of 12 citrus tatter leaf virus isolates for the development of a detection assay. PLoS ONE 2019, 14, e0223958. [Google Scholar] [CrossRef]
- Mason, M.G.; Botella, J.R. Rapid (30-second), equipment-free purification of nucleic acids using easy-to-make dipsticks. Nat. Protoc. 2020, 15, 3663–3677. [Google Scholar] [CrossRef] [PubMed]
- Alaeddini, R. Forensic implications of PCR inhibition—A review. Forensic Sci. Int. Genet. 2012, 6, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qi, J.; Xiao, D.; Wang, Z.; Tian, K. A re-evaluation of dilution for eliminating PCR inhibition in soil DNA samples. Soil Biol. Biochem. 2017, 106, 109–118. [Google Scholar] [CrossRef]
- Ambers, A.; Wiley, R.; Novroski, N.; Budowle, B. Direct PCR amplification of DNA from human bloodstains, saliva, and touch samples collected with microFLOQ® swabs. Forensic Sci. Int. Genet. 2018, 32, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Merindol, N.; Pépin, G.; Marchand, C.; Rheault, M.; Peterson, C.; Poirier, A.; Houle, C.; Germain, H.; Danylo, A. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J. Clin. Virol. 2020, 128, 104423. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Mirza, F.; Al-Hail, H.; Sundararaju, S.; Xaba, T.; Iqbal, M.; Alhussain, H.; Yassine, H.M.; Perez-Lopez, A.; Tang, P. Detection of SARS-CoV-2 RNA by direct RT-qPCR on nasopharyngeal specimens without extraction of viral RNA. PLoS ONE 2020, 15, e0236564. [Google Scholar]
- Wee, S.K.; Sivalingam, S.P.; Yap, E.P.H. Rapid direct nucleic acid amplification test without RNA extraction for SARS-CoV-2 using a portable PCR thermocycler. Genes 2020, 11, 664. [Google Scholar] [CrossRef] [PubMed]
- Heikrujam, J.; Kishor, R.; Mazumder, P.B. The chemistry behind plant DNA isolation protocols. Biochem. Anal. Tools–Methods Bio-Mol. Stud 2020, 8, 131–141. [Google Scholar]
- Madhad, V.J.; Sentheil, K. The Rapid & Non-Enzymatic isolation of DNA from the Human peripheral whole blood suitable for Genotyping. Eur. J. Biotechnol. Biosci. 2014, 1, 1–16. [Google Scholar]
- Gan, W.; Gu, Y.; Han, J.; Li, C.-x.; Sun, J.; Liu, P. Chitosan-modified filter paper for nucleic acid extraction and “in situ PCR” on a thermoplastic microchip. Anal. Chem. 2017, 89, 3568–3575. [Google Scholar] [CrossRef]
- Cheng, Y.-J.; Guo, W.-W.; Yi, H.-L.; Pang, X.-M.; Deng, X. An efficient protocol for genomic DNA extraction from Citrus species. Plant Mol. Biol. Report. 2003, 21, 177–178. [Google Scholar] [CrossRef]


| Category | Source Number | Source Plant | Citrus Pathogen/Isolate |
|---|---|---|---|
| Single infection | 2987-32 | Pineapple sweet orange | CTV/T517 |
| Not available | Unknown | CPsV/P202 | |
| 3069-1 | Sweet orange | CLBV | |
| 2765-1 | Madam vinus sweet orange | CEVd | |
| 2765-4 | Madam vinus sweet orange | CVd-IIa a/E818 | |
| 3200-1 | Madam vinus sweet orange | CVd-IV b | |
| Not available | Pineapple sweet orange | S. citri/C189 | |
| Multiple infection | 3300-7 | Madam vinus sweet orange | CLBV + CTV + CPsV |
| 3210-44 | Madam vinus sweet orange | CEVd + CVdII + CVdIV | |
| 3210-46 | Madam vinus sweet orange | CEVd + CVdII + CVdIV | |
| 3207-8 | Madam vinus sweet orange | CEVd + CBCVd + HSVd + CBLVd + CDVd + CVd-V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-W.; Bodaghi, S.; Vidalakis, G.; Tsutsui, H. Quick Plant Sample Preparation Methods Using a Micro-Homogenizer for the Detection of Multiple Citrus Pathogens. Chemosensors 2024, 12, 105. https://doi.org/10.3390/chemosensors12060105
Liu C-W, Bodaghi S, Vidalakis G, Tsutsui H. Quick Plant Sample Preparation Methods Using a Micro-Homogenizer for the Detection of Multiple Citrus Pathogens. Chemosensors. 2024; 12(6):105. https://doi.org/10.3390/chemosensors12060105
Chicago/Turabian StyleLiu, Chia-Wei, Sohrab Bodaghi, Georgios Vidalakis, and Hideaki Tsutsui. 2024. "Quick Plant Sample Preparation Methods Using a Micro-Homogenizer for the Detection of Multiple Citrus Pathogens" Chemosensors 12, no. 6: 105. https://doi.org/10.3390/chemosensors12060105
APA StyleLiu, C.-W., Bodaghi, S., Vidalakis, G., & Tsutsui, H. (2024). Quick Plant Sample Preparation Methods Using a Micro-Homogenizer for the Detection of Multiple Citrus Pathogens. Chemosensors, 12(6), 105. https://doi.org/10.3390/chemosensors12060105






