Impact of Physical Activity and Exercise on the Epigenome in Skeletal Muscle and Effects on Systemic Metabolism
Abstract
:1. Introduction
2. Methods
3. Physical Activity and Exercise and the Epigenetic Changes in Skeletal Muscle
3.1. Skeletal Muscle Physiology and Adaptations to Exercise
3.2. Epigenetics and Physical Activity/Exercise in Skeletal Muscle
3.2.1. DNA Methylation
| Endurance Exercise | |||||||
|---|---|---|---|---|---|---|---|
| Reference | Sample Size | Age and Sex | Participant Profiles | Exercise Doses | Biopsy Times | Technology | Methylation Changes and Gene Expression |
| Barres et al., (2012) [60] | n = 14 | 25 ± 1 years, men and women | Sedentary | Acute session, Intensity: 80% VO2, Volume: until 1.674 kJ | Before, after and 3 h post-exercise | Pyrosequencing | Hypomethylation of PGC-1α, TFAM, MEF2A Y PDK4 after exercise. Hypomethylation of PPAR-d 3 h post exercise |
| Bajpeyi et al., (2017) [72] | n = 11 | 24 ± 1 years, men | High and low responders | Acute session, Intensity: 50% VO2, Volume: until 650 kcal | Before and after exercise | Pyrosequencing | Hypomethylation of PGC1α and higher mRNA levels in responders to exercise |
| Lane et al., (2015) [73] | n = 7 | 29 ± 5 years, men | Cyclists completing two trials receiving isoenergetic diets differing in the timing of ingestion | Acute session, Intensity: 50% VO2, Duration: 120 min | Before and after exercise | Pyrosequencing | Hypermethylation of COX411 y FABP3 4 h after the training session. DNA methylation of PPARs increased only in the fasting group |
| Nitert et al., (2012) [74] | n = 28 | 37.5 ± 5.2 years, men | Individuals with/without familiar diabetes history | 6 months of endurance exercise and spinning (2–3 sessions per week, 1 h) | Before and after exercise | MeDIP-Chip | Hypomethylation of RUNX1, MEF2A, THADA y NDUFC2 |
| Alibegovic et al., (2010) [70] | n = 20 | 25 ± 1 years, men | Healthy Caucasian without Type 2 diabetes antecedents | 4 weeks, 6 days per week, Volume: 30 min, Intensity: 70% VO2 max | Before and after exercise | EZ DNA Methylation kit | Hypermethylation of PPARGC1A after 8 bed rest. Hypomethylation of PPARGC1A after a training program |
| Robinson et al., (2017) [75] | n = 34 | 29 ± 5 years, men and women | Adults | 12 weeks, 3 times per week, 4 × 4, 90% VO2, 3 min active rest, 3 days of treadmill, walking (45 min 70%) | Before and after exercise | 450 K array (Illumina) | Shifts in the DNA methylation less than 10% |
| Lindholm et al., (2014) [76] | n = 23 | 27 ± 0.8, men and women | Young people (without practicing exercise) | 3 months (resistance to 1 leg), 4 sessions, 45 min | Resting, before and after | 450 K array (Illumina) | Changes in DNA methylation in 5000 sites and different gene expression in 4000 genes. |
| Turner et al., (2020) [77] | n = 30 | 27 ± 4.4 years, men | Young adults | 1-6 sessions per week for 6 months | - | 850 K Array (Illumina) | Hypomethylation of HOXB1 y HOXA3. |
| Sailani et al., (2019) [78] | n = 8 | 62.1 ± 1.3 years, men | Healthy individuals, (performing regular exercise or remained sedentary their entire lives) | More than 3 times per week | - | 850 K Array (Illumina) | Hypomethylation in 714 promoters of the physically active than inactive men. Promoters for genes encoding critical insulin-responsive enzymes in glycogen metabolism, glycolysis and TCA cycle were hypomethylated in active relative to inactive men. |
| Rowlands et al., (2014) [79] | n = 8 | 49 ± 5 years, Men and women | Individuals with type 2 diabetes and obesity | 16 weeks, 3 days per week, 40–60 min per session | Before and after exercise | 450 K array (Illumina) | Hypermethylation in NRF1 y SLC27A4 |
| Stephens et al., (2018) [80] | n = 17 | 50.7 ± 1.9 years, women | Individuals with type 2 diabetes | 10 weeks, 4 days per week, Progressive intensity | Before and after exercise | 450 K array (Illumina) | Hypermethylation in responders compared to non-responders. |
| Maasar et al., (2021) [68] | n = 5 | 26 ± 2 years, men | Sport team members | (1) change of direction (COD) versus; (2) straight line (ST), running exercise. Wash-out period of at least 2 weeks between trials. | Before (30 min) and 24 h after exercise | 850 K Array (Illumina) | Hypomethylation after 30 min, mainly in AMPK, MAPK, protein binding, insulin, and axon guidance pathways. Hypermethylation of VEGFA, PPARGC1A, NR4A3 |
| Resistance Exercise | |||||||
| Reference | Sample Size | Age and Sex | Participants Profile | Exercise Doses | Biopsies Time | Technology | Methylation Changes and Gene Expression |
| Rowlands et al., (2014) [79] | n = 9 | 49 ± 5 years, men and women | Type 2 diabetes and obesity | 16 weeks, 3 days per week, Participants were randomized into endurance or resistance exercise groups comprising supervised progressive-loading exercise sessions 3 ×/week on non-consecutive days | Before and after exercise | 450 K Array (Illumina) | Hypomethylation of 409 CpGs sites and hypermethylation of 146 CpGs sites. |
| Seaborne et al., (2018) [81] | n = 8 | 27.6 ± 2.4 years, men | Adults (non-trained) | An acute bout of resistance exercise (acute RE), followed by 7 weeks (3d/week) of resistance exercise (loading), 7 weeks of exercise cessation (unloading) and a further period of 7 weeks (3d/week) resistance exercise (re-loading). | Before the first training session, after acute exercise, after a period of 7 weeks of resistance exercise (loading), exercise cessation (unloading) and a subsequent second period of 7 weeks resistance exercise (reloading). | DNA microarray | Hypomethylation of AXIN1, GRIK2, CAMK-IV, TRAF1, UBR5, RPL35a, HEG1, PLA2G16 y SETD3 |
| Bagley et al., (2020) [66] | n = 11 and n = 8 | 26.2 ± 0.1 years, and 22.9 ±1.1 years | Trained vs sedentary young individuals | 3 × 10 repeats, 70% RM, press and leg extension | Before and 4 h after exercise | PCR | Global DNA hypomethylation in trained individuals compared to sedentary. Hypermethylation of GPAM y SREBF2 in trained individuals and hypomethylation of SREBF2 in sedentary individuals. No changes in DNA methylation of genes are associated with hypertrophy and inflammation. |
3.2.2. Histone Modifications
Acetylation
Methylation
Phosphorylation
Lactylation
3.2.3. Micro-RNAs
| Endurance Exercise | |||||||
|---|---|---|---|---|---|---|---|
| Reference | Sample Size | Age and Sex | Participant Profiles | Exercise Doses | Biopsy Times | Technology | Epigenetic Changes and Gene Expression |
| Russel et al., (2013) [116] | n = 9 | 23 ± 5 years, men | Healthy people (less than 2 h of exercise per week) | Acute: 60 min 70%VO2max, Chronic (10 days), Progression: from 45 a 90 min to 75%VO2, 4 days of HIIT: 6 × 5 min (90–100% VO2), 2 min of resting | Before and after intervention | TaqMan qRT-PCR | Acute: up-regulation of miR-1, −133a, 133b, −181 and down-regulation of miR-9, −23a, −23b, y −31. Chronic: up-regulation of miR-29b and down-regulation of miR-31 |
| Keller et al., (2011) [119] | n = 8 | 29 ± 6 years, men | Sedentary healthy individuals | 4 days/week, 70% VO2max, 45 min | Before and after intervention | TaqMan RT-PCR | Lower expression of miRNAs (14 vs 7), Lower levels of miR-1, miR-133, miR-101 y miR-455. |
| Nielsen et al., (2010) [120] | n = 10 | 30.5 ± 5.5 years | Trained individuals | Acute: 60 min, 65% Pmax, Chronic (12 weeks), 5 days per week, 55–91% Pmax, 60–150 min | Before, 1 h before and 3 h after intervention | TaqMan RT-PCR | Acute: Higher expression of miR-1 and −133a, Chronic: all miRNAs were lower and restored after 2 weeks of intervention |
| Fyfe, J.J. et al., (2016) [121] | n = 8 | 27± 4 years, Men | Active young individuals | 2 × 10 min, 1 min rest, 120% lactic umbral | Before, 1 h before and 3 h after intervention | TaqMan RT-PCR | Lower expression of miR-133a, miR-378 y miR-486 |
| Margolis, L.M. et al., (2017) [122] | n = 25 | 18–40 years, Men and women | 90 min, 2.2 ± 0.1 L/min, | Before and 3 h after intervention | TaqMan RT-PCR | Lower expression of myomiR in the highest loaded group (miR-1-3p, miR-206, miR-208a-5p, y miR-499), Higher expression of myomiR in the endurance group | |
| Resistance Exercise | |||||||
| Reference | Sample Size | Age and Sex | Participants Profile | Exercise Doses | Biopsies Time | Technology | Epigenetic Changes and Gene Expression |
| Davidsen et al., (2011) [123] | n = 56 | 18–30 years, men | Active individuals | 12 weeks 5 days/week 60 min per session 20 sets by muscle group | Before and after intervention | TaqMan RT-PCR | 17 miRNAs were detected, and miR-78, miR-29a, miR-26a, and miR-451 were lower in the low-responders. miR-451 was up-regulated. |
| Rivas et al., (2014) [124] | n = 8 | 22 ± 1 years, 74 ± 2 years, men | Adults, Young and old people | 3 series of 10 repetitions, 80% Maximun repetition, 2 types of exercises | Before and 6 h after intervention | PCR-Array | 17 miRNAs were differentially expressed in young people and no changes were found in old individuals. Only miR-423-5p was up-regulated in both young and old individuals. |
| Ogasawara et al., (2016) [125] | n = 18 | 21.4 ± 1.1 years, men | Healthy and trained (resistance) individuals | 12 weeks: 10 repetitions at 70% of 1 repetition maximum (RM) for 3 sets with 2 min rest intervals. 3 days per week on alternative days for 6 week. | Before and 3 h after intervention, 12 weeks after | Multiplexed NanoString nCounter human miRNA expression assay | 26 miRNAs were different between high and low responders, miRNA-136-5p and miRNA-376a-3p were up-regulated both in the acute and chronic treatment |
| Mueller et al., (2011) [126] | n = 28 | 80.1 ± 3.7 years, men and women | Old individuals | 2 sessions per week for 12 weeks of training with two weekly resistance exercise sessions or eccentric ergometer sessions | Before and 12 weeks after intervention | miRNA analysis by custom-designed low-density PCR arrays | Lower expression of miRNA 1 |
3.2.4. Epitranscriptomics
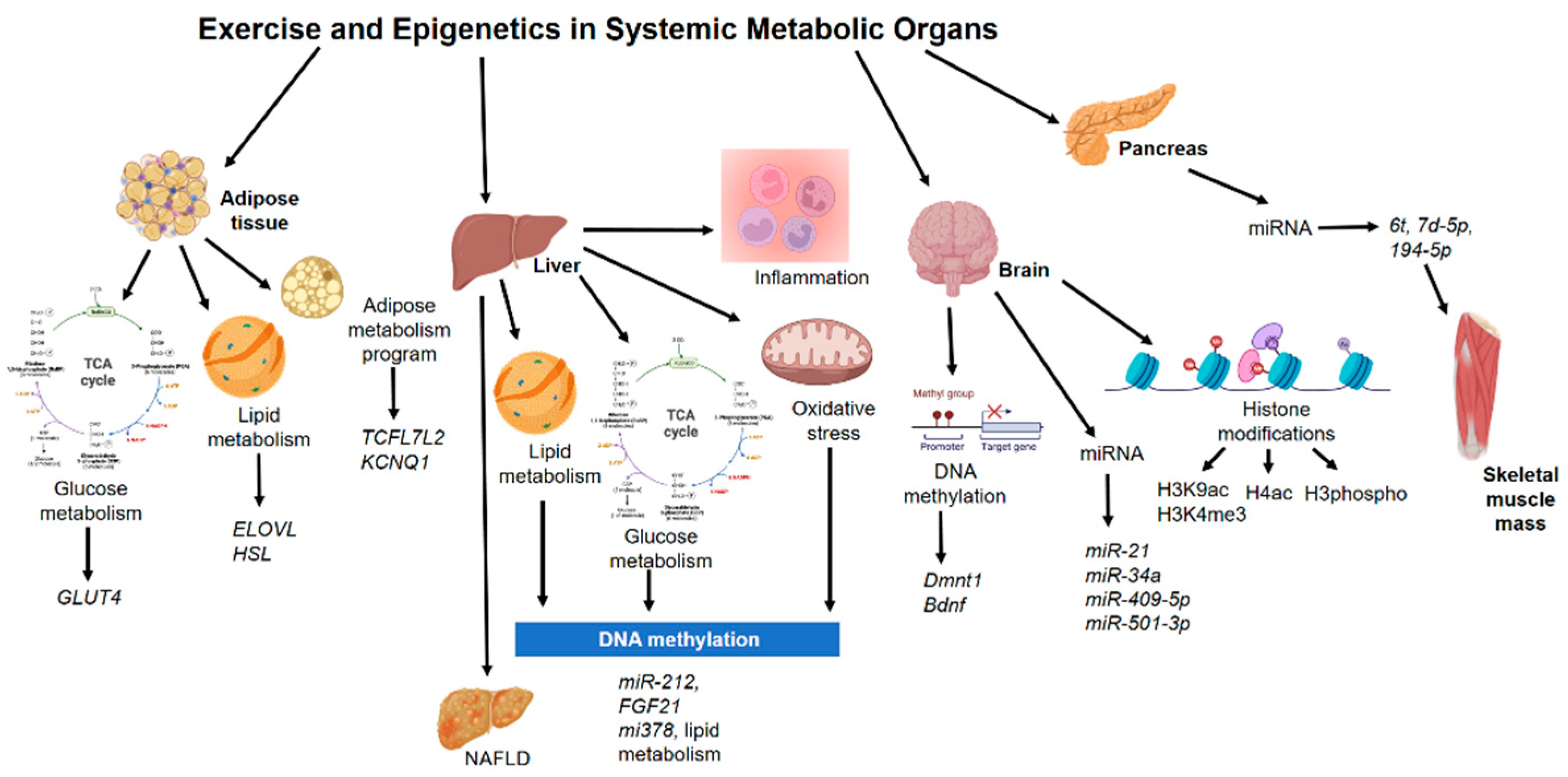
4. Epigenetics Mechanisms in Other Non-Muscle Tissue during Exercise
4.1. Adipose Tissue
4.2. Liver
4.3. Pancreas
4.4. Brain
5. Effects of Metabolism and Its Metabolites on Epigenetic Modifications during Exercise
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Latorre-Román, P.Á.; Guzmán-Guzmán, I.P.; Delgado-Floody, P.; Herrador Sanchez, J.; Aragón-Vela, J.; García Pinillos, F.; Párraga Montilla, J.A. Protective role of physical activity patterns prior to COVID-19 confinement with the severity/duration of respiratory pathologies consistent with COVID-19 symptoms in Spanish populations. Res. Sports Med. 2021, 1–12. [Google Scholar] [CrossRef]
- Caspersen, C.J.; Powell, K.E.; Christenson, G.M. Physical activity, exercise, and physical fitness: Definitions and distinctions for health-related research. Public Health Rep. 1985, 100, 126. [Google Scholar]
- Aragon-Vela, J.; Solis-Urra, P.; Ruiz-Ojeda, F.J.; Alvarez-Mercado, A.I.; Olivares-Arancibia, J.; Plaza-Diaz, J. Impact of Exercise on Gut Microbiota in Obesity. Nutrients 2021, 13, 3999. [Google Scholar] [CrossRef]
- Cox, C.E. Role of Physical Activity for Weight Loss and Weight Maintenance. Diabetes Spectr. 2017, 30, 157–160. [Google Scholar] [CrossRef] [Green Version]
- Noakes, T.; Spedding, M. Olympics: Run for your life. Nature 2012, 487, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Booth, F.W.; Roberts, C.K.; Laye, M.J. Lack of exercise is a major cause of chronic diseases. Compr. Physiol. 2012, 2, 1143–1211. [Google Scholar] [CrossRef] [Green Version]
- Hawley, J.A.; Hargreaves, M.; Joyner, M.J.; Zierath, J.R. Integrative biology of exercise. Cell 2014, 159, 738–749. [Google Scholar] [CrossRef] [Green Version]
- Coffey, V.G.; Hawley, J.A. The molecular bases of training adaptation. Sports Med. 2007, 37, 737–763. [Google Scholar] [CrossRef]
- Baar, K. Training for endurance and strength: Lessons from cell signaling. Med. Sci. Sports Exerc. 2006, 38, 1939–1944. [Google Scholar] [CrossRef] [Green Version]
- Karp, J.R. Muscle fiber types and training. Strength Cond. J. 2001, 23, 21. [Google Scholar] [CrossRef]
- Hughes, D.C.; Ellefsen, S.; Baar, K. Adaptations to Endurance and Strength Training. Cold Spring Harb. Perspect. Med. 2018, 8, a029769. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Brown, A.M.; Frontera, W.R. Principles of exercise physiology: Responses to acute exercise and long-term adaptations to training. Pm&r 2012, 4, 797–804. [Google Scholar] [CrossRef]
- Hawley, J.A. Molecular responses to strength and endurance training: Are they incompatible? Appl. Physiol. Nutr. Metab. 2009, 34, 355–361. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, N.J.; Parker, B.L.; Chaudhuri, R.; Fisher-Wellman, K.H.; Kleinert, M.; Humphrey, S.J.; Yang, P.; Holliday, M.; Trefely, S.; Fazakerley, D.J.; et al. Global Phosphoproteomic Analysis of Human Skeletal Muscle Reveals a Network of Exercise-Regulated Kinases and AMPK Substrates. Cell Metab. 2015, 22, 922–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowans, G.J.; Hawley, S.A.; Ross, F.A.; Hardie, D.G. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013, 18, 556–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardie, D.G. AMP—Ksensing energy while talking to other signaling pathways. Cell Metab. 2014, 20, 939–952. [Google Scholar] [CrossRef] [Green Version]
- Viollet, B. The Energy Sensor AMPK: Adaptations to Exercise, Nutritional and Hormonal Signals. In Hormones, Metabolism and the Benefits of Exercise; Spiegelman, B., Ed.; Springer Nature: Cham, Switzerland, 2017; pp. 13–24. [Google Scholar] [CrossRef] [Green Version]
- Chin, E.R. Intracellular Ca2+ signaling in skeletal muscle: Decoding a complex message. Exerc. Sport Sci. Rev. 2010, 38, 76–85. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Zhang, C.L.; Olson, E.N. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc. Natl. Acad. Sci. USA 2000, 97, 14400–14405. [Google Scholar] [CrossRef] [Green Version]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [Green Version]
- Rakyan, V.K.; Down, T.A.; Maslau, S.; Andrew, T.; Yang, T.P.; Beyan, H.; Whittaker, P.; McCann, O.T.; Finer, S.; Valdes, A.M.; et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010, 20, 434–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seaborne, R.A.; Sharples, A.P. The Interplay Between Exercise Metabolism, Epigenetics, and Skeletal Muscle Remodeling. Exerc. Sport Sci. Rev. 2020, 48, 188–200. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.L.; Hargreaves, M. Epigenetics and Exercise. Trends Endocrinol. Metab. 2019, 30, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Stephens, K.E.; Miaskowski, C.A.; Levine, J.D.; Pullinger, C.R.; Aouizerat, B.E. Epigenetic regulation and measurement of epigenetic changes. Biol. Res. Nurs. 2013, 15, 373–381. [Google Scholar] [CrossRef] [Green Version]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [Green Version]
- Handy, D.E.; Castro, R.; Loscalzo, J. Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 2011, 123, 2145–2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Tiffon, C. The Impact of Nutrition and Environmental Epigenetics on Human Health and Disease. Int. J. Mol. Sci. 2018, 19, 3425. [Google Scholar] [CrossRef] [Green Version]
- van Dongen, J.; Nivard, M.G.; Willemsen, G.; Hottenga, J.J.; Helmer, Q.; Dolan, C.V.; Ehli, E.A.; Davies, G.E.; van Iterson, M.; Breeze, C.E.; et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat. Commun. 2016, 7, 11115. [Google Scholar] [CrossRef] [Green Version]
- Widmann, M.; Niess, A.M.; Munz, B. Physical Exercise and Epigenetic Modifications in Skeletal Muscle. Sports Med. 2019, 49, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Jacques, M.; Hiam, D.; Craig, J.; Barres, R.; Eynon, N.; Voisin, S. Epigenetic changes in healthy human skeletal muscle following exercise—A systematic review. Epigenetics 2019, 14, 633–648. [Google Scholar] [CrossRef]
- Rasmussen, M.; Zierath, J.R.; Barres, R. Dynamic epigenetic responses to muscle contraction. Drug Discov. Today 2014, 19, 1010–1014. [Google Scholar] [CrossRef]
- Santos, J.M.; Tewari, S.; Benite-Ribeiro, S.A. The effect of exercise on epigenetic modifications of PGC1: The impact on type 2 diabetes. Med. Hypotheses 2014, 82, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Bonnet, M.; Lefaucheur, L.; Picard, B.; Bugeon, J. How Muscle Structure and Composition Influence Meat and Flesh Quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef] [Green Version]
- Atherton, P.J.; Smith, K. Muscle protein synthesis in response to nutrition and exercise. J. Physiol. 2012, 590, 1049–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Bottinelli, R.; Reggiani, C. Human skeletal muscle fibres: Molecular and functional diversity. Prog. Biophys. Mol. Biol. 2000, 73, 195–262. [Google Scholar] [CrossRef]
- Taylor, J.L.; Amann, M.; Duchateau, J.; Meeusen, R.; Rice, C.L. Neural Contributions to Muscle Fatigue: From the Brain to the Muscle and Back Again. Med. Sci. Sports Exerc. 2016, 48, 2294–2306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyner, M.J.; Casey, D.P. Regulation of increased blood flow (hyperemia) to muscles during exercise: A hierarchy of competing physiological needs. Physiol. Rev. 2015, 95, 549–601. [Google Scholar] [CrossRef] [Green Version]
- Radak, Z.; Zhao, Z.; Koltai, E.; Ohno, H.; Atalay, M. Oxygen consumption and usage during physical exercise: The balance between oxidative stress and ROS-dependent adaptive signaling. Antioxid. Redox Signal. 2013, 18, 1208–1246. [Google Scholar] [CrossRef] [Green Version]
- McGee, S.L.; Hargreaves, M. Exercise adaptations: Molecular mechanisms and potential targets for therapeutic benefit. Nat. Rev. Endocrinol. 2020, 16, 495–505. [Google Scholar] [CrossRef]
- Aon, M.A.; Bhatt, N.; Cortassa, S.C. Mitochondrial and cellular mechanisms for managing lipid excess. Front. Physiol. 2014, 5, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazgir, B.; Fathi, R.; Rezazadeh Valojerdi, M.; Mozdziak, P.; Asgari, A. Satellite Cells Contribution to Exercise Mediated Muscle Hypertrophy and Repair. Cell J. 2017, 18, 473–484. [Google Scholar] [CrossRef]
- Roberts, M.D.; Haun, C.T.; Vann, C.G.; Osburn, S.C.; Young, K.C. Sarcoplasmic Hypertrophy in Skeletal Muscle: A Scientific “Unicorn” or Resistance Training Adaptation? Front. Physiol. 2020, 11, 816. [Google Scholar] [CrossRef] [PubMed]
- Krzysztofik, M.; Wilk, M.; Wojdala, G.; Golas, A. Maximizing Muscle Hypertrophy: A Systematic Review of Advanced Resistance Training Techniques and Methods. Int. J. Environ. Res. Public Health 2019, 16, 4897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangine, G.T.; Hoffman, J.R.; Gonzalez, A.M.; Townsend, J.R.; Wells, A.J.; Jajtner, A.R.; Beyer, K.S.; Boone, C.H.; Miramonti, A.A.; Wang, R.; et al. The effect of training volume and intensity on improvements in muscular strength and size in resistance-trained men. Physiol. Rep. 2015, 3, e12472. [Google Scholar] [CrossRef] [Green Version]
- de Freitas, M.C.; Gerosa-Neto, J.; Zanchi, N.E.; Lira, F.S.; Rossi, F.E. Role of metabolic stress for enhancing muscle adaptations: Practical applications. World J. Methodol. 2017, 7, 46–54. [Google Scholar] [CrossRef]
- Phillips, B.E.; Williams, J.P.; Gustafsson, T.; Bouchard, C.; Rankinen, T.; Knudsen, S.; Smith, K.; Timmons, J.A.; Atherton, P.J. Molecular networks of human muscle adaptation to exercise and age. PLoS Genet. 2013, 9, e1003389. [Google Scholar] [CrossRef]
- McGlory, C.; Devries, M.C.; Phillips, S.M. Skeletal muscle and resistance exercise training; the role of protein synthesis in recovery and remodeling. J. Appl. Physiol. 2017, 122, 541–548. [Google Scholar] [CrossRef]
- Korthuis, R.J. Exercise hyperemia and regulation of tissue oxygenation during muscular activity. In Skeletal Muscle Circulation; Morgan & Claypool Life Sciences; Morgan & Claypool: San Rafael, CA, USA, 2011. [Google Scholar]
- Egan, B.; Zierath, J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013, 17, 162–184. [Google Scholar] [CrossRef] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef]
- Feng, J.; Chang, H.; Li, E.; Fan, G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci. Res. 2005, 79, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Goto, K.; Numata, M.; Komura, J.I.; Ono, T.; Bestor, T.H.; Kondo, H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation 1994, 56, 39–44. [Google Scholar] [CrossRef]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [Green Version]
- Illingworth, R.S.; Gruenewald-Schneider, U.; Webb, S.; Kerr, A.R.; James, K.D.; Turner, D.J.; Smith, C.; Harrison, D.J.; Andrews, R.; Bird, A.P. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010, 6, e1001134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hervouet, E.; Peixoto, P.; Delage-Mourroux, R.; Boyer-Guittaut, M.; Cartron, P.F. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin. Epigenetics 2018, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.; Molloy, P.L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988, 2, 1136–1143. [Google Scholar] [CrossRef] [Green Version]
- Barres, R.; Yan, J.; Egan, B.; Treebak, J.T.; Rasmussen, M.; Fritz, T.; Caidahl, K.; Krook, A.; O’Gorman, D.J.; Zierath, J.R. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012, 15, 405–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King-Himmelreich, T.S.; Schramm, S.; Wolters, M.C.; Schmetzer, J.; Moser, C.V.; Knothe, C.; Resch, E.; Peil, J.; Geisslinger, G.; Niederberger, E. The impact of endurance exercise on global and AMPK gene-specific DNA methylation. Biochem. Biophys. Res. Commun. 2016, 474, 284–290. [Google Scholar] [CrossRef]
- Baker, J.S.; McCormick, M.C.; Robergs, R.A. Interaction among Skeletal Muscle Metabolic Energy Systems during Intense Exercise. J. Nutr. Metab. 2010, 2010, 905612. [Google Scholar] [CrossRef] [Green Version]
- Kietzmann, T.; Petry, A.; Shvetsova, A.; Gerhold, J.M.; Gorlach, A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br. J. Pharmacol. 2017, 174, 1533–1554. [Google Scholar] [CrossRef] [PubMed]
- Petracci, I.; Gabbianelli, R.; Bordoni, L. The Role of Nutri(epi)genomics in Achieving the Body’s Full Potential in Physical Activity. Antioxidants 2020, 9, 498. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.P. Interleukin-6 in acute exercise and training: What is the biological relevance? Exerc. Immunol. Rev. 2006, 12, 6–33. [Google Scholar]
- Bagley, J.R.; Burghardt, K.J.; McManus, R.; Howlett, B.; Costa, P.B.; Coburn, J.W.; Arevalo, J.A.; Malek, M.H.; Galpin, A.J. Epigenetic Responses to Acute Resistance Exercise in Trained vs. Sedentary Men. J. Strength Cond. Res. 2020, 34, 1574–1580. [Google Scholar] [CrossRef]
- Voisin, S.; Eynon, N.; Yan, X.; Bishop, D.J. Exercise training and DNA methylation in humans. Acta Physiol. 2015, 213, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Maasar, M.F.; Turner, D.C.; Gorski, P.P.; Seaborne, R.A.; Strauss, J.A.; Shepherd, S.O.; Cocks, M.; Pillon, N.J.; Zierath, J.R.; Hulton, A.T.; et al. The Comparative Methylome and Transcriptome after Change of Direction Compared to Straight Line Running Exercise in Human Skeletal Muscle. Front. Physiol. 2021, 12, 619447. [Google Scholar] [CrossRef] [PubMed]
- Laker, R.C.; Lillard, T.S.; Okutsu, M.; Zhang, M.; Hoehn, K.L.; Connelly, J.J.; Yan, Z. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1alpha gene and age-dependent metabolic dysfunction in the offspring. Diabetes 2014, 63, 1605–1611. [Google Scholar] [CrossRef] [Green Version]
- Alibegovic, A.C.; Sonne, M.P.; Hojbjerre, L.; Bork-Jensen, J.; Jacobsen, S.; Nilsson, E.; Faerch, K.; Hiscock, N.; Mortensen, B.; Friedrichsen, M.; et al. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E752–E763. [Google Scholar] [CrossRef] [Green Version]
- Theilen, N.T.; Kunkel, G.H.; Tyagi, S.C. The Role of Exercise and TFAM in Preventing Skeletal Muscle Atrophy. J. Cell. Physiol. 2017, 232, 2348–2358. [Google Scholar] [CrossRef]
- Bajpeyi, S.; Covington, J.D.; Taylor, E.M.; Stewart, L.K.; Galgani, J.E.; Henagan, T.M. Skeletal Muscle PGC1alpha −1 Nucleosome Position and −260 nt DNA Methylation Determine Exercise Response and Prevent Ectopic Lipid Accumulation in Men. Endocrinology 2017, 158, 2190–2199. [Google Scholar] [CrossRef]
- Lane, S.C.; Camera, D.M.; Lassiter, D.G.; Areta, J.L.; Bird, S.R.; Yeo, W.K.; Jeacocke, N.A.; Krook, A.; Zierath, J.R.; Burke, L.M.; et al. Effects of sleeping with reduced carbohydrate availability on acute training responses. J. Appl. Physiol. 2015, 119, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Nitert, M.D.; Dayeh, T.; Volkov, P.; Elgzyri, T.; Hall, E.; Nilsson, E.; Yang, B.T.; Lang, S.; Parikh, H.; Wessman, Y.; et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 2012, 61, 3322–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.M.; Dasari, S.; Konopka, A.R.; Johnson, M.L.; Manjunatha, S.; Esponda, R.R.; Carter, R.E.; Lanza, I.R.; Nair, K.S. Enhanced Protein Translation Underlies Improved Metabolic and Physical Adaptations to Different Exercise Training Modes in Young and Old Humans. Cell Metab. 2017, 25, 581–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindholm, M.E.; Marabita, F.; Gomez-Cabrero, D.; Rundqvist, H.; Ekstrom, T.J.; Tegner, J.; Sundberg, C.J. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics 2014, 9, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020, 10, 15360. [Google Scholar] [CrossRef] [PubMed]
- Sailani, M.R.; Halling, J.F.; Moller, H.D.; Lee, H.; Plomgaard, P.; Pilegaard, H.; Snyder, M.P.; Regenberg, B. Lifelong physical activity is associated with promoter hypomethylation of genes involved in metabolism, myogenesis, contractile properties and oxidative stress resistance in aged human skeletal muscle. Sci. Rep. 2019, 9, 3272. [Google Scholar] [CrossRef] [Green Version]
- Rowlands, D.S.; Page, R.A.; Sukala, W.R.; Giri, M.; Ghimbovschi, S.D.; Hayat, I.; Cheema, B.S.; Lys, I.; Leikis, M.; Sheard, P.W.; et al. Multi-omic integrated networks connect DNA methylation and miRNA with skeletal muscle plasticity to chronic exercise in Type 2 diabetic obesity. Physiol. Genomics 2014, 46, 747–765. [Google Scholar] [CrossRef]
- Stephens, N.A.; Brouwers, B.; Eroshkin, A.M.; Yi, F.; Cornnell, H.H.; Meyer, C.; Goodpaster, B.H.; Pratley, R.E.; Smith, S.R.; Sparks, L.M. Exercise Response Variations in Skeletal Muscle PCr Recovery Rate and Insulin Sensitivity Relate to Muscle Epigenomic Profiles in Individuals With Type 2 Diabetes. Diabetes Care 2018, 41, 2245–2254. [Google Scholar] [CrossRef] [Green Version]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; Someren, K.A.V.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Methylome of human skeletal muscle after acute & chronic resistance exercise training, detraining & retraining. Sci. Data 2018, 5, 180213. [Google Scholar] [CrossRef] [Green Version]
- Solsona, R.; Sanchez, A.M.J. Exercise and ribosome biogenesis in skeletal muscle hypertrophy: Impact of genetic and epigenetic factors. J. Physiol. 2021, 599, 3803–3805. [Google Scholar] [CrossRef]
- Figueiredo, V.C.; Wen, Y.; Alkner, B.; Fernandez-Gonzalo, R.; Norrbom, J.; Vechetti, I.J., Jr.; Valentino, T.; Mobley, C.B.; Zentner, G.E.; Peterson, C.A.; et al. Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J. Physiol. 2021, 599, 3363–3384. [Google Scholar] [CrossRef]
- Sanchez, A.M.; Candau, R.; Bernardi, H. Recent Data on Cellular Component Turnover: Focus on Adaptations to Physical Exercise. Cells 2019, 8, 542. [Google Scholar] [CrossRef] [Green Version]
- Ruple, B.A.; Godwin, J.S.; Mesquita, P.H.C.; Osburn, S.C.; Vann, C.G.; Lamb, D.A.; Sexton, C.L.; Candow, D.G.; Forbes, S.C.; Fruge, A.D.; et al. Resistance training rejuvenates the mitochondrial methylome in aged human skeletal muscle. FASEB J. 2021, 35, e21864. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.E.; Scott, M.P. Communicating with Hedgehogs. Nat. Rev. Mol. Cell Biol. 2005, 6, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.; Litingtung, Y.; Lee, E.; Young, K.E.; Corden, J.L.; Westphal, H.; Beachy, P.A. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 1996, 383, 407–413. [Google Scholar] [CrossRef]
- Pepinsky, R.B.; Zeng, C.; Wen, D.; Rayhorn, P.; Baker, D.P.; Williams, K.P.; Bixler, S.A.; Ambrose, C.M.; Garber, E.A.; Miatkowski, K.; et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 1998, 273, 14037–14045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roessler, E.; Belloni, E.; Gaudenz, K.; Jay, P.; Berta, P.; Scherer, S.W.; Tsui, L.C.; Muenke, M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 1996, 14, 357–360. [Google Scholar] [CrossRef]
- Palma, V.; Ruiz i Altaba, A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 2004, 131, 337–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nusslein-Volhard, C.; Wieschaus, E. Mutations affecting segment number and polarity in Drosophila. Nature 1980, 287, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087. [Google Scholar] [CrossRef] [Green Version]
- Torroja, C.; Gorfinkiel, N.; Guerrero, I. Mechanisms of Hedgehog gradient formation and interpretation. J. Neurobiol. 2005, 64, 334–356. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.; Carrasquilla, G.D.; Ingerslev, L.R.; Hochreuter, M.Y.; Hansson, S.; Pillon, N.J.; Donkin, I.; Versteyhe, S.; Zierath, J.R.; Kilpelainen, T.O.; et al. Epigenetic rewiring of skeletal muscle enhancers after exercise training supports a role in whole-body function and human health. Mol. Metab. 2021, 53, 101290. [Google Scholar] [CrossRef] [PubMed]
- McGee, S.L.; Fairlie, E.; Garnham, A.P.; Hargreaves, M. Exercise-induced histone modifications in human skeletal muscle. J. Physiol. 2009, 587, 5951–5958. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Stepto, N.K.; Chibalin, A.V.; Fryer, L.G.; Carling, D.; Krook, A.; Hawley, J.A.; Zierath, J.R. Metabolic and mitogenic signal transduction in human skeletal muscle after intense cycling exercise. J. Physiol. 2003, 546, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, E.; Sznajder, J.I. Epigenetic regulation of muscle phenotype and adaptation: A potential role in COPD muscle dysfunction. J. Appl. Physiol. 2013, 114, 1263–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Clayton, A.L.; Hebbes, T.R.; Thorne, A.W.; Crane-Robinson, C. Histone acetylation and gene induction in human cells. FEBS Lett. 1993, 336, 23–26. [Google Scholar] [CrossRef] [Green Version]
- Pogo, B.G.; Allfrey, V.G.; Mirsky, A.E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc. Natl. Acad. Sci. USA 1966, 55, 805–812. [Google Scholar] [CrossRef] [Green Version]
- Gorisch, S.M.; Wachsmuth, M.; Toth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118, 5825–5834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015, 16, 178–189. [Google Scholar] [CrossRef]
- Lim, C.; Shimizu, J.; Kawano, F.; Kim, H.J.; Kim, C.K. Adaptive responses of histone modifications to resistance exercise in human skeletal muscle. PLoS ONE 2020, 15, e0231321. [Google Scholar] [CrossRef] [Green Version]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [Green Version]
- Murray, K. The Occurrence of Epsilon-N-Methyl Lysine in Histones. Biochemistry 1964, 3, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef]
- North, J.A.; Simon, M.; Ferdinand, M.B.; Shoffner, M.A.; Picking, J.W.; Howard, C.J.; Mooney, A.M.; van Noort, J.; Poirier, M.G.; Ottesen, J.J. Histone H3 phosphorylation near the nucleosome dyad alters chromatin structure. Nucleic Acids Res. 2014, 42, 4922–4933. [Google Scholar] [CrossRef] [Green Version]
- Schaffer, B.E.; Levin, R.S.; Hertz, N.T.; Maures, T.J.; Schoof, M.L.; Hollstein, P.E.; Benayoun, B.A.; Banko, M.R.; Shaw, R.J.; Shokat, K.M.; et al. Identification of AMPK Phosphorylation Sites Reveals a Network of Proteins Involved in Cell Invasion and Facilitates Large-Scale Substrate Prediction. Cell Metab. 2015, 22, 907–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awad, S.; Kunhi, M.; Little, G.H.; Bai, Y.; An, W.; Bers, D.; Kedes, L.; Poizat, C. Nuclear CaMKII enhances histone H3 phosphorylation and remodels chromatin during cardiac hypertrophy. Nucleic Acids Res. 2013, 41, 7656–7672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, W.S.; Trievel, R.C.; Rojas, J.R.; Duggan, L.; Hsu, J.Y.; Allis, C.D.; Marmorstein, R.; Berger, S.L. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 2000, 5, 917–926. [Google Scholar] [CrossRef]
- Latham, T.; Mackay, L.; Sproul, D.; Karim, M.; Culley, J.; Harrison, D.J.; Hayward, L.; Langridge-Smith, P.; Gilbert, N.; Ramsahoye, B.H. Lactate, a product of glycolytic metabolism, inhibits histone deacetylase activity and promotes changes in gene expression. Nucleic Acids Res. 2012, 40, 4794–4803. [Google Scholar] [CrossRef] [Green Version]
- Liberti, M.V.; Locasale, J.W. Histone Lactylation: A New Role for Glucose Metabolism. Trends Biochem. Sci. 2020, 45, 179–182. [Google Scholar] [CrossRef]
- Izzo, L.T.; Wellen, K.E. Histone lactylation links metabolism and gene regulation. Nature 2019, 574, 492–493. [Google Scholar] [CrossRef] [PubMed]
- Soci, U.P.R.; Melo, S.F.S.; Gomes, J.L.P.; Silveira, A.C.; Nobrega, C.; de Oliveira, E.M. Exercise Training and Epigenetic Regulation: Multilevel Modification and Regulation of Gene Expression. Adv. Exp. Med. Biol. 2017, 1000, 281–322. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.P.; Lamon, S.; Boon, H.; Wada, S.; Guller, I.; Brown, E.L.; Chibalin, A.V.; Zierath, J.R.; Snow, R.J.; Stepto, N.; et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short-term endurance training. J. Physiol. 2013, 591, 4637–4653. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mooren, F.C.; Viereck, J.; Kruger, K.; Thum, T. Circulating microRNAs as potential biomarkers of aerobic exercise capacity. Am J. Physiol. Heart Circ. Physiol. 2014, 306, H557–H563. [Google Scholar] [CrossRef]
- Keller, P.; Vollaard, N.B.; Gustafsson, T.; Gallagher, I.J.; Sundberg, C.J.; Rankinen, T.; Britton, S.L.; Bouchard, C.; Koch, L.G.; Timmons, J.A. A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J. Appl. Physiol. 2011, 110, 46–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, S.; Scheele, C.; Yfanti, C.; Akerstrom, T.; Nielsen, A.R.; Pedersen, B.K.; Laye, M.J. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle. J. Physiol. 2010, 588, 4029–4037. [Google Scholar] [CrossRef] [PubMed]
- Fyfe, J.J.; Bishop, D.J.; Zacharewicz, E.; Russell, A.P.; Stepto, N.K. Concurrent exercise incorporating high-intensity interval or continuous training modulates mTORC1 signaling and microRNA expression in human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 310, R1297–R1311. [Google Scholar] [CrossRef]
- Margolis, L.M.; McClung, H.L.; Murphy, N.E.; Carrigan, C.T.; Pasiakos, S.M. Skeletal Muscle myomiR Are Differentially Expressed by Endurance Exercise Mode and Combined Essential Amino Acid and Carbohydrate Supplementation. Front. Physiol. 2017, 8, 182. [Google Scholar] [CrossRef] [Green Version]
- Davidsen, P.K.; Gallagher, I.J.; Hartman, J.W.; Tarnopolsky, M.A.; Dela, F.; Helge, J.W.; Timmons, J.A.; Phillips, S.M. High responders to resistance exercise training demonstrate differential regulation of skeletal muscle microRNA expression. J. Appl. Physiol. 2011, 110, 309–317. [Google Scholar] [CrossRef] [Green Version]
- Rivas, D.A.; Lessard, S.J.; Rice, N.P.; Lustgarten, M.S.; So, K.; Goodyear, L.J.; Parnell, L.D.; Fielding, R.A. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF-1 signaling. FASEB J. 2014, 28, 4133–4147. [Google Scholar] [CrossRef] [Green Version]
- Ogasawara, R.; Akimoto, T.; Umeno, T.; Sawada, S.; Hamaoka, T.; Fujita, S. MicroRNA expression profiling in skeletal muscle reveals different regulatory patterns in high and low responders to resistance training. Physiol. Genomics 2016, 48, 320–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, M.; Breil, F.A.; Lurman, G.; Klossner, S.; Fluck, M.; Billeter, R.; Dapp, C.; Hoppeler, H. Different molecular and structural adaptations with eccentric and conventional strength training in elderly men and women. Gerontology 2011, 57, 528–538. [Google Scholar] [CrossRef] [Green Version]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, L.T.; Xie, S.J.; Lei, H.; Qiu, X.S.; Huang, M.C.; Tao, S.; Hou, Y.R.; Hu, Y.X.; Sun, Y.J.; Zhang, Q.; et al. METTL3 regulates skeletal muscle specific miRNAs at both transcriptional and post-transcriptional levels. Biochem. Biophys. Res. Commun. 2021, 552, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.J.; Lei, H.; Yang, B.; Diao, L.T.; Liao, J.Y.; He, J.H.; Tao, S.; Hu, Y.X.; Hou, Y.R.; Sun, Y.J.; et al. Dynamic m(6)A mRNA Methylation Reveals the Role of METTL3/14-m(6)A-MNK2-ERK Signaling Axis in Skeletal Muscle Differentiation and Regeneration. Front. Cell Dev. Biol. 2021, 9, 744171. [Google Scholar] [CrossRef]
- Li, J.; Pei, Y.; Zhou, R.; Tang, Z.; Yang, Y. Regulation of RNA N(6)-methyladenosine modification and its emerging roles in skeletal muscle development. Int. J. Biol. Sci. 2021, 17, 1682–1692. [Google Scholar] [CrossRef] [PubMed]
- Campion, J.; Milagro, F.; Martinez, J.A. Epigenetics and obesity. Prog. Mol. Biol. Transl. Sci. 2010, 94, 291–347. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-W.; Friso, S. Epigenetics: A new bridge between nutrition and health. Adv. Nutr. 2010, 1, 8–16. [Google Scholar] [CrossRef]
- McGee, S.L.; Walder, K.R. Exercise and the Skeletal Muscle Epigenome. Cold Spring Harb. Perspect. Med. 2017, 7, a029876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoene, M.; Weigert, C. The stress response of the liver to physical exercise. Exerc. Immunol. Rev. 2010, 16, 163–183. [Google Scholar]
- Chandramohan, Y.; Droste, S.K.; Arthur, J.S.C.; Reul, J.M. The forced swimming-induced behavioural immobility response involves histone H3 phospho-acetylation and c-Fos induction in dentate gyrus granule neurons via activation of the N-methyl-d-aspartate/extracellular signal-regulated kinase/mitogen-and stress-activated kinase signalling pathway. Eur. J. Neurosci. 2008, 27, 2701–2713. [Google Scholar] [PubMed]
- Gomez-Pinilla, F.; Zhuang, Y.; Feng, J.; Ying, Z.; Fan, G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur. J. Neurosci. 2011, 33, 383–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, J.-S.; Oh, A.-R.; Lee, H.-J.; Ahn, Y.-H.; Cha, J.-Y. Hepatic Elovl6 gene expression is regulated by the synergistic action of ChREBP and SREBP-1c. Biochem. Biophys. Res. Commun. 2016, 478, 1060–1066. [Google Scholar] [CrossRef]
- Morigny, P.; Houssier, M.; Mairal, A.; Ghilain, C.; Mouisel, E.; Benhamed, F.; Masri, B.; Recazens, E.; Denechaud, P.D.; Tavernier, G.; et al. Interaction between hormone-sensitive lipase and ChREBP in fat cells controls insulin sensitivity. Nat. Metab. 2019, 1, 133–146. [Google Scholar] [CrossRef] [Green Version]
- Chadt, A.; Al-Hasani, H. Glucose transporters in adipose tissue, liver, and skeletal muscle in metabolic health and disease. Pflug. Arch. 2020, 472, 1273–1298. [Google Scholar] [CrossRef]
- Wheatley, K.E.; Nogueira, L.M.; Perkins, S.N.; Hursting, S.D. Differential effects of calorie restriction and exercise on the adipose transcriptome in diet-induced obese mice. J. Obes. 2011, 2011, 265417. [Google Scholar] [CrossRef] [Green Version]
- Ronn, T.; Volkov, P.; Davegardh, C.; Dayeh, T.; Hall, E.; Olsson, A.H.; Nilsson, E.; Tornberg, A.; Dekker Nitert, M.; Eriksson, K.F.; et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013, 9, e1003572. [Google Scholar] [CrossRef]
- Fabre, O.; Ingerslev, L.R.; Garde, C.; Donkin, I.; Simar, D.; Barres, R. Exercise training alters the genomic response to acute exercise in human adipose tissue. Epigenomics 2018, 10, 1033–1050. [Google Scholar] [CrossRef] [Green Version]
- Stevanovic, J.; Beleza, J.; Coxito, P.; Ascensao, A.; Magalhaes, J. Physical exercise and liver “fitness”: Role of mitochondrial function and epigenetics-related mechanisms in non-alcoholic fatty liver disease. Mol. Metab. 2020, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Hlady, R.A.; Schafer, M.J.; White, T.A.; Liu, C.; Choi, J.H.; Miller, J.D.; Roberts, L.R.; LeBrasseur, N.K.; Robertson, K.D. High fat diet and exercise lead to a disrupted and pathogenic DNA methylome in mouse liver. Epigenetics 2017, 12, 55–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Bei, Y.; Liu, J.; Dimitrova-Shumkovska, J.; Kuang, D.; Zhou, Q.; Li, J.; Yang, Y.; Xiang, Y.; Wang, F. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J. Cell. Mol. Med. 2016, 20, 204–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.-L.; Jing, W.; Feng, L.-S.; Zhang, L.; Xu, J.-F.; You, T.-J.; Zhao, J. Effects of hypoxic exercise training on microRNA expression and lipid metabolism in obese rat livers. J. Zhejiang Univ.-Sci. B 2014, 15, 820–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McPherson, N.O.; Lane, M.; Sandeman, L.; Owens, J.A.; Fullston, T. An exercise-only intervention in obese fathers restores glucose and insulin regulation in conjunction with the rescue of pancreatic islet cell morphology and microRNA expression in male offspring. Nutrients 2017, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Delezie, J.; Handschin, C. Endocrine Crosstalk Between Skeletal Muscle and the Brain. Front. Neurol. 2018, 9, 698. [Google Scholar] [CrossRef]
- Collins, A.; Hill, L.E.; Chandramohan, Y.; Whitcomb, D.; Droste, S.K.; Reul, J.M. Exercise improves cognitive responses to psychological stress through enhancement of epigenetic mechanisms and gene expression in the dentate gyrus. PLoS ONE 2009, 4, e4330. [Google Scholar] [CrossRef] [Green Version]
- Cosin-Tomas, M.; Alvarez-Lopez, M.J.; Sanchez-Roige, S.; Lalanza, J.F.; Bayod, S.; Sanfeliu, C.; Pallas, M.; Escorihuela, R.M.; Kaliman, P. Epigenetic alterations in hippocampus of SAMP8 senescent mice and modulation by voluntary physical exercise. Front. Aging Neurosci. 2014, 6, 51. [Google Scholar] [CrossRef]
- Li, X.; Inoue, T.; Hayashi, M.; Maejima, H. Exercise enhances the expression of brain-derived neurotrophic factor in the hippocampus accompanied by epigenetic alterations in senescence-accelerated mice prone 8. Neurosci. Lett. 2019, 706, 176–181. [Google Scholar] [CrossRef]
- Maejima, H.; Kanemura, N.; Kokubun, T.; Murata, K.; Takayanagi, K. Exercise enhances cognitive function and neurotrophin expression in the hippocampus accompanied by changes in epigenetic programming in senescence-accelerated mice. Neurosci. Lett. 2018, 665, 67–73. [Google Scholar] [CrossRef]
- Maejima, H.; Ninuma, S.; Okuda, A.; Inoue, T.; Hayashi, M. Exercise and low-level GABAA receptor inhibition modulate locomotor activity and the expression of BDNF accompanied by changes in epigenetic regulation in the hippocampus. Neurosci. Lett. 2018, 685, 18–23. [Google Scholar] [CrossRef]
- Bao, T.H.; Miao, W.; Han, J.H.; Yin, M.; Yan, Y.; Wang, W.W.; Zhu, Y.H. Spontaneous running wheel improves cognitive functions of mouse associated with miRNA expressional alteration in hippocampus following traumatic brain injury. J. Mol. Neurosci. 2014, 54, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Torma, F.; Bori, Z.; Koltai, E.; Felszeghy, K.; Vacz, G.; Koch, L.; Britton, S.; Boldogh, I.; Radak, Z. Eating habits modulate short term memory and epigenetical regulation of brain derived neurotrophic factor in hippocampus of low- and high running capacity rats. Brain Res. Bull. 2014, 107, 54–60. [Google Scholar] [CrossRef] [Green Version]
- Cechinel, L.R.; Basso, C.G.; Bertoldi, K.; Schallenberger, B.; de Meireles, L.C.F.; Siqueira, I.R. Treadmill exercise induces age and protocol-dependent epigenetic changes in prefrontal cortex of Wistar rats. Behav. Brain Res. 2016, 313, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Abel, J.L.; Rissman, E.F. Running-induced epigenetic and gene expression changes in the adolescent brain. Int. J. Dev. Neurosci. 2013, 31, 382–390. [Google Scholar] [CrossRef] [Green Version]
- de Meireles, L.C.; Bertoldi, K.; Cechinel, L.R.; Schallenberger, B.L.; da Silva, V.K.; Schroder, N.; Siqueira, I.R. Treadmill exercise induces selective changes in hippocampal histone acetylation during the aging process in rats. Neurosci. Lett. 2016, 634, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Elsner, V.R.; Lovatel, G.A.; Moyses, F.; Bertoldi, K.; Spindler, C.; Cechinel, L.R.; Muotri, A.R.; Siqueira, I.R. Exercise induces age-dependent changes on epigenetic parameters in rat hippocampus: A preliminary study. Exp. Gerontol. 2013, 48, 136–139. [Google Scholar] [CrossRef] [Green Version]
- Lovatel, G.A.; Elsner, V.R.; Bertoldi, K.; Vanzella, C.; Moyses Fdos, S.; Vizuete, A.; Spindler, C.; Cechinel, L.R.; Netto, C.A.; Muotri, A.R.; et al. Treadmill exercise induces age-related changes in aversive memory, neuroinflammatory and epigenetic processes in the rat hippocampus. Neurobiol. Learn. Mem. 2013, 101, 94–102. [Google Scholar] [CrossRef]
- Spindler, C.; Cechinel, L.R.; Basso, C.; Moyses, F.; Bertoldi, K.; Roesler, R.; Lovatel, G.A.; Rostirola Elsner, V.; Siqueira, I.R. Treadmill exercise alters histone acetyltransferases and histone deacetylases activities in frontal cortices from wistar rats. Cell. Mol. Neurobiol. 2014, 34, 1097–1101. [Google Scholar] [CrossRef]
- Kashimoto, R.K.; Toffoli, L.V.; Manfredo, M.H.F.; Volpini, V.L.; Martins-Pinge, M.C.; Pelosi, G.G.; Gomes, M.V. Physical exercise affects the epigenetic programming of rat brain and modulates the adaptive response evoked by repeated restraint stress. Behav. Brain Res. 2016, 296, 286–289. [Google Scholar] [CrossRef]
- Jessop, P.; Toledo-Rodriguez, M. Hippocampal TET1 and TET2 Expression and DNA Hydroxymethylation Are Affected by Physical Exercise in Aged Mice. Front. Cell Dev. Biol. 2018, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, J.; Vieira, A.S.; Kramer-Soares, J.C.; Da Silva, E.A.; Lee, K.S.; Lopes-Cendes, I.; Arida, R.M. Hippocampal microRNA-mRNA regulatory network is affected by physical exercise. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1711–1720. [Google Scholar] [CrossRef]
- de Meireles, L.C.F.; Galvao, F., Jr.; Walker, D.M.; Cechinel, L.R.; de Souza Grefenhagen, A.I.; Andrade, G.; Palazzo, R.P.; Lovatel, G.A.; Basso, C.G.; Nestler, E.J.; et al. Exercise Modalities Improve Aversive Memory and Survival Rate in Aged Rats: Role of Hippocampal Epigenetic Modifications. Mol. Neurobiol. 2019, 56, 8408–8419. [Google Scholar] [CrossRef] [PubMed]
- MacKay, H.; Scott, C.A.; Duryea, J.D.; Baker, M.S.; Laritsky, E.; Elson, A.E.; Garland, T., Jr.; Fiorotto, M.L.; Chen, R.; Li, Y.; et al. DNA methylation in AgRP neurons regulates voluntary exercise behavior in mice. Nat. Commun. 2019, 10, 5364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maejima, H.; Kitahara, M.; Takamatsu, Y.; Mani, H.; Inoue, T. Effects of exercise and pharmacological inhibition of histone deacetylases (HDACs) on epigenetic regulations and gene expressions crucial for neuronal plasticity in the motor cortex. Brain Res. 2021, 1751, 147191. [Google Scholar] [CrossRef] [PubMed]
- Tomiga, Y.; Sakai, K.; Ra, S.G.; Kusano, M.; Ito, A.; Uehara, Y.; Takahashi, H.; Kawanaka, K.; Soejima, H.; Higaki, Y. Short-term running exercise alters DNA methylation patterns in neuronal nitric oxide synthase and brain-derived neurotrophic factor genes in the mouse hippocampus and reduces anxiety-like behaviors. FASEB J. 2021, 35, e21767. [Google Scholar] [CrossRef]
- Goldberg, M.; Islam, M.R.; Kerimoglu, C.; Lancelin, C.; Gisa, V.; Burkhardt, S.; Kruger, D.M.; Marquardt, T.; Malchow, B.; Schmitt, A.; et al. Exercise as a model to identify microRNAs linked to human cognition: A role for microRNA-409 and microRNA-501. Transl. Psychiatry 2021, 11, 514. [Google Scholar] [CrossRef]
- Rodrigues, G.M., Jr.; Toffoli, L.V.; Manfredo, M.H.; Francis-Oliveira, J.; Silva, A.S.; Raquel, H.A.; Martins-Pinge, M.C.; Moreira, E.G.; Fernandes, K.B.; Pelosi, G.G.; et al. Acute stress affects the global DNA methylation profile in rat brain: Modulation by physical exercise. Behav. Brain Res. 2015, 279, 123–128. [Google Scholar] [CrossRef]
- Gibb, A.A.; Epstein, P.N.; Uchida, S.; Zheng, Y.; McNally, L.A.; Obal, D.; Katragadda, K.; Trainor, P.; Conklin, D.J.; Brittian, K.R.; et al. Exercise-Induced Changes in Glucose Metabolism Promote Physiological Cardiac Growth. Circulation 2017, 136, 2144–2157. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Karnib, N.; El-Ghandour, R.; El Hayek, L.; Nasrallah, P.; Khalifeh, M.; Barmo, N.; Jabre, V.; Ibrahim, P.; Bilen, M.; Stephan, J.S.; et al. Lactate is an antidepressant that mediates resilience to stress by modulating the hippocampal levels and activity of histone deacetylases. Neuropsychopharmacology 2019, 44, 1152–1162. [Google Scholar] [CrossRef] [PubMed]
- Longo, N.; Frigeni, M.; Pasquali, M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta 2016, 1863, 2422–2435. [Google Scholar] [CrossRef]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, J.C.; Verdin, E. Ketone bodies as signaling metabolites. Trends Endocrinol. Metab. 2014, 25, 42–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sleiman, S.F.; Henry, J.; Al-Haddad, R.; El Hayek, L.; Abou Haidar, E.; Stringer, T.; Ulja, D.; Karuppagounder, S.S.; Holson, E.B.; Ratan, R.R.; et al. Exercise promotes the expression of brain derived neurotrophic factor (BDNF) through the action of the ketone body beta-hydroxybutyrate. eLife 2016, 5, e15092. [Google Scholar] [CrossRef] [PubMed]
- Marosi, K.; Kim, S.W.; Moehl, K.; Scheibye-Knudsen, M.; Cheng, A.; Cutler, R.; Camandola, S.; Mattson, M.P. 3-Hydroxybutyrate regulates energy metabolism and induces BDNF expression in cerebral cortical neurons. J. Neurochem. 2016, 139, 769–781. [Google Scholar] [CrossRef]
- Chriett, S.; Dabek, A.; Wojtala, M.; Vidal, H.; Balcerczyk, A.; Pirola, L. Prominent action of butyrate over beta-hydroxybutyrate as histone deacetylase inhibitor, transcriptional modulator and anti-inflammatory molecule. Sci. Rep. 2019, 9, 742. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Morvan, F.; Morozzi, G.; Jourde, B.; Minetti, G.C.; Kahle, P.; Rivet, H.; Brebbia, P.; Toussaint, G.; Glass, D.J.; et al. ATP Citrate Lyase Regulates Myofiber Differentiation and Increases Regeneration by Altering Histone Acetylation. Cell Rep. 2017, 21, 3003–3011. [Google Scholar] [CrossRef] [Green Version]
- Swulius, M.T.; Waxham, M.N. Ca(2+)/calmodulin-dependent protein kinases. Cell. Mol. Life Sci. 2008, 65, 2637–2657. [Google Scholar] [CrossRef] [Green Version]
- Severinsen, M.C.K.; Pedersen, B.K. Muscle-Organ Crosstalk: The Emerging Roles of Myokines. Endocr. Rev. 2020, 41, 594–609. [Google Scholar] [CrossRef]
- Vechetti, I.J., Jr.; Valentino, T.; Mobley, C.B.; McCarthy, J.J. The role of extracellular vesicles in skeletal muscle and systematic adaptation to exercise. J. Physiol. 2021, 599, 845–861. [Google Scholar] [CrossRef] [PubMed]

| Resistance and Endurance Exercise | |||||||
|---|---|---|---|---|---|---|---|
| Reference | Sample Size | Age and Sex | Participant Profiles | Exercise Doses | Biopsy Times | Technology | Epigenetic Changes and Gene Expression |
| McGee et al., (2009) [95] | n = 9 | 23 ± 1 years, men | Healthy adults | Volume: 60 min, 72 ± 2% VO2 max | Before and after exercise | Electrotransference | Higher global acetylation of H3K36 |
| Yu et al., (2003) [96] | n = 9 | 27 ± 2 years, men | Trained and non-trained individuals | Intensity: 85% VO2 max, Rest: 60 s | Before and after exercise | Electrotransference | Higher phosphorylation of H3Histones |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plaza-Diaz, J.; Izquierdo, D.; Torres-Martos, Á.; Baig, A.T.; Aguilera, C.M.; Ruiz-Ojeda, F.J. Impact of Physical Activity and Exercise on the Epigenome in Skeletal Muscle and Effects on Systemic Metabolism. Biomedicines 2022, 10, 126. https://doi.org/10.3390/biomedicines10010126
Plaza-Diaz J, Izquierdo D, Torres-Martos Á, Baig AT, Aguilera CM, Ruiz-Ojeda FJ. Impact of Physical Activity and Exercise on the Epigenome in Skeletal Muscle and Effects on Systemic Metabolism. Biomedicines. 2022; 10(1):126. https://doi.org/10.3390/biomedicines10010126
Chicago/Turabian StylePlaza-Diaz, Julio, David Izquierdo, Álvaro Torres-Martos, Aiman Tariq Baig, Concepción M. Aguilera, and Francisco Javier Ruiz-Ojeda. 2022. "Impact of Physical Activity and Exercise on the Epigenome in Skeletal Muscle and Effects on Systemic Metabolism" Biomedicines 10, no. 1: 126. https://doi.org/10.3390/biomedicines10010126
APA StylePlaza-Diaz, J., Izquierdo, D., Torres-Martos, Á., Baig, A. T., Aguilera, C. M., & Ruiz-Ojeda, F. J. (2022). Impact of Physical Activity and Exercise on the Epigenome in Skeletal Muscle and Effects on Systemic Metabolism. Biomedicines, 10(1), 126. https://doi.org/10.3390/biomedicines10010126








