Advances in Antimicrobial Resistance Monitoring Using Sensors and Biosensors: A Review
Abstract
:1. Antimicrobial Resistance
2. Sensors and Biosensors for AMR Detection
2.1. Sensors and Biosensors for Phenotypic AMR Detection
2.1.1. AST Magnetic, Mass, and Mechanical (Bio)Sensors

2.1.2. AST Optical (Bio)Sensors
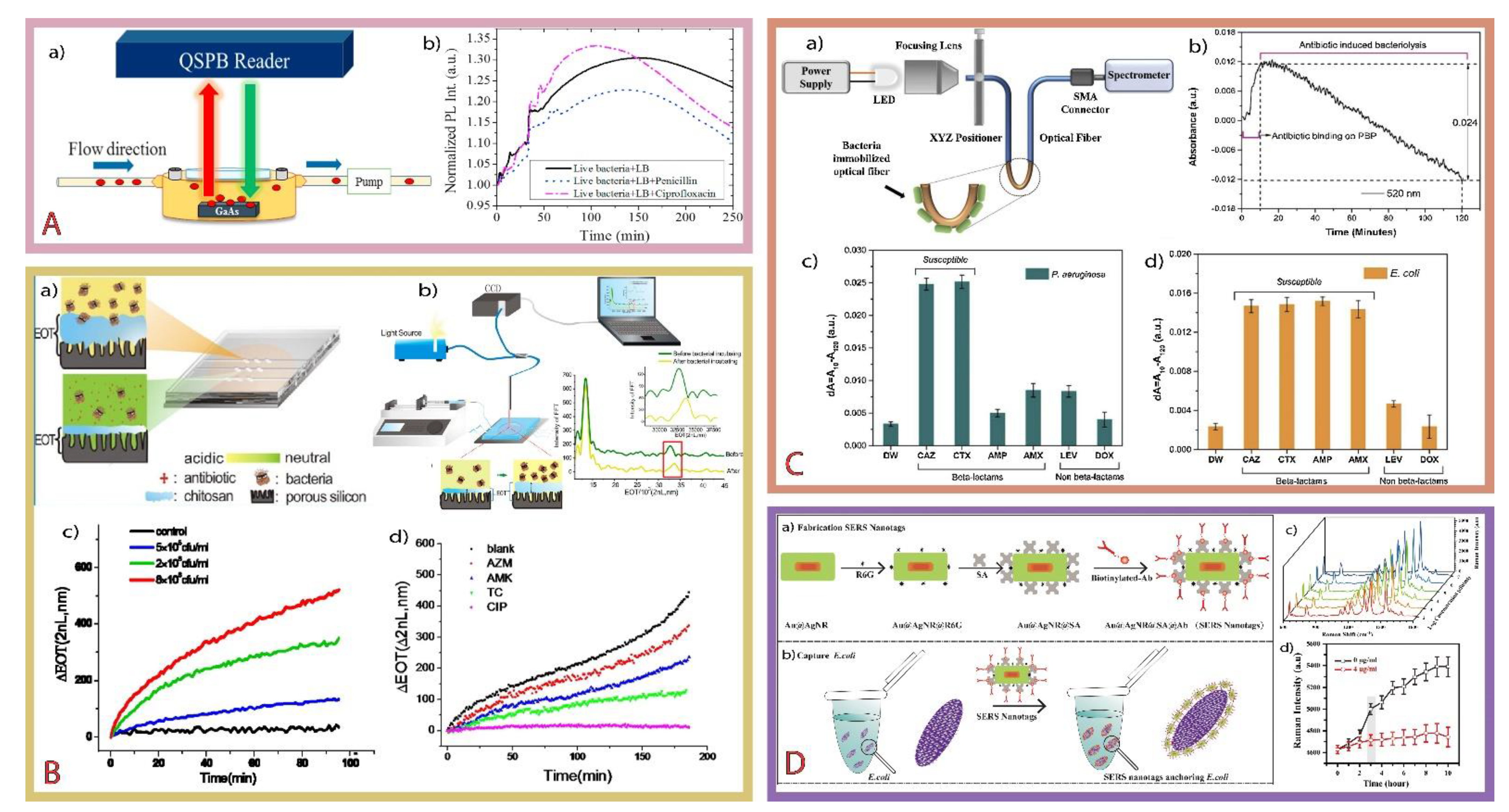
2.1.3. AST Electrochemical (Bio)Sensors

2.2. Sensors and Biosensors for Genotypic Antimicrobial Resistance Detection
3. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [Green Version]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- OECD. Stemming the Superbug Tide: Just A Few Dollars More. In OECD Health Policy Studies; OECD Health Policy Studies, OECD Publishing: Paris, France, 2018; ISBN 9789264307582. [Google Scholar]
- Sims, N.; Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef] [PubMed]
- Gelband, H.; Laxminarayan, R. Tackling antimicrobial resistance at global and local scales. Trends Microbiol. 2015, 23, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Roope, L.S.J.; Smith, R.D.; Pouwels, K.B.; Buchanan, J.; Abel, L.; Eibich, P.; Butler, C.C.; Tan, P.S.; Sarah Walker, A.; Robotham, J.V.; et al. The challenge of antimicrobial resistance: What economics can contribute. Science 2019, 364, eaau4679. [Google Scholar] [CrossRef]
- Smith, R.; Coast, J. The true cost of antimicrobial resistance. BMJ Br. Med. J. 2013, 346, f1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Global Action Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Boolchandani, M.; D’Souza, A.W.; Dantas, G. Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 2019, 20, 356–370. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; The Review on Antimicrobial Resistance: London, UK, 2016; ISBN 9789241564748. [Google Scholar]
- AMR. Action Found the AMR Innovation Challenge. Available online: https://amractionfund.com/amr-innovation-challenge/ (accessed on 17 August 2021).
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global Contributors to Antibiotic Resistance. J. Glob. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [PubMed]
- Vikesland, P.; Garner, E.; Gupta, S.; Kang, S.; Maile-Moskowitz, A.; Zhu, N. Differential Drivers of Antimicrobial Resistance across the World. Acc. Chem. Res. 2019, 52, 916–924. [Google Scholar] [CrossRef] [PubMed]
- WHO. Critically Important Antimicrobials for Human Medicine; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.ecdc.europa.eu/en/news-events/who-publishes-list-bacteria-which-new-antibiotics-are-urgently-needed (accessed on 17 August 2021).
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern Tools for Rapid Diagnostics of Antimicrobial Resistance. Front. Cell. Infect. Microbiol. 2020, 10, 308. [Google Scholar] [CrossRef] [PubMed]
- Salimiyan Rizi, K.; Aryan, E.; Meshkat, Z.; Ranjbar, G.; Sankian, M.; Ghazvini, K.; Farsiani, H.; Pourianfar, H.R.; Rezayi, M. The overview and perspectives of biosensors and Mycobacterium tuberculosis: A systematic review. J. Cell. Physiol. 2021, 236, 1730–1750. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeter, G.; Giske, C.G.; Kirn, T.J.; Sharp, S.E. Point-Counterpoint: Differences between the European Committee on Antimicrobial Susceptibility Testing and Clinical and Laboratory Standards Institute Recommendations for Reporting Antimicrobial Susceptibility Results. J. Clin. Microbiol. 2019, 57, e01129-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davenport, M.; Mach, K.E.; Shortliffe, L.M.D.; Banaei, N.; Wang, T.H.; Liao, J.C. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 2017, 14, 298–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based Methods for Detection of Antibiotic Resistance in Agroecosystems: Advantages, Challenges, and Gaps in Knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Edouard, S.; Pagnier, I.; Mediannikov, O.; Drancourt, M.; Raoult, D. Current and Past Strategies for Bacterial Culture in Clinical Microbiology. Clin. Microbiol. Rev. 2015, 28, 208–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, A.W.; Perry, D.M.; Kirby, W.M.M. Single-Disk Antibiotic-Sensitivity Testing of Staphylococci: An Analysis of Technique and Results. AMA Arch. Intern. Med. 1959, 104, 208–216. [Google Scholar] [CrossRef]
- Pitruzzello, G.; Conteduca, D.; Krauss, T.F. Nanophotonics for bacterial detection and antimicrobial susceptibility testing. Nanophotonics 2020, 9, 4447–4472. [Google Scholar] [CrossRef]
- Benkova, M.; Soukup, O.; Marek, J. Antimicrobial susceptibility testing: Currently used methods and devices and the near future in clinical practice. J. Appl. Microbiol. 2020, 129, 806–822. [Google Scholar] [CrossRef]
- Rotilie, C.A.; Fass, R.J.; Prior, R.B.; Perkins, R.L. Microdilution Technique for Antimicrobial Susceptibility Testing of Anaerobic Bacteria. Antimicrob. Agents Chemother. 1975, 7, 311–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietvorst, J.; Vilaplana, L.; Uria, N.; Marco, M.-P.; Muñoz-Berbel, X. Current and near-future technologies for antibiotic susceptibility testing and resistant bacteria detection. TrAC Trends Anal. Chem. 2020, 127, 115891. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Zhao, W. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. SLAS Technol. 2017, 22, 585–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, J.W.; Munier, G.K.; Johnson, C.L. Direct comparison of the BD phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 2008, 46, 2327–2333. [Google Scholar] [CrossRef] [Green Version]
- Whistler, T.; Sangwichian, O.; Jorakate, P.; Sawatwong, P.; Surin, U.; Piralam, B.; Thamthitiwat, S.; Promkong, C.; Peruski, L. Identification of Gram negative nonfermentative Bacteria: How hard can it be? PLoS Negl. Trop. Dis. 2019, 13, e0007729. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Schmit, T.; Klomp, M.; Khan, M.N. An Overview of Flow Cytometry: Its Principles and Applications in Allergic Disease Research. Anim. Models Allerg. Dis. 2021, 2223, 169–182. [Google Scholar]
- Entenza, J.M.; Bétrisey, B.; Manuel, O.; Giddey, M.; Sakwinska, O.; Laurent, F.; Bizzini, A. Rapid Detection of Staphylococcus aureus Strains with Reduced Susceptibility to Vancomycin by Isothermal Microcalorimetry. J. Clin. Microbiol. 2014, 52, 180–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, M.; Wirz, D.; Daniels, A.U.; Braissant, O. Application of a Microcalorimetric Method for Determining Drug Susceptibility in Mycobacterium Species. J. Clin. Microbiol. 2012, 50, 16–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braissant, O.; Wirz, D.; Göpfert, B.; Daniels, A.U. Biomedical Use of Isothermal Microcalorimeters. Sensors 2010, 10, 9369–9383. [Google Scholar] [CrossRef] [PubMed]
- Butini, M.E.; Gonzalez Moreno, M.; Czuban, M.; Koliszak, A.; Tkhilaishvili, T.; Trampuz, A.; Di Luca, M. Real-Time Antimicrobial Susceptibility Assay of Planktonic and Biofilm Bacteria by Isothermal Microcalorimetry. Adv. Exp. Med. Biol. 2019, 1214, 61–77. [Google Scholar]
- Burnham, C.A.D.; Leeds, J.; Nordmann, P.; O’Grady, J.; Patel, J. Diagnosing antimicrobial resistance. Nat. Rev. Microbiol. 2017, 15, 697–703. [Google Scholar] [CrossRef]
- Oviaño, M.; Bou, G. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for the Rapid Detection of Antimicrobial Resistance Mechanisms and Beyond. Clin. Microbiol. Rev. 2019, 32, e00037-18. [Google Scholar] [CrossRef] [Green Version]
- Rentschler, S.; Kaiser, L.; Deigner, H.-P. Emerging Options for the Diagnosis of Bacterial Infections and the Characterization of Antimicrobial Resistance. Int. J. Mol. Sci. 2021, 22, 456. [Google Scholar] [CrossRef] [PubMed]
- Anjum, M.F.; Zankari, E.; Hasman, H. Molecular Methods for Detection of Antimicrobial Resistance. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Schwarz, S., Cavaco, L.M., Shen, J., Eds.; ASM Press: Washington, DC, USA, 2018; pp. 35–50. [Google Scholar]
- Leva-Bueno, J.; Peyman, S.A.; Millner, P.A. A review on impedimetric immunosensors for pathogen and biomarker detection. Med. Microbiol. Immunol. 2020, 209, 343–362. [Google Scholar] [CrossRef] [Green Version]
- Luby, E.; Mark Ibekwe, A.; Zilles, J.; Pruden, A. Molecular methods for assessment of antibiotic resistance in agricultural ecosystems: Prospects and challenges. J. Environ. Qual. 2016, 45, 441–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yee, R.; Simner, P.J. Next-Generation Sequencing Approaches to Predicting Antimicrobial Susceptibility Testing Results. Adv. Mol. Pathol. 2019, 2, 99–110. [Google Scholar] [CrossRef]
- Gillespie, S. Chapter 3—Current status of molecular microbiological techniques for the analysis of drinking water. In Molecular Microbial Diagnostic Methods; Cook, N., D’Agostino, M., Thompson, K.C., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 39–58. ISBN 978012416999-9. [Google Scholar]
- Laschi, S.; Palchetti, I.; Marrazza, G.; Mascini, M. Enzyme-amplified electrochemical hybridization assay based on PNA, LNA and DNA probe-modified micro-magnetic beads. Bioelectrochemistry 2009, 76, 214–220. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M.; Berg, R.H.; Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991, 254, 1497–1500. [Google Scholar] [CrossRef]
- Gupta, A.; Mishra, A.; Puri, N. Peptide nucleic acids: Advanced tools for biomedical applications. J. Biotechnol. 2017, 259, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wu, C. Recent Advances in Peptide Nucleic Acids for Rapid Detection of Foodborne Pathogens. Food Anal. Methods 2020, 13, 1956–1972. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification. Anal. Lett. 2001, 34, 635–659. [Google Scholar] [CrossRef] [Green Version]
- Reynoso, E.C.; Torres, E.; Bettazzi, F.; Palchetti, I. Trends and perspectives in immunosensors for determination of currently-used pesticides: The case of glyphosate, organophosphates, and neonicotinoids. Biosensors 2019, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Palchetti, I.; Mascini, M. Nucleic acid biosensors for environmental pollution monitoring. Analyst 2008, 133, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Palchetti, I.; Mascini, M. Biosensor technology: A brief history. In Sensors and Microsystems; Springer: Dordrecht, The Netherlands, 2010; Volume 54, ISBN 9789048136056. [Google Scholar]
- Leonard, H.; Colodner, R.; Halachmi, S.; Segal, E. Recent Advances in the Race to Design a Rapid Diagnostic Test for Antimicrobial Resistance. ACS Sens. 2018, 3, 2202–2217. [Google Scholar] [CrossRef] [PubMed]
- Van Belkum, A.; Burnham, C.A.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Bettazzi, F.; Palchetti, I. Nanotoxicity assessment: A challenging application for cutting edge electroanalytical tools. Anal. Chim. Acta 2019, 1072, 61–74. [Google Scholar] [CrossRef]
- Ensafi, A.A. Chapter 1—An introduction to sensors and biosensors. In Electrochemical Biosensors; Ensafi, A.A., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–10. ISBN 9780128164914. [Google Scholar]
- Bonini, A.; Poma, N.; Vivaldi, F.; Kirchhain, A.; Salvo, P.; Bottai, D.; Tavanti, A.; Di Francesco, F. Advances in biosensing: The CRISPR/Cas system as a new powerful tool for the detection of nucleic acids. J. Pharm. Biomed. Anal. 2021, 192, 113645. [Google Scholar] [CrossRef] [PubMed]
- Ceylan Koydemir, H.; Külah, H.; Özgen, C.; Alp, A.; Hasçelik, G. MEMS biosensors for detection of methicillin resistant Staphylococcus aureus. Biosens. Bioelectron. 2011, 29, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liang, W.; Wen, Y.; Wang, L.; Yang, X.; Ren, S.; Jia, N.; Zuo, X.; Liu, G. An ultrasensitive electrochemical biosensor for the detection of mecA gene in methicillin-resistant Staphylococcus aureus. Biosens. Bioelectron. 2018, 99, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Bhardwaj, S.K.; Mehta, J.; Mohanta, G.C.; Deep, A. Bacteriophage immobilized graphene electrodes for impedimetric sensing of bacteria (Staphylococcus arlettae). Anal. Biochem. 2016, 505, 18–25. [Google Scholar] [CrossRef]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 2019, 141, 111435. [Google Scholar] [CrossRef]
- Hu, J.; Ghosh, M.; Miller, M.J.; Bohn, P.W. Whole-cell biosensing by siderophore-based molecular recognition and localized surface plasmon resonance. Anal. Methods 2019, 11, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial Peptides: Powerful Biorecognition Elements to Detect Bacteria in Biosensing Technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mach, K.E.; Wong, P.K.; Liao, J.C. Biosensor diagnosis of urinary tract infections: A path to better treatment? Trends Pharmacol. Sci. 2011, 32, 330–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reder-Christ, K.; Bendas, G. Biosensor Applications in the Field of Antibiotic Research—A Review of Recent Developments. Sensors 2011, 11, 9450–9466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.M.; Lee, S.Y. Optical Biosensors for the Detection of Pathogenic Microorganisms. Trends Biotechnol. 2016, 34, 7–25. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.K.; Alam, K.K.; Verosloff, M.S.; Capdevila, D.A.; Desmau, M.; Clauer, P.R.; Lee, J.W.; Nguyen, P.Q.; Pastén, P.A.; Matiasek, S.J.; et al. Cell-free biosensors for rapid detection of water contaminants. Nat. Biotechnol. 2020, 38, 1451–1459. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Pan, Y.; Ge, S.; Coulon, F.; Yang, Z. Rapid methods for antimicrobial resistance diagnosis in contaminated soils for effective remediation strategy. TrAC Trends Anal. Chem. 2021, 137, 116203. [Google Scholar] [CrossRef]
- Li, Z.; Liu, C.; Sarpong, V.; Gu, Z. Multisegment nanowire/nanoparticle hybrid arrays as electrochemical biosensors for simultaneous detection of antibiotics. Biosens. Bioelectron. 2019, 126, 632–639. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [Green Version]
- Argudín, M.A.; Deplano, A.; Meghraoui, A.; Dodémont, M.; Heinrichs, A.; Denis, O.; Nonhoff, C.; Roisin, S. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes. Antibiotics 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Behera, B.; Anil Vishnu, G.K.; Chatterjee, S.; Sitaramgupta, V.S.N.; Sreekumar, N.; Nagabhushan, A.; Rajendran, N.; Prathik, B.H.; Pandya, H.J. Emerging technologies for antibiotic susceptibility testing. Biosens. Bioelectron. 2019, 142, 111552. [Google Scholar] [CrossRef] [PubMed]
- Ferapontova, E.E. Electrochemical assays for microbial analysis: How far they are from solving microbiota and microbiome challenges. Curr. Opin. Electrochem. 2020, 19, 153–161. [Google Scholar] [CrossRef]
- Pujol-Vila, F.; Villa, R.; Alvarez, M. Nanomechanical Sensors as a Tool for Bacteria Detection and Antibiotic Susceptibility Testing. Front. Mech. Eng. 2020, 6, 44. [Google Scholar] [CrossRef]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xing, Y.; Zhou, X.; Chen, G.Y.; Shi, H. Light-sheet skew rays enhanced U-shaped fiber-optic fluorescent immunosensor for Microcystin-LR. Biosens. Bioelectron. 2021, 176, 112902. [Google Scholar] [CrossRef] [PubMed]
- Cardenosa-Rubio, M.C.; Robison, H.M.; Bailey, R.C. Recent advances in environmental and clinical analysis using microring resonator-based sensors. Curr. Opin. Environ. Sci. Heal. 2019, 10, 38–46. [Google Scholar] [CrossRef]
- Gupta, B.D.; Shrivastav, A.M.; Usha, S.P. Optical Sensors for Biomedical Diagnostics and Environmental Monitoring, 1st ed.; CRC Press: Boca Raton, FL, USA, 2017; ISBN 9781315156033. [Google Scholar]
- Berneschi, S.; Bettazzi, F.; Giannetti, A.; Baldini, F.; Nunzi Conti, G.; Pelli, S.; Palchetti, I. Optical whispering gallery mode resonators for label-free detection of water contaminants. TrAC Trends Anal. Chem. 2020, 126, 115856. [Google Scholar] [CrossRef]
- Miyazaki, C.M.; Shimizu, F.M.; Ferreira, M. 6—Surface Plasmon Resonance (SPR) for Sensors and Biosensors. In Nanocharacterization Techniques; Da Róz, A.L., Ferreira, M., de Lima Leite, F., Oliveira, O.N.J., Eds.; William Andrew Publishing: Norwich, NY, USA, 2017; pp. 183–200. ISBN 9780323497787. [Google Scholar]
- Labuda, J.; Oliveira Brett, A.M.; Evtugyn, G.; Fojta, M.; Mascini, M.; Ozsoz, M.; Palchetti, I.; Paleček, E.; Wang, J. Electrochemical nucleic acid-based biosensors: Concepts, terms, and methodology (IUPAC Technical Report). Pure Appl. Chem. 2010, 82, 1161–1187. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Bettazzi, F.; Palchetti, I. Photoelectrochemical genosensors for the determination of nucleic acid cancer biomarkers. Curr. Opin. Electrochem. 2018, 12, 51–59. [Google Scholar] [CrossRef]
- Voccia, D.; Sosnowska, M.; Bettazzi, F.; Roscigno, G.; Fratini, E.; De Franciscis, V.; Condorelli, G.; Chitta, R.; D’Souza, F.; Kutner, W.; et al. Direct determination of small RNAs using a biotinylated polythiophene impedimetric genosensor. Biosens. Bioelectron. 2017, 87, 1012–1019. [Google Scholar] [CrossRef] [Green Version]
- Sinn, I.; Kinnunen, P.; Albertson, T.; McNaughton, B.H.; Newton, D.W.; Burns, M.A.; Kopelman, R. Asynchronous magnetic bead rotation (AMBR) biosensor in microfluidic droplets for rapid bacterial growth and susceptibility measurements. Lab. Chip 2011, 11, 2604–2611. [Google Scholar] [CrossRef]
- Kinnunen, P.; Sinn, I.; McNaughton, B.H.; Kopelman, R. High frequency asynchronous magnetic bead rotation for improved biosensors. Appl. Phys. Lett. 2010, 97, 223701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinnunen, P.; McNaughton, B.H.; Albertson, T.; Sinn, I.; Mofakham, S.; Elbez, R.; Newton, D.W.; Hunt, A.; Kopelman, R. Self-Assembled Magnetic Bead Biosensor for Measuring Bacterial Growth and Antimicrobial Susceptibility Testing. Small 2012, 8, 2477–2482. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-C.; Chi, S.-W.; Yang, T.-H.; Chuang, H.-S. Label-Free Monitoring of Microorganisms and Their Responses to Antibiotics Based on Self-Powered Microbead Sensors. ACS Sens. 2018, 3, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Xu, K.; Xu, C.; Xu, B. Biofunctional magnetic nanoparticles for protein separation and pathogen detection. Chem. Commun. 2006, 941–949. [Google Scholar] [CrossRef]
- Jha, S.N. Chapter 5—Biosensor. In Rapid Detection of Food Adulterants and Contaminants; Jha, S.N., Ed.; Academic Press: San Diego, CA, USA, 2016; pp. 125–145. ISBN 9780124200845. [Google Scholar]
- Ragavan, K.V.; Neethirajan, S. Chapter 7—Nanoparticles as Biosensors for Food Quality and Safety Assessment. In Nanomaterials for Food Applications; López Rubio, A., Fabra Rovira, M.J., Martínez Sanz, M., Gómez-Mascaraque, L.G., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 147–202. ISBN 9780128141304. [Google Scholar]
- Plácido, A.; Amaral, J.S.; Costa, J.; Fernandes, T.J.R.; Oliveira, M.B.P.P.; Delerue-Matos, C.; Mafra, I. Chapter 12—Novel Strategies for Genetically Modified Organism Detection. In Genetically Modified Organisms in Food; Watson, R.R., Preedy, V.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 119–131. ISBN 9780128022597. [Google Scholar]
- Reyes, P.I.; Yang, K.; Zheng, A.; Li, R.; Li, G.; Lu, Y.; Tsang, C.K.; Zheng, S.X.F. Dynamic monitoring of antimicrobial resistance using magnesium zinc oxide nanostructure-modified quartz crystal microbalance. Biosens. Bioelectron. 2017, 93, 189–197. [Google Scholar] [CrossRef] [Green Version]
- Guliy, O.I.; Zaitsev, B.D.; Borodina, I.A. New approach for determination of antimicrobial susceptibility to antibiotics by an acoustic sensor. Appl. Microbiol. Biotechnol. 2020, 104, 1283–1290. [Google Scholar] [CrossRef]
- Jin, Y.; Joshi, S.G. Propagation of a quasi-shear horizontal acoustic wave in Z-X lithium niobate plates [and conductivity sensor application. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 1996, 43, 491–494. [Google Scholar] [CrossRef]
- Kasas, S.; Malovichko, A.; Villalba, M.I.; Vela, M.E.; Yantorno, O.; Willaert, R.G. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics 2021, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Dragoman, M.; Dragoman, D. Microelectromechanical Systems. In Encyclopedia of Condensed Matter Physics; Bassani, F., Liedl, G.L., Wyder, P., Eds.; Elsevier: Oxford, UK, 2005; pp. 415–423. ISBN 9780123694010. [Google Scholar]
- Bennett, I.; Pyne, A.L.B.; McKendry, R.A. Cantilever Sensors for Rapid Optical Antimicrobial Sensitivity Testing. ACS Sens. 2020, 5, 3133–3139. [Google Scholar] [CrossRef] [PubMed]
- Sinha Ray, S. 4—Techniques for characterizing the structure and properties of polymer nanocomposites. In Woodhead Publishing Series in Composites Science and Engineering; Sinha Ray, S., Ed.; Woodhead Publishing: Sawston, UK, 2013; pp. 74–88. ISBN 9780857097774. [Google Scholar]
- Stupar, P.; Opota, O.; Longo, G.; Prod’hom, G.; Dietler, G.; Greub, G.; Kasas, S. Nanomechanical sensor applied to blood culture pellets: A fast approach to determine the antibiotic susceptibility against agents of bloodstream infections. Clin. Microbiol. Infect. 2017, 23, 400–405. [Google Scholar] [CrossRef] [Green Version]
- Kinnunen, P.; Sinn, I.; McNaughton, B.H.; Newton, D.W.; Burns, M.A.; Kopelman, R. Monitoring the growth and drug susceptibility of individual bacteria using asynchronous magnetic bead rotation sensors. Biosens. Bioelectron. 2011, 26, 2751–2755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinn, I.; Albertson, T.; Kinnunen, P.; Breslauer, D.N.; McNaughton, B.H.; Burns, M.A.; Kopelman, R. Asynchronous Magnetic Bead Rotation Microviscometer for Rapid, Sensitive, and Label-Free Studies of Bacterial Growth and Drug Sensitivity. Anal. Chem. 2012, 84, 5250–5256. [Google Scholar] [CrossRef] [Green Version]
- Kinnunen, P.; Carey, M.E.; Craig, E.; Brahmasandra, S.N.; McNaughton, B.H. Rapid bacterial growth and antimicrobial response using self-assembled magnetic bead sensors. Sens. Actuators B Chem. 2014, 190, 265–269. [Google Scholar] [CrossRef]
- Chung, C.-Y.; Wang, J.-C.; Chuang, H.-S. Rapid Bead-Based Antimicrobial Susceptibility Testing by Optical Diffusometry. PLoS ONE 2016, 11, e0148864. [Google Scholar] [CrossRef]
- He, F.; Zhou, J. A new antimicrobial susceptibility testing method of Escherichia coli against ampicillin by MSPQC. J. Microbiol. Methods 2007, 68, 563–567. [Google Scholar] [CrossRef]
- Wu, Y.; Li, G.; Hong, Y.; Zhao, X.; Reyes, P.I.; Lu, Y. Rapid and dynamic detection of antimicrobial treatment response using spectral amplitude modulation in MZO nanostructure-modified quartz crystal microbalance. J. Microbiol. Methods 2020, 178, 106071. [Google Scholar] [CrossRef]
- Ma, F.; Rehman, A.; Sims, M.; Zeng, X. Antimicrobial Susceptibility Assays Based on the Quantification of Bacterial Lipopolysaccharides via a Label Free Lectin Biosensor. Anal. Chem. 2015, 87, 4385–4393. [Google Scholar] [CrossRef]
- Ren, J.; Ma, L.; Li, Z.; lin, Q.; Huang, H.; Yi, S. Simultaneous and early detection of Mycobacterium tuberculosis resistance to antituberculosis drugs using an indirect series piezoelectric system. Biosens. Bioelectron. 2013, 43, 115–119. [Google Scholar] [CrossRef]
- Wan, Y.; Zhang, D.; Hou, B. Determination of sulphate-reducing bacteria based on vancomycin-functionalised magnetic nanoparticles using a modification-free quartz crystal microbalance. Biosens. Bioelectron. 2010, 25, 1847–1850. [Google Scholar] [CrossRef]
- Mustazzolu, A.; Venturelli, L.; Dinarelli, S.; Brown, K.; Floto, R.A.; Dietler, G.; Fattorini, L.; Kasas, S.; Girasole, M.; Longo, G. A Rapid Unraveling of the Activity and Antibiotic Susceptibility of Mycobacteria. Antimicrob. Agents Chemother. 2019, 63, e02194-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Khan, M.F.; Kaur, K.; Thundat, T. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat. Commun. 2016, 7, 12947. [Google Scholar] [CrossRef] [PubMed]
- Villalba, M.I.; Stupar, P.; Chomicki, W.; Bertacchi, M.; Dietler, G.; Arnal, L.; Vela, M.E.; Yantorno, O.; Kasas, S. Nanomotion Detection Method for Testing Antibiotic Resistance and Susceptibility of Slow-Growing Bacteria. Small 2018, 14, 1702671. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.; Niazi, J.H. Biosensors for detecting viral and bacterial infections using host biomarkers: A review. Analyst 2020, 145, 7825–7848. [Google Scholar] [CrossRef]
- Moran, K.L.M.; Fitzgerald, J.; McPartlin, D.A.; Loftus, J.H.; O’Kennedy, R. Chapter 4—Biosensor-Based Technologies for the Detection of Pathogens and Toxins. In Biosensors for Sustainable Food—New Opportunities and Technical Challenges; Scognamiglio, V., Rea, G., Arduini, F., Palleschi, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 74, pp. 93–120. ISBN 0166526X. [Google Scholar]
- Ye, Y.; Wu, T.; Jiang, X.; Cao, J.; Ling, X.; Mei, Q.; Chen, H.; Han, D.; Xu, J.-J.; Shen, Y. Portable Smartphone-Based QDs for the Visual Onsite Monitoring of Fluoroquinolone Antibiotics in Actual Food and Environmental Samples. ACS Appl. Mater. Interfaces 2020, 12, 14552–14562. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.-E.; Kaushik, A.; Hsieh, K.; Chang, E.; Chen, L.; Zhang, P.; Wang, T.-H. Toward Decentralizing Antibiotic Susceptibility Testing via Ready-to-Use Microwell Array and Resazurin-Aided Colorimetric Readout. Anal. Chem. 2021, 93, 1260–1265. [Google Scholar] [CrossRef]
- Sun, J.; Warden, A.R.; Huang, J.; Wang, W.; Ding, X. Colorimetric and Electrochemical Detection of Escherichia coli and Antibiotic Resistance Based on a p-Benzoquinone-Mediated Bioassay. Anal. Chem. 2019, 91, 7524–7530. [Google Scholar] [CrossRef] [PubMed]
- Mocan, T.; Matea, C.T.; Pop, T.; Mosteanu, O.; Buzoianu, A.D.; Puia, C.; Iancu, C.; Mocan, L. Development of nanoparticle-based optical sensors for pathogenic bacterial detection. J. Nanobiotechnol. 2017, 15, 25. [Google Scholar] [CrossRef] [Green Version]
- Kadlec, M.W.; You, D.; Liao, J.C.; Wong, P.K. A Cell Phone-Based Microphotometric System for Rapid Antimicrobial Susceptibility Testing. J. Lab. Autom. 2014, 19, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazemi, E.; Hassen, W.M.; Frost, E.H.; Dubowski, J.J. Monitoring growth and antibiotic susceptibility of Escherichia coli with photoluminescence of GaAs/AlGaAs quantum well microstructures. Biosens. Bioelectron. 2017, 93, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhen, L.; Liu, J.; Wu, J. Rapid antibiotic susceptibility testing in a microfluidic pH sensor. Anal. Chem. 2013, 85, 2787–2794. [Google Scholar] [CrossRef]
- Nag, P.; Sadani, K.; Mukherji, S.; Mukherji, S. Beta-lactam antibiotics induced bacteriolysis on LSPR sensors for assessment of antimicrobial resistance and quantification of antibiotics. Sens. Actuators B Chem. 2020, 311, 127945. [Google Scholar] [CrossRef]
- Adya, A.K.; Canetta, E. Chapter 16—Nanotechnology and its applications to animal biotechnology. In Animal Biotechnology, 2nd ed.; Verma, A.S., Singh, A., Eds.; Academic Press: Boston, MA, USA, 2020; pp. 309–326. ISBN 9780128117101. [Google Scholar]
- Uzunoglu, D.; Altunbek, M.; Kuku, G.; Culha, M. Chapter 8—Single-Cell Omics in Noninvasive Diagnosis and Testing—Surface-Enhanced Raman Spectroscopy-Based Approach. In Single-Cell Omics; Barh, D., Azevedo, V.B., Eds.; Academic Press: Boston, MA, USA, 2019; pp. 153–177. ISBN 9780128149195. [Google Scholar]
- Wood, B.R.; Kochan, K.; Marzec, K.M. Chapter 13—Resonance Raman spectroscopy of hemoglobin in red blood cells. In Vibrational Spectroscopy in Protein Research; Ozaki, Y., Baranska, M., Lednev, I.K., Wood, B.R., Eds.; Academic Press: Boston, MA, USA, 2020; pp. 375–414. ISBN 9780128186107. [Google Scholar]
- Bi, L.; Wang, X.; Cao, X.; Liu, L.; Bai, C.; Zheng, Q.; Choo, J.; Chen, L. SERS-active Au@Ag core-shell nanorod (Au@AgNR) tags for ultrasensitive bacteria detection and antibiotic-susceptibility testing. Talanta 2020, 220, 121397. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Bian, X.; Zhang, H.; Wen, Y.; Chen, Q.; Yan, Y.; Li, L.; Liu, G.; Yan, J. Controllable design of a nano-bio aptasensing interface based on tetrahedral framework nucleic acids in an integrated microfluidic platform. Biosens. Bioelectron. 2021, 176, 112943. [Google Scholar] [CrossRef]
- Thrift, W.J.; Ronaghi, S.; Samad, M.; Wei, H.; Nguyen, D.G.; Cabuslay, A.S.; Groome, C.E.; Santiago, P.J.; Baldi, P.; Hochbaum, A.I.; et al. Deep Learning Analysis of Vibrational Spectra of Bacterial Lysate for Rapid Antimicrobial Susceptibility Testing. ACS Nano 2020, 14, 15336–15348. [Google Scholar] [CrossRef]
- He, P.J.W.; Katis, I.N.; Kumar, A.J.U.; Bryant, C.A.; Keevil, C.W.; Somani, B.K.; Mahobia, N.; Eason, R.W.; Sones, C.L. Laser-patterned paper-based sensors for rapid point-of-care detection and antibiotic-resistance testing of bacterial infections. Biosens. Bioelectron. 2020, 152, 112008. [Google Scholar] [CrossRef] [PubMed]
- Ucak Ozkaya, G.; Durak, M.Z.; Akyar, I.; Karatuna, O. Antimicrobial Susceptibility Test for the Determination of Resistant and Susceptible S. aureus and Enterococcus spp. Using a Multi-Channel Surface Plasmon Resonance Device. Diagnostics 2019, 9, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gahlaut, S.K.; Kalyani, N.; Sharan, C.; Mishra, P.; Singh, J.P. Smartphone based dual mode in situ detection of viability of bacteria using Ag nanorods array. Biosens. Bioelectron. 2019, 126, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.M.; Hsieh, K.; Chen, L.; Shin, D.J.; Liao, J.C.; Wang, T.-H. Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform. Biosens. Bioelectron. 2017, 97, 260–266. [Google Scholar] [CrossRef]
- Sabhachandani, P.; Sarkar, S.; Zucchi, P.C.; Whitfield, B.A.; Kirby, J.E.; Hirsch, E.B.; Konry, T. Integrated microfluidic platform for rapid antimicrobial susceptibility testing and bacterial growth analysis using bead-based biosensor via fluorescence imaging. Microchim. Acta 2017, 184, 4619–4628. [Google Scholar] [CrossRef]
- Wang, P.; Pang, S.; Zhang, H.; Fan, M.; He, L. Characterization of Lactococcus lactis response to ampicillin and ciprofloxacin using surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2016, 408, 933–941. [Google Scholar] [CrossRef]
- Mohan, R.; Mukherjee, A.; Sevgen, S.E.; Sanpitakseree, C.; Lee, J.; Schroeder, C.M.; Kenis, P.J.A. A multiplexed microfluidic platform for rapid antibiotic susceptibility testing. Biosens. Bioelectron. 2013, 49, 118–125. [Google Scholar] [CrossRef]
- Chiang, Y.-L.; Lin, C.-H.; Yen, M.-Y.; Su, Y.-D.; Chen, S.-J.; Chen, H. Innovative antimicrobial susceptibility testing method using surface plasmon resonance. Biosens. Bioelectron. 2009, 24, 1905–1910. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, X.; Wang, T.; Li, Z.; Han, D.; Yu, C.; Yang, C.; Qu, H.; Chi, H.; Wang, Y.; et al. A sensitive and rapid bacterial antibiotic susceptibility test method by surface enhanced Raman spectroscopy. Braz. J. Microbiol. 2020, 51, 875–881. [Google Scholar] [CrossRef]
- Bernatová, S.; Rebrošová, K.; Pilát, Z.; Šerý, M.; Gjevik, A.; Samek, O.; Ježek, J.; Šiler, M.; Kizovský, M.; Klementová, T.; et al. Rapid detection of antibiotic sensitivity of Staphylococcus aureus by Raman tweezers. Eur. Phys. J. Plus 2021, 136, 233. [Google Scholar] [CrossRef]
- Simoska, O.; Stevenson, K.J. Electrochemical sensors for rapid diagnosis of pathogens in real time. Analyst 2019, 144, 6461–6478. [Google Scholar] [CrossRef] [PubMed]
- Nemr, C.R.; Smith, S.J.; Liu, W.; Mepham, A.H.; Mohamadi, R.M.; Labib, M.; Kelley, S.O. Nanoparticle-Mediated Capture and Electrochemical Detection of Methicillin-Resistant Staphylococcus aureus. Anal. Chem. 2019, 91, 2847–2853. [Google Scholar] [CrossRef] [PubMed]
- Karbelkar, A.A.; Furst, A.L. Electrochemical Diagnostics for Bacterial Infectious Diseases. ACS Infect. Dis. 2020, 6, 1567–1571. [Google Scholar] [CrossRef]
- Liang, B.; Li, L.; Tang, X.; Lang, Q.; Wang, H.; Li, F.; Shi, J.; Shen, W.; Palchetti, I.; Mascini, M.; et al. Microbial surface display of glucose dehydrogenase for amperometric glucose biosensor. Biosens. Bioelectron. 2013, 45, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Farabullini, F.; Lucarelli, F.; Palchetti, I.; Marrazza, G.; Mascini, M. Disposable electrochemical genosensor for the simultaneous analysis of different bacterial food contaminants. Biosens. Bioelectron. 2007, 22, 1544–1549. [Google Scholar] [CrossRef] [PubMed]
- Voccia, D.; Bettazzi, F.; Fratini, E.; Berti, D.; Palchetti, I. Improving impedimetric nucleic acid detection by using enzyme-decorated liposomes and nanostructured screen-printed electrodes. Anal. Bioanal. Chem. 2016, 408, 7271–7281. [Google Scholar] [CrossRef] [PubMed]
- Beitollahi, H.; Mohammadi, S.Z.; Safaei, M.; Tajik, S. Applications of electrochemical sensors and biosensors based on modified screen-printed electrodes: A review. Anal. Methods 2020, 12, 1547–1560. [Google Scholar] [CrossRef]
- Hannah, S.; Dobrea, A.; Lasserre, P.; Blair, E.O.; Alcorn, D.; Hoskisson, P.A.; Corrigan, D.K. Development of a Rapid, Antimicrobial Susceptibility Test for E. coli Based on Low-Cost, Screen-Printed Electrodes. Biosensors 2020, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Altobelli, E.; Mohan, R.; Mach, K.E.; Sin, M.L.Y.; Anikst, V.; Buscarini, M.; Wong, P.K.; Gau, V.; Banaei, N.; Liao, J.C. Integrated Biosensor Assay for Rapid Uropathogen Identification and Phenotypic Antimicrobial Susceptibility Testing. Eur. Urol. Focus 2017, 3, 293–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Jović, M.; Lesch, A.; Tissières Lovey, L.; Prudent, M.; Pick, H.; Girault, H.H. Immuno-affinity Amperometric Detection of Bacterial Infections. Angew. Chem. Int. Ed. 2018, 57, 14942–14946. [Google Scholar] [CrossRef]
- Mach, K.E.; Ruchika, M.; Jo, B.E.; Mei-Chiung, S.; Vincent, G.; Kin, W.P.; Liao, J.C. A Biosensor Platform for Rapid Antimicrobial Susceptibility Testing Directly From Clinical Samples. J. Urol. 2011, 185, 148–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, J.C.; Mastali, M.; Gau, V.; Suchard, M.A.; Møller, A.K.; Bruckner, D.A.; Babbitt, J.T.; Li, Y.; Gornbein, J.; Landaw, E.M.; et al. Use of Electrochemical DNA Biosensors for Rapid Molecular Identification of Uropathogens in Clinical Urine Specimens. J. Clin. Microbiol. 2006, 44, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Vu, Q.K.; Tran, Q.H.; Vu, N.P.; Anh, T.-L.; Le Dang, T.T.; Matteo, T.; Nguyen, T.H.H. A label-free electrochemical biosensor based on screen-printed electrodes modified with gold nanoparticles for quick detection of bacterial pathogens. Mater. Today Commun. 2021, 26, 101726. [Google Scholar] [CrossRef]
- Shi, X.; Kadiyala, U.; VanEpps, J.S.; Yau, S.-T. Culture-free bacterial detection and identification from blood with rapid, phenotypic, antibiotic susceptibility testing. Sci. Rep. 2018, 8, 3416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mascini, M.; Palchetti, I.; Tombelli, S. Nucleic acid and peptide aptamers: Fundamentals and bioanalytical aspects. Angew. Chem. Int. Ed. 2012, 51, 1316–1332. [Google Scholar] [CrossRef]
- Palchetti, I.; Mascini, M. Electrochemical nanomaterial-based nucleic acid aptasensors. Anal. Bioanal. Chem. 2012, 402, 3103–3114. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.R.; Pulingam, T.; Appaturi, J.N.; Zifruddin, A.N.; Teh, S.J.; Lim, T.W.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 2018, 554, 34–43. [Google Scholar] [CrossRef]
- Muniandy, S.; Dinshaw, I.J.; Teh, S.J.; Lai, C.W.; Ibrahim, F.; Thong, K.L.; Leo, B.F. Graphene-based label-free electrochemical aptasensor for rapid and sensitive detection of foodborne pathogen. Anal. Bioanal. Chem. 2017, 409, 6893–6905. [Google Scholar] [CrossRef] [PubMed]
- Jo, N.; Kim, B.; Lee, S.-M.; Oh, J.; Park, I.H.; Jin Lim, K.; Shin, J.-S.; Yoo, K.-H. Aptamer-functionalized capacitance sensors for real-time monitoring of bacterial growth and antibiotic susceptibility. Biosens. Bioelectron. 2018, 102, 164–170. [Google Scholar] [CrossRef]
- Fu, W.; Jiang, L.; van Geest, E.P.; Lima, L.M.C.; Schneider, G.F. Sensing at the Surface of Graphene Field-Effect Transistors. Adv. Mater. 2017, 29, 1603610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, J.; Vishnubhotla, R.; Vrudhula, A.; Johnson, A.T.C. Scalable Production of High-Sensitivity, Label-Free DNA Biosensors Based on Back-Gated Graphene Field Effect Transistors. ACS Nano 2016, 10, 8700–8704. [Google Scholar] [CrossRef]
- Thakur, B.; Zhou, G.; Chang, J.; Pu, H.; Jin, B.; Sui, X.; Yuan, X.; Yang, C.-H.; Magruder, M.; Chen, J. Rapid detection of single E. coli bacteria using a graphene-based field-effect transistor device. Biosens. Bioelectron. 2018, 110, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Dai, Z.; Tang, X.; Lin, Z.; Lo, P.K.; Meyyappan, M.; Lai, K.W.C. Graphene Field-Effect Transistors for the Sensitive and Selective Detection of Escherichia coli Using Pyrene-Tagged DNA Aptamer. Adv. Healthc. Mater. 2017, 6, 1700736. [Google Scholar] [CrossRef]
- Kumar, N.; Wang, W.; Ortiz-Marquez, J.C.; Catalano, M.; Gray, M.; Biglari, N.; Hikari, K.; Ling, X.; Gao, J.; van Opijnen, T.; et al. Dielectrophoresis assisted rapid, selective and single cell detection of antibiotic resistant bacteria with G-FETs. Biosens. Bioelectron. 2020, 156, 112123. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Ji, J.; Sun, J.; Pi, F.; Zhang, Y.; Sun, X. Rapid detection of antibiotic resistance in Salmonella with screen printed carbon electrodes. J. Solid State Electrochem. 2020, 24, 1539–1549. [Google Scholar] [CrossRef]
- Lee, K.-S.; Lee, S.-M.; Oh, J.; Park, I.H.; Song, J.H.; Han, M.; Yong, D.; Lim, K.J.; Shin, J.-S.; Yoo, K.-H. Electrical antimicrobial susceptibility testing based on aptamer-functionalized capacitance sensor array for clinical isolates. Sci. Rep. 2020, 10, 13709. [Google Scholar] [CrossRef]
- Hannah, S.; Addington, E.; Alcorn, D.; Shu, W.; Hoskisson, P.A.; Corrigan, D.K. Rapid antibiotic susceptibility testing using low-cost, commercially available screen-printed electrodes. Biosens. Bioelectron. 2019, 145, 111696. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, D.; Mishra, K.P.; Kaur, G.; Dhull, N.; Tomar, M.; Gupta, V.; Kumar, B.; Ganju, L. Rapid antibiotic susceptibility testing by resazurin using thin film platinum as a bio-electrode. J. Microbiol. Methods 2019, 162, 69–76. [Google Scholar] [CrossRef]
- Rueda, D.; Furukawa, R.; Fuentes, P.; Comina, G.; Rey De Castro, N.G.; Requena, D.; Gilman, R.H.; Sheen, P.; Zimic, M. A novel inexpensive electrochemical sensor for pyrazinoic acid as a potential tool for the identification of pyrazinamide-resistant Mycobacterium tuberculosis. Int. J. Mycobacteriol. 2018, 7, 275–281. [Google Scholar]
- Sitkov, N.O.; Zimina, T.M.; Soloviev, A.V. Development of impedimetric sensor for E. coli M-17 antibiotic susceptibility testing. In Proceedings of the 2018 IEEE Conference of Russian Young Researchers in Electrical and Electronic Engineering (EIConRus), Moscow and St. Petersburg, Russia, 29 January–1 February 2018; pp. 1227–1230. [Google Scholar]
- Ibarlucea, B.; Rim, T.; Baek, C.K.; de Visser, J.A.G.M.; Baraban, L.; Cuniberti, G. Nanowire sensors monitor bacterial growth kinetics and response to antibiotics. Lab. Chip 2017, 17, 4283–4293. [Google Scholar] [CrossRef] [PubMed]
- Niyomdecha, S.; Limbut, W.; Numnuam, A.; Asawatreratanakul, P.; Kanatharana, P.; Thavarungkul, P. Capacitive antibacterial susceptibility screening test with a simple renewable sensing surface. Biosens. Bioelectron. 2017, 96, 84–88. [Google Scholar] [CrossRef]
- Safavieh, M.; Pandya, H.J.; Venkataraman, M.; Thirumalaraju, P.; Kanakasabapathy, M.K.; Singh, A.; Prabhakar, D.; Chug, M.K.; Shafiee, H. Rapid Real-Time Antimicrobial Susceptibility Testing with Electrical Sensing on Plastic Microchips with Printed Electrodes. ACS Appl. Mater. Interfaces 2017, 9, 12832–12840. [Google Scholar] [CrossRef] [Green Version]
- Abeyrathne, C.D.; Huynh, D.H.; Mcintire, T.W.; Nguyen, T.C.; Nasr, B.; Zantomio, D.; Chana, G.; Abbott, I.; Choong, P.; Catton, M.; et al. Lab on a chip sensor for rapid detection and antibiotic resistance determination of Staphylococcus aureus. Analyst 2016, 141, 1922–1929. [Google Scholar] [CrossRef]
- Török, M.E.; Cooke, F.J.; Moran, E. Basics of Antimicrobials. In Oxford Handbook of Infectious Diseases and Microbiology; Torok, M.E., Moran, E., Cooke, F., Eds.; Oxford University Press: Oxford, UK, 2016; pp. 3–30. ISBN 9780199671328. [Google Scholar]
- Vikesland, P.J.; Pruden, A.; Alvarez, P.J.J.; Aga, D.; Bu, H.; Li, X.; Manaia, C.M.; Nambi, I.; Wigginton, K.; Zhang, T. Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance. Environ. Sci. Technol. 2017, 51, 13061–13069. [Google Scholar] [CrossRef] [Green Version]
- Haddaoui, M.; Sola, C.; Raouafi, N.; Korri-Youssoufi, H. E-DNA detection of rpoB gene resistance in Mycobacterium tuberculosis in real samples using Fe3O4/polypyrrole nanocomposite. Biosens. Bioelectron. 2019, 128, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Bizid, S.; Blili, S.; Mlika, R.; Haj Said, A.; Korri-Youssoufi, H. Direct E-DNA sensor of Mycobacterium tuberculosis mutant strain based on new nanocomposite transducer (Fc-ac-OMPA/MWCNTs). Talanta 2018, 184, 475–483. [Google Scholar] [CrossRef]
- Rachkov, A.; Patskovsky, S.; Soldatkin, A.; Meunier, M. Surface plasmon resonance detection of oligonucleotide sequences of the rpoB genes of Mycobacterium tuberculosis. Talanta 2011, 85, 2094–2099. [Google Scholar] [CrossRef]
- Matsishin, M.; Rachkov, A.; Lopatynskyi, A.; Chegel, V.; Soldatkin, A.; El’skaya, A. Selective Amplification of SPR Biosensor Signal for Recognition of rpoB Gene Fragments by Use of Gold Nanoparticles Modified by Thiolated DNA. Nanoscale Res. Lett. 2017, 12, 252. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.; Kalsi, S.; Zeimpekis, I.; Sun, K.; Ashburn, P.; Turner, C.; Sutton, J.M.; Morgan, H. Ultra-fast electronic detection of antimicrobial resistance genes using isothermal amplification and Thin Film Transistor sensors. Biosens. Bioelectron. 2017, 96, 281–287. [Google Scholar] [CrossRef]
- Sadat Mousavi, P.; Smith, S.J.; Chen, J.B.; Karlikow, M.; Tinafar, A.; Robinson, C.; Liu, W.; Ma, D.; Green, A.A.; Kelley, S.O.; et al. A multiplexed, electrochemical interface for gene-circuit-based sensors. Nat. Chem. 2020, 12, 48–55. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Z.; Li, Y.; Xie, G. Amplified electrochemical detection of mecA gene in methicillin-resistant Staphylococcus aureus based on target recycling amplification and isothermal strand-displacement polymerization reaction. Sens. Actuators B Chem. 2015, 221, 148–154. [Google Scholar] [CrossRef]
- Huang, J.M.-Y.; Henihan, G.; Macdonald, D.; Michalowski, A.; Templeton, K.; Gibb, A.P.; Schulze, H.; Bachmann, T.T. Rapid Electrochemical Detection of New Delhi Metallo-beta-lactamase Genes To Enable Point-of-Care Testing of Carbapenem-Resistant Enterobacteriaceae. Anal. Chem. 2015, 87, 7738–7745. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Kainz, K.; Barisic, I.; Hainberger, R. An Impedimetric Sensor for Real-Time Detection of Antibiotic Resistance Genes Employing Rolling Circle Amplification. Int. J. Electrochem. Sci. 2015, 10, 2026–2034. [Google Scholar]
- Maldonado, J.; González-Guerrero, A.B.; Fernández-Gavela, A.; González-López, J.J.; Lechuga, L.M. Ultrasensitive Label-Free Detection of Unamplified Multidrug-Resistance Bacteria Genes with a Bimodal Waveguide Interferometric Biosensor. Diagnostics 2020, 10, 845. [Google Scholar] [CrossRef]
- Hu, S.; Niu, L.; Zhao, F.; Yan, L.; Nong, J.; Wang, C.; Gao, N.; Zhu, X.; Wu, L.; Bo, T.; et al. Identification of Acinetobacter baumannii and its carbapenem-resistant gene blaOXA-23-like by multiple cross displacement amplification combined with lateral flow biosensor. Sci. Rep. 2019, 9, 17888. [Google Scholar] [CrossRef]
- Lu, S.; Du, J.; Sun, Z.; Jing, C. Hairpin-Structured Magnetic SERS Sensor for Tetracycline Resistance Gene tetA Detection. Anal. Chem. 2020, 92, 16229–16235. [Google Scholar] [CrossRef]
- Meena, G.G.; Wall, T.A.; Stott, M.A.; Brown, O.; Robison, R.; Hawkins, A.R.; Schmidt, H. 7× multiplexed, optofluidic detection of nucleic acids for antibiotic-resistance bacterial screening. Opt. Express 2020, 28, 33019–33027. [Google Scholar] [CrossRef]
- Bizid, S.; Blili, S.; Mlika, R.; Haj Said, A.; Korri-Youssoufi, H. Direct Electrochemical DNA Sensor based on a new redox oligomer modified with ferrocene and carboxylic acid: Application to the detection of Mycobacterium tuberculosis mutant strain. Anal. Chim. Acta 2017, 994, 10–18. [Google Scholar] [CrossRef]
- Bengtson, H.N.; Homolka, S.; Niemann, S.; Reis, A.J.; da Silva, P.E.; Gerasimova, Y.V.; Kolpashchikov, D.M.; Rohde, K.H. Multiplex detection of extensively drug resistant tuberculosis using binary deoxyribozyme sensors. Biosens. Bioelectron. 2017, 94, 176–183. [Google Scholar] [CrossRef]
- Zribi, B.; Roy, E.; Pallandre, A.; Chebil, S.; Koubaa, M.; Mejri, N.; Magdinier Gomez, H.; Sola, C.; Korri-Youssoufi, H.; Haghiri-Gosnet, A.-M. A microfluidic electrochemical biosensor based on multiwall carbon nanotube/ferrocene for genomic DNA detection of Mycobacterium tuberculosis in clinical isolates. Biomicrofluidics 2016, 10, 14115. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Hedström, M.; Chen, D.; Fan, X.; Mattiasson, B. A capacitive DNA sensor-based test for simple and sensitive analysis of antibiotic resistance in field setting. Biosens. Bioelectron. 2015, 64, 255–259. [Google Scholar] [CrossRef]
- Peng, H.-P.; Hu, Y.; Liu, P.; Deng, Y.-N.; Wang, P.; Chen, W.; Liu, A.-L.; Chen, Y.-Z.; Lin, X.-H. Label-free electrochemical DNA biosensor for rapid detection of mutidrug resistance gene based on Au nanoparticles/toluidine blue-graphene oxide nanocomposites. Sens. Actuators B Chem. 2015, 207, 269–276. [Google Scholar] [CrossRef]
- Liong, M.; Hoang, A.N.; Chung, J.; Gural, N.; Ford, C.B.; Min, C.; Shah, R.R.; Ahmad, R.; Fernandez-Suarez, M.; Fortune, S.M.; et al. Magnetic barcode assay for genetic detection of pathogens. Nat. Commun. 2013, 4, 1752. [Google Scholar] [CrossRef] [Green Version]
- Bengtson, H.N.; Kolpashchikov, D.M. A Differential Fluorescent Receptor for Nucleic Acid Analysis. ChemBioChem 2014, 15, 228–231. [Google Scholar] [CrossRef] [Green Version]
- Rachkov, A.; Patskovsky, S.; Soldatkin, A.; Meunier, M. Discrimination of single base mismatched oligonucleotides related to the rpoB gene of Mycobacterium tuberculosis using a surface plasmon resonance biosensor. Biotechnol. Appl. Biochem. 2013, 60, 453–458. [Google Scholar] [CrossRef]
- Lam, B.; Das, J.; Holmes, R.D.; Live, L.; Sage, A.; Sargent, E.H.; Kelley, S.O. Solution-based circuits enable rapid and multiplexed pathogen detection. Nat. Commun. 2013, 4, 2001. [Google Scholar] [CrossRef] [Green Version]
- Strohsahl, C.M.; Miller, B.L.; Krauss, T.D. Detection of methicillin-resistant Staphylococcus aureus (MRSA) using the NanoLantern Biosensor. In Proceedings of the SPIE BiOS, San Jose, CA, USA, 28 February 2008; Volume 7167. [Google Scholar]
- Koets, M.; van der Wijk, T.; van Eemeren, J.T.W.M.; van Amerongen, A.; Prins, M.W.J. Rapid DNA multi-analyte immunoassay on a magneto-resistance biosensor. Biosens. Bioelectron. 2009, 24, 1893–1898. [Google Scholar] [CrossRef]
- Kara, P.; Cavusoglu, C.; Cavdar, S.; Ozsoz, M. Direct electrochemical genosensing for multiple point mutation detection of Mycobacterium tuberculosis during the development of rifampin resistance. Biosens. Bioelectron. 2009, 24, 1796–1800. [Google Scholar] [CrossRef]
- Tombelli, S.; Minunni, M.; Santucci, A.; Spiriti, M.M.; Mascini, M. A DNA-based piezoelectric biosensor: Strategies for coupling nucleic acids to piezoelectric devices. Talanta 2006, 68, 806–812. [Google Scholar] [CrossRef]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.-G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.-C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef]
- Metzger, S.; Frobel, R.A.; Dunne, W.M. Rapid simultaneous identification and quantitation of Staphylococcus aureus and Pseudomonas aeruginosa directly from bronchoalveolar lavage specimens using automated microscopy. Diagn. Microbiol. Infect. Dis. 2014, 79, 160–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredborg, M.; Andersen, K.R.; Jørgensen, E.; Droce, A.; Olesen, T.; Jensen, B.B.; Rosenvinge, F.S.; Sondergaard, T.E. Real-time optical antimicrobial susceptibility testing. J. Clin. Microbiol. 2013, 51, 2047–2053. [Google Scholar] [CrossRef] [Green Version]


| Antimicrobial Group | Antimicrobial Agent(s) | Some of Resistance Gene(s) | Resistance Mechanism |
|---|---|---|---|
| Aminocoumarins | Novobiocin, Coumermycin, Clorobiocin | gyrB, parE, parY | Target modification |
| Aminoglycosides | Amikacin, Dibekacin, framycetin, gentamicin, kanamycin, neomycin, netilmicin, plazomicin, sisomicin, spectinomycin, streptomycin, tobramycin | aacA-aphD, aadD, aadE, aadY, ant(4′)-Ia, aphA3, armA, rmtA, rmtB, rmtC, str | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Amphenicol | Azidamfenicol, chloramphenicol, florfenicol, thiamphenicol | agmR, catA1, cmlA1, floR, ttgABC | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Ansamycins | Rifampicin, rifamixin | dnaA, rbpA, rpoB | Target modification, antibiotic modification, reduced permeability |
| Carbaphenems | Doripenem, ertapenem, Imipenem | blaOXA-23, blaOXA-58, blaOXA-497, blaVIM-1 | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Cephalosporins | Cefacetrile, cefalexin, cefalotin, cefapyrin, cefazolin, cefiximine, cefotaxime, ceftriaxone, cefuroxime | ampC, bla-genes | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Ethambutol | Ethambutol | embB, embC | Expression changes, target modification |
| Fluoroquinolones | Ciprofloxacin, clinafloxacin, levofloxacin, moxifloxacin, ofloxacin, pazufloxacin, sarafloxacin | gyrA, gyrB, parC, parE | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Fosfomycin | Fosfomycin | fomA, fomB, fosC, fosA, fosB, fosX | Antibiotic modification, target modification |
| Fusidanes | Fusidic acid | fusB, fusC | Reduced permeability, target protection |
| Glycopeptides | Corbomycin, ramoplanin, telavancin, teicoplanin, vancomycin | vanA, vanB, vanD, vanR, vanS | Reduced permeability, target modification |
| Isoniazid | Isoniazid (INH) | ahpC, inhA, katG | Expression changes, target modification |
| Lincosamides | clindamycin, lincomycin, pirlimycin | ermA, ermB, erm(31), ein, lnu(A), lnu(B), lsa(B), sal(A) | Antibiotic efflux, antibiotic modification, target modification |
| Lipopeptides | Bacillomycin, caspofungin, daptomycin, mycosubtilin, surfactin, surotomicyn | cdsA, rpoB | Antibiotic modification, target modification |
| Macrolides | Azithromycin, erythromycin, oleandomycin, josamycin, roxithromycin, spiramycin, | cfr, ermA, ermB, erm(31), ereA, ereB, gimA, mefA, mefE, mel, mgt, ole | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Mupirocin | Mupirocin | mupA, mupB | Target modification |
| Nitroimidazoles | Azanidazole, benzinidazole, dimetridazole, megazol, metronidazole, nimorazole, ornidazole, pretomanid, tinidazole | nimA, nimB, nimC, nimD, nimE | Antibiotic modification |
| Nitrofurans | Furazolidone, nifuroxazide, nifurtimox, nifurtoinol, nitrofural, nitrofurantoin, nifurzide | nfsA, nfsB | Antibiotic efflux, target modification |
| Oxazolidinones | Linezolid, posizolid, radezolid, tedizolid | cfr, optrA, poxtA | Antibiotic efflux, target modification |
| Penicillins | Amoxicillin, ampicillin, benzylpenicillin, cloxacillin, penicillin, phenethicillin | ampC, blaZ, mecA, mecZ | Reduced permeability, antibiotic efflux, antibiotic modification, target modification |
| Polymixins | Bacitracin, colistin | mcr-genes, mgrB, pmrA, pmrB, pmrE | Antibiotic efflux, target modification, expression changes |
| Pyrazinamide | Pyrazinamide | clpC1, pncA, rpsA | Target modification |
| Streptogramins | Quinupristin/dalfopristin, pristinamycin, virginiamycin | cfr, erm-genes, lsa, msrA, vga, vgaB, vatA, vatB, vatC, vatD, vatE | Antibiotic efflux, antibiotic modification, target modification |
| Sulfonamides | Sulfadiazine, sulfadimethoxine, sulfadimidine, sulfafurazole, sulfamerazine, sulfamethoxazole, sulfanilamide, sulfapyridine | sul1, sul2, sul3, sul4 | Reduced permeability, expression changes, target modification |
| Tetracyclines | Chlortetracycline, doxycycline, omadacycline, oxytetracycline, tetracycline | tetA, tetB, tetC, tetM, tetO, tetQ, tetX, tet30, tet31, tet32, tet36 | Reduced permeability, antibiotic efflux, target protection, target modification |
| Trimethoprim | Trimethoprim | dfrA, dfrD, dfrG, dfrK | Target modification |
| Technic/Signal/Type | Target | Antibiotic | MIC (µg/L) | Limit of Detection | Time | Reference |
|---|---|---|---|---|---|---|
| Asynchronous magnetic bead rotation | E. coli | Gentamicin | 1 | Single bacterium binding events | 15 min | [87] |
| Atomic force microscope cantilevers | E. coli | Ampicillin | 12.5–50 | NM, 1 × 105 CFU/mL ** | 45 min | [100] |
| Asynchronous magnetic bead rotation | E. coli | Ampicillin | 8 | Single bacterium binding events | 1.5 h | [103] |
| Asynchronous magnetic bead rotation | E. coli | Gentamicin | 1 | 50 cells per drop | 100 min | [104] |
| Asynchronous magnetic bead rotation | E. coli | Gentamicin | 2 | Single bacterium binding events | 91 min | [105] |
| Brownian motion | P. aeruginosa | Gentamicin | 2 | One bacterium | 2 h | [106] |
| Multi-channel series piezoelectric quartz crystal | E. coli | Ampicillin | 32 | 5 × 105 CFU/mL ** | 5–8 h | [107] |
| Spectral amplitude modulation MZO- QCM | S. epidermidis | Ciprofloxacin Oxacillin Ciprofloxacin | 0.5 1 1 | 1 × 105 CFU/mL | 1.5 h | [108] |
| Orthogonal quartz crystal microbalance | E. coli | Ciprofloxacin Ceftriaxone Tetracycline | 12.5 15 150 | 5 × 108 CFU/mL** | 1 h | [109] |
| Indirect series piezoelectric | M. tuberculosis | Isoniazid Rifampin Ethambutol Streptomycin Capreomycin p-Aminosalicylic acid Ethionamide Rifabutin | 0.1 1.0 2.5 2.0 10 2.0 5.0 0.5 | 1 × 103 CFU/mL ** | >1 day | [110] |
| QCM under an external magnetic field | D. desulfotomaculum | Vancomycin | NR | 1.8 × 104 CFU/mL ** | 30 min | [111] |
| Cantiliver NMS under an external magnetic field | M. bovis, M. abscessus | Amikacin Rifampin Isoniazid | 1.7 0.15 0.17 | 100 bacterial cells | 30 min | [112] |
| Atomic force microscope cantilevers | E. coli, S. aureus | Ampicillin Kanamycin | 2.0 70.0 | 4.6 ± 0.5 bacteria/100 µm2 | 30–40 min | [113] |
| biomaterial microcantilever with an embedded microfluidic channel | E. coli | Ampicillin Kanamycin | NM | 1 × 105 CFU/mL ** | 30 min | [114] |
| Atomic force microscope cantilevers | B. Pertussis | Erythromycin Clarithromycin | 0.06 0.12 | NM | 20–40 min | [115] |
| Technique | Recognition Probe | Target | Antibiotic | MIC (µg/L) | Limit of Detection | Time | Reference |
|---|---|---|---|---|---|---|---|
| Colorimetric | Tetrazolium salts-8 | E. coli | Ampicillin | 128 | 10 CFU/mL | 2 h | [122] |
| Fluorescence | TDN-aptamer/SYTO 9 Green | E. coli | Kanamycin Streptomycin Ofloxacin Norfloxacin Chloramphenicol | 4.0 8.0 0.5 1.0 2.0 | 10 CFU/mL | 5 h | [130] |
| SPR | 2PAC—Au nanosphere/block copolymer templates (PS-b-PMMA) | E. coli, P. aeruginosa | Carbenicillin Gentamicin Rifampicin | 100 1 Resistant | NR | 30 min | [131] |
| laser-patterned paper-based | Chromogenic agar CHROMagar/photopolymer DeSolite® | E. coli | Amoxicillin | 30 | 2.5 × 109 CFU/mL | 18 h | [132] |
| SPR | Poly-L-lysine/glass slide coated with gold sensor chip | MRSAMSSA | Cefoxitin | 32 to >128 1 to 4 | 5 × 105 CFU/mL * | 3 h | [133] |
| Colorimetric | Endogenous H2S/AgNRs | E. coli | Ampicillin | 100 (MBC) | 102 cell/mL * | 4–6 h | [134] |
| Fluorescence | Resazurin | E. coli | Gentamicin | 4 | Single cell | 1 h | [135] |
| Fluorescence imaging | anti-E.coli antibody/streptavidin-coated polystyrene microsphere | E. coli | Ceftazidime Levofloxacin | 4 32 | Single cell | 30 min | [136] |
| SERS | Gold nanoparticles | L. lactis | Ampicillin Ciprofloxacin | NR | NR | 1.5 h | [137] |
| Fluorescence | PDMS/TLFM | E. coli | Ampicillin Cefalexin Chloramphenicol Tetracycline | 8 12 8 2 | Single cell | 2–4 h | [138] |
| SPR | Poly-L-lysine/Au thin film | E. coli S. epidermidis | Ampicillin Tetracycline | 3 10 | NR | 2 h | [139] |
| SERS | Bacteria-aptamer/AgNPs | E. coli S aureus | Tigecycline Vancomycin | 0.02 0.2 | 5 × 103 CFU/mL * | 2 h | [140] |
| Raman tweezers | fused-silica microfluidic chip | S. aureus | Oxacillin | 2000 | 1012 cells/L | 4 h | [141] |
| Electrode—Recognizing Element | Respond | Target | Antibiotic | MIC (µg/L) | Limit of Detection | Time | Reference |
|---|---|---|---|---|---|---|---|
| Au SPE—antibody alkaline phosphatase | DPV | S. aureus | MRSA strain | Nm | 845 CFU/mL | 4.5 h | [143] |
| SPE—Thiolated oligonucleotide capture probes | Amperometric | E. coli | Ciprofloxacin | 2 | 103 CFU/mL | 5 h | [150] |
| G-FET—peptide probes | Dirac voltage | S. aureus, A. baumannii | colistin resistant strain | NM | 104 cells/mL | 5 min | [165] |
| SPCE/MWCNTs/AuNPs—reduction of resazurin | DPV | S. gallinarum | Ofloxacin Penicillin | 32 16 | 102 CFU/mL | 1 h | [166] |
| Au—aptamers | Capacitance | E. coli *, A. baumannii, P. aeruginosa, K. pneumoniae, S. aureus, E. faecalis | Amikacin Ampicillin Aztreonam Cefepime Cefotaxime Ceftazidime Gentamicin | ≤2 ≥32 ≤1 ≤1 ≤4 ≤1 ≤1 | 105 CFU/mL ** | 6 h | [167] |
| Au SPE—agarose-based hydrogel deposit | EIS | S. aureus | Amoxicillin Oxacillin | 8 in both | 107 CFU/mL (50,000 CFUs) ** | 45 min | [168] |
| Pt deposited over a glass substrate—reduction of resazurin | DPV | E. coli, K. pneumoniae | Ampicillin Kanamycin Tetracycline | NM | 104 cells/mL | 4 h | [169] |
| Two working electrodes (Au and Pt)—POA detection | CV | M. tuberculosis | Pyrazinamide | NM | 40 µM of POA | NR | [170] |
| Array of interdigital electrodes—FD of the impedance of living and dead microorganisms | Impedance | E. coli | Ceftazidime Ceftriaxone Benzylpenicillin | NR | NR | 2 h | [171] |
| Silicon nanowire FETs | Current caused by varying pH values | E. coli | Kanamycin Cefotaxime Ofloxacin | 1–4 0.1–6 5 | Single cells | 6 h | [172] |
| 3-APBA modified electrode bind with cis-diol groups on the cell wall | Capacitance | E. coli, S. thyphi, P. aeruginosa, S. epidermidis, S. aureus, B. subtilis | Ceftriaxone Q. infectoria Ampicillin Vancomycin Rhodomyrtone | 0.03 20 0.5 1.25 0.5 | 108 CFU/mL | 2.5 h | [173] |
| SPE plastic-based microchips—antibodies | Impedance | E. coli, S. aureus | Erythromycin | 0.1 | 103 CFU/mL ** | 1.5 h | [174] |
| Interdigital electrodes—antibodies | Impedance | S. aureus | Flucloxacillin | 100 | 104 cells/mL ** | 2 h | [175] |
| Technic | Recognition Element | Target | Type of Resistance | Limit of Detection | Previous Amplification | Reference |
|---|---|---|---|---|---|---|
| Electrochemical—EIS and CV | DNA probe | rpoB | Rifampicin resistance | 0.08 fmol/L | Yes | [179] |
| Optic—fluorescence | DNA probe | CTX-MNDM-1 | Cephalosporins resistanceCarbapenems resistance | <10 copies of the gene | Yes | [182] |
| Optic—SERS | Hairpin-structured | tetA | Tetracycline Resistance | 25 copies/μL | No | [189] |
| Optic—fluorescence | Fluorescent nucleic acid probe | VIM NDM IMP KPC | carbapenem antibiotic resistance genes | (1.8 ± 0.7) × 106 beads/mL per target | No | [190] |
| Electrochemical—EIS and CV | DNA probe | rpoB | Rifampicin resistance | 0.2 fM | Yes | [191] |
| Optic—fluorescence | Binary deoxyribozyme | rpoB katG inhA | Rifampin resistanceIsoniazid resistanceFluoroquinolone resistance | 5 fg–15.6 pg | Yes | [192] |
| Electrochemical—EIS and CV | DNA probe | rpoB | Rifampicin resistance | 0.1 fM–1 pM | No | [193] |
| Electrochemical—capacitance | DNA probe | ampR | Ampicillin resistance | 1–4 pM | No | [194] |
| Electrochemical—DPV | DNA probe | MDR1 | Multidrug resistance | 2.95 × 10−12 M | No | [195] |
| Optic—fluorescence | DNA probe | rpoB | Rifampicin resistance | 1 nM ssDNA in 1 mL sample volume | Yes | [196] |
| Optic—fluorescence | DNA probe | rpoB | Rifampicin resistance | 100 nM | Yes | [197] |
| Optic—SPR | DNA probe | rpoB | Rifampicin resistance | NR | No | [198] |
| Electrochemical—DPV | PNA probe | rpoB | Rifampicin resistance | 1 CFU/ml | No | [199] |
| Optic—fluorescence | Fluorescence DNA hairpin | mecR | Methicillin resistance | 1 nM | Yes | [200] |
| Optic—fluorescence | Ab-DNA probe | lamB | Increases resistance to chlortetracycline, ciprofloxacin, balofloxacin and nalidixic acid | 4–250 pM amplicon concentrations | Yes | [201] |
| Electrochemical—DPV | DNA probe | rpoB | Rifampicin resistance | 20 fM | Yes | [202] |
| Mechanic—piezoelectric | DNA probe | mecR | Methicillin resistance | 0.125 µM | Yes | [203] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reynoso, E.C.; Laschi, S.; Palchetti, I.; Torres, E. Advances in Antimicrobial Resistance Monitoring Using Sensors and Biosensors: A Review. Chemosensors 2021, 9, 232. https://doi.org/10.3390/chemosensors9080232
Reynoso EC, Laschi S, Palchetti I, Torres E. Advances in Antimicrobial Resistance Monitoring Using Sensors and Biosensors: A Review. Chemosensors. 2021; 9(8):232. https://doi.org/10.3390/chemosensors9080232
Chicago/Turabian StyleReynoso, Eduardo C., Serena Laschi, Ilaria Palchetti, and Eduardo Torres. 2021. "Advances in Antimicrobial Resistance Monitoring Using Sensors and Biosensors: A Review" Chemosensors 9, no. 8: 232. https://doi.org/10.3390/chemosensors9080232
APA StyleReynoso, E. C., Laschi, S., Palchetti, I., & Torres, E. (2021). Advances in Antimicrobial Resistance Monitoring Using Sensors and Biosensors: A Review. Chemosensors, 9(8), 232. https://doi.org/10.3390/chemosensors9080232







