Enoxaparin and Pentosan Polysulfate Bind to the SARS-CoV-2 Spike Protein and Human ACE2 Receptor, Inhibiting Vero Cell Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Recombinant Protein Production
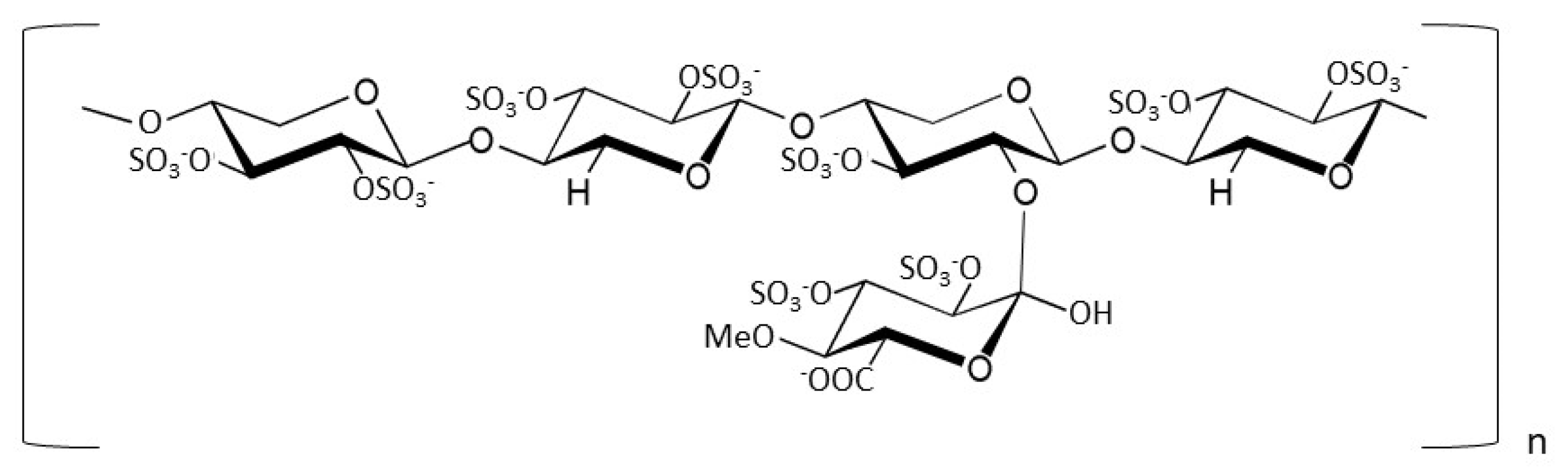
2.2. Preparative Size-Exclusion Chromatography of Enoxaparin Sodium
2.3. Isothermal Fluorescence Titration
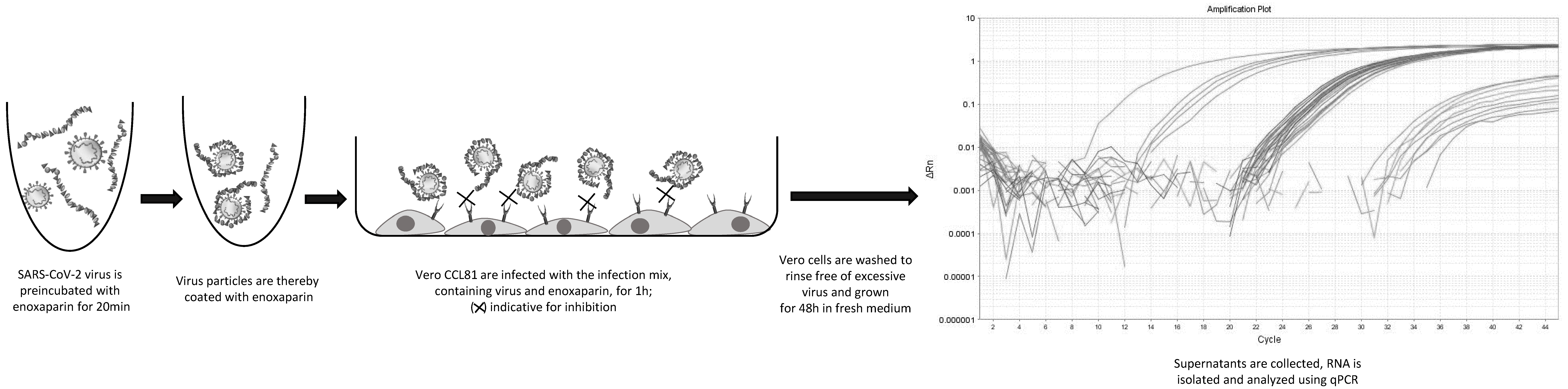
2.4. Inhibition Experiments and RNA Analysis
3. Results
3.1. Spike FL and Spike RBD Bind GAGs and PPS with Different Affinities and Discriminate Regarding Chain-Length and Modification
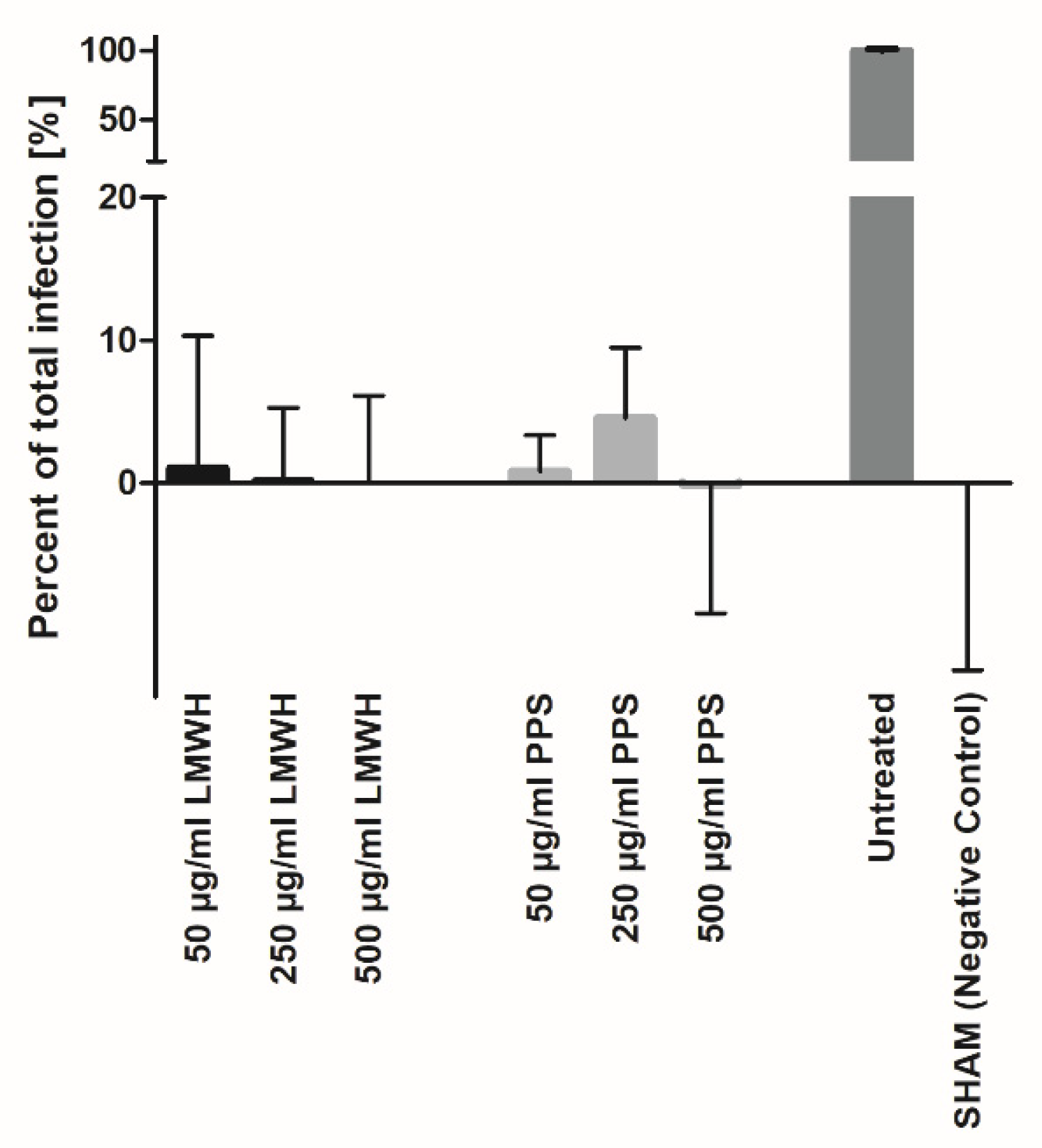
3.2. Heparin and PPS Inhibit SARS-CoV-2 Infection In Vitro
3.3. Heparin and PPS Inhibit SARS-CoV-2 Infection in a Biomatrix-like Environment
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Q.; Chen, C.Z.; Swaroop, M.; Xu, M.; Wang, L.; Lee, J.; Wang, A.Q.; Pradhan, M.; Hagen, N.; Chen, L.; et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020, 6, 270. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Painter, C.D.; Thacker, B.E.; Glass, C.A.; Narayanan, A.; Majowicz, S.A.; Zhang, Y.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057. [Google Scholar] [CrossRef]
- Da Rosa Mesquita, R.; Francelino Silva Junior, L.C.; Santos Santana, F.M.; Farias de Oliveira, T.; Campos Alcântara, R.; Monteiro Arnozo, G.; Da Rodrigues Silva Filho, E.; Galdino Dos Santos, A.G.; Da Oliveira Cunha, E.J.; Salgueiro de Aquino, S.H.; et al. Clinical manifestations of COVID-19 in the general population: Systematic review. Wien. Klin. Wochenschr. 2021, 133, 377–382. [Google Scholar] [CrossRef]
- Pum, A.; Ennemoser, M.; Adage, T.; Kungl, A.J. Cytokines and Chemokines in SARS-CoV-2 Infections-Therapeutic Strategies Targeting Cytokine Storm. Biomolecules 2021, 11, 91. [Google Scholar] [CrossRef]
- Shen, W.-X.; Luo, R.-C.; Wang, J.-Q.; Chen, Z.-S. Features of Cytokine Storm Identified by Distinguishing Clinical Manifestations in COVID-19. Front. Public Health 2021, 9, 671788. [Google Scholar] [CrossRef]
- Al Adem, K.; Shanti, A.; Stefanini, C.; Lee, S. Inhibition of SARS-CoV-2 Entry into Host Cells Using Small Molecules. Pharmaceuticals 2020, 13, 447. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Tortorici, M.A.; Walls, A.C.; Lang, Y.; Wang, C.; Li, Z.; Koerhuis, D.; Boons, G.-J.; Bosch, B.-J.; Rey, F.A.; de Groot, R.J.; et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019, 26, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Rabenstein, D.L. Heparin and heparan sulfate: Structure and function. Nat. Prod. Rep. 2002, 19, 312–331. [Google Scholar] [CrossRef]
- Soares da Costa, D.; Reis, R.L.; Pashkuleva, I. Sulfation of Glycosaminoglycans and Its Implications in Human Health and Disorders. Annu. Rev. Biomed. Eng. 2017, 19, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Jin, W.; Sood, A.; Montgomery, D.W.; Grant, O.C.; Fuster, M.M.; Fu, L.; Dordick, J.S.; Woods, R.J.; Zhang, F.; et al. Glycosaminoglycan Binding Motif at S1/S2 Proteolytic Cleavage Site on Spike Glycoprotein May Facilitate Novel Coronavirus (SARS-CoV-2) Host Cell Entry; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2020. [Google Scholar]
- Mycroft-West, C.J.; Su, D.; Pagani, I.; Rudd, T.R.; Elli, S.; Gandhi, N.S.; Guimond, S.E.; Miller, G.J.; Meneghetti, M.C.Z.; Nader, H.B.; et al. Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin. Thromb. Haemost. 2020, 120, 1700–1715. [Google Scholar] [CrossRef]
- Varki, A. Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2009; ISBN 9780879697709. [Google Scholar]
- Gozzo, L.; Viale, P.; Longo, L.; Vitale, D.C.; Drago, F. The Potential Role of Heparin in Patients With COVID-19: Beyond the Anticoagulant Effect. A Review. Front. Pharmacol. 2020, 11, 1307. [Google Scholar] [CrossRef]
- Tree, J.A.; Turnbull, J.E.; Buttigieg, K.R.; Elmore, M.J.; Coombes, N.; Hogwood, J.; Mycroft-West, C.J.; Lima, M.A.; Skidmore, M.A.; Karlsson, R.; et al. Unfractionated heparin inhibits live wild-type SARS-CoV-2 cell infectivity at therapeutically relevant concentrations. Br. J. Pharmacol. 2021, 178, 626–635. [Google Scholar] [CrossRef]
- Mulloy, B.; Hogwood, J.; Gray, E.; Lever, R.; Page, C.P. Pharmacology of Heparin and Related Drugs. Pharmacol. Rev. 2016, 68, 76–141. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Arnold, K.; Pawlinski, R.; Key, N.S. Using heparin molecules to manage COVID-2019. Res. Pract. Thromb. Haemost. 2020, 4, 518–523. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Mu, S.; Li, X.; Liang, Y.; Wang, L.; Ma, X. Unfractionated Heparin Alleviates Sepsis-Induced Acute Lung Injury by Protecting Tight Junctions. J. Surg. Res. 2019, 238, 175–185. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Pang, H.; Li, S.J. Heparin interacts with the main protease of SARS-CoV-2 and inhibits its activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 267, 120595. [Google Scholar] [CrossRef]
- Pereyra, D.; Heber, S.; Schrottmaier, W.C.; Santol, J.; Pirabe, A.; Schmuckenschlager, A.; Kammerer, K.; Ammon, D.; Sorz, T.; Fritsch, F.; et al. Low molecular weight heparin use in COVID-19 is associated with curtailed viral persistence—A retrospective multicenter observational study. Cardiovasc. Res. 2021, 117, 2807–2820. [Google Scholar] [CrossRef]
- Anderson, V.R.; Perry, C.M. Pentosan polysulfate: A review of its use in the relief of bladder pain or discomfort in interstitial cystitis. Drugs 2006, 66, 821–835. [Google Scholar] [CrossRef]
- Fellström, B.; Björklund, U.; Danielson, B.G.; Eriksson, H.; Odlind, B.; Tengblad, A. Pentosan polysulphate (Elmiron): Pharmacokinetics and effects on the urinary inhibition of crystal growth. In Pathogenese und Klinik der Harnsteine XII; Bericht über Das Symposium in Bonn vom 20–22 March 1986; Vahlensieck, W., Gasser, G., Eds.; Steinkopff: Heidelberg, Germany, 1987; pp. 340–344. ISBN 978-3-642-72399-5. [Google Scholar]
- Tandon, R.; Sharp, J.S.; Zhang, F.; Pomin, V.H.; Ashpole, N.M.; Mitra, D.; Jin, W.; Liu, H.; Sharma, P.; Linhardt, R.J. Effective Inhibition of SARS-CoV-2 Entry by Heparin and Enoxaparin Derivatives. J. Virol. 2021, 95, e01987-20. [Google Scholar] [CrossRef]
- Hao, W.; Ma, B.; Li, Z.; Wang, X.; Gao, X.; Li, Y.; Qin, B.; Shang, S.; Cui, S.; Tan, Z. Binding of the SARS-CoV-2 Spike Protein to Glycans. Sci. Bull. 2020, 66, 1205–1214. [Google Scholar] [CrossRef]
- Liu, L.; Chopra, P.; Li, X.; Wolfert, M.A.; Tompkins, S.M.; Boons, G.-J. SARS-CoV-2 spike protein binds heparan sulfate in a length- and sequence-dependent manner. ACS Cent Sci. 2021, 7, 1009–1018. [Google Scholar] [CrossRef]
- Kitic, N.; Gschwandtner, M.; Derler, R.; Gerlza, T.; Kungl, A.J. Preparation and Characterization of Glycosaminoglycan Chemokine Coreceptors. Methods Enzymol. 2016, 570, 517–538. [Google Scholar] [CrossRef]
- Gunay, N.S.; Linhardt, R.J. Capillary electrophoretic separation of heparin oligosaccharides under conditions amenable to mass spectrometric detection. J. Chromatogr. A 2003, 1014, 225–233. [Google Scholar] [CrossRef]
- Gerlza, T.; Hecher, B.; Jeremic, D.; Fuchs, T.; Gschwandtner, M.; Falsone, A.; Gesslbauer, B.; Kungl, A.J. A combinatorial approach to biophysically characterise chemokine-glycan binding affinities for drug development. Molecules 2014, 19, 10618–10634. [Google Scholar] [CrossRef] [Green Version]
- Frevert, C.W.; Sannes, P.L. Matrix proteoglycans as effector molecules for epithelial cell function. Eur. Respir. Rev. 2005, 14, 137–144. [Google Scholar] [CrossRef]
- Schuurs, Z.P.; Hammond, E.; Elli, S.; Rudd, T.R.; Mycroft-West, C.J.; Lima, M.A.; Skidmore, M.A.; Karlsson, R.; Chen, Y.-H.; Bagdonaite, I.; et al. Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain. Comput. Struct. Biotechnol. J. 2021, 19, 2806–2818. [Google Scholar] [CrossRef]
- Paiardi, G.; Richter, S.; Rusnati, M.; Wade, R.C. Mechanism of inhibition of SARS-CoV-2 infection by the interaction of the spike glycoprotein with heparin. J. Biol. Chem. 2021, 101507. [Google Scholar] [CrossRef]
- Mousa, S.A.; Zhang, F.; Aljada, A.; Chaturvedi, S.; Takieddin, M.; Zhang, H.; Chi, L.; Castelli, M.C.; Friedman, K.; Goldberg, M.M.; et al. Pharmacokinetics and Pharmacodynamics of Oral Heparin Solid Dosage Form in Healthy Human Subjects. J. Clin. Pharmacol. 2007, 47, 1508–1520. [Google Scholar] [CrossRef] [Green Version]



| Spike FL | Spike RBD | |
|---|---|---|
| Enoxaparin | 604.3 ± 67.4 | 678.4 ± 116.1 |
| HS | 680.3 ± 66.8 | 600 ± 78.6 |
| DS | 784.8 ± 65.6 | 912.5 ± 163.4 |
| PSS | 930 ± 95.7 | 655 ± 118.5 |
| Dp4 | 1054.7 ± 122.9 | 1063.3 ± 180.1 |
| Dp6 | 646.6 ± 65.2 | 619.2 ± 80.4 |
| Dp8 | 648.8 ± 64.6 | 795.6 ± 137.3 |
| Dp10 | 718.5 ± 66.6 | 768.7 ± 140.7 |
| Dp12 | 857.9 ± 88.1 | 1287 ± 213 |
| 6O-des | 870.3 ± 92.2 | 781.4 ± 175.7 |
| 2O-des | 940.2 ± 121.7 | 681.9 ± 175.7 |
| N-des | 682.2 ± 55.9 | 878.5 ± 96.4 |
| ACE2 | |
|---|---|
| Enoxaparin | 1232.6 ± 174.9 |
| HS | 1130 ± 145.3 |
| DS | 1057.7 ± 142.1 |
| PSS | 585 ± 41 |
| Dp4 | 531.6 ± 38.6 |
| Dp6 | 590.1 ± 56 |
| Dp8 | 509.9 ± 35 |
| Dp10 | 692.9 ± 65 |
| Dp12 | 460.7 ± 33 |
| 6O-des | 1034 ± 116 |
| 2O-des | 908 ± 90 |
| N-des | 647 ± 40 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ennemoser, M.; Rieger, J.; Muttenthaler, E.; Gerlza, T.; Zatloukal, K.; Kungl, A.J. Enoxaparin and Pentosan Polysulfate Bind to the SARS-CoV-2 Spike Protein and Human ACE2 Receptor, Inhibiting Vero Cell Infection. Biomedicines 2022, 10, 49. https://doi.org/10.3390/biomedicines10010049
Ennemoser M, Rieger J, Muttenthaler E, Gerlza T, Zatloukal K, Kungl AJ. Enoxaparin and Pentosan Polysulfate Bind to the SARS-CoV-2 Spike Protein and Human ACE2 Receptor, Inhibiting Vero Cell Infection. Biomedicines. 2022; 10(1):49. https://doi.org/10.3390/biomedicines10010049
Chicago/Turabian StyleEnnemoser, Maria, Julia Rieger, Eva Muttenthaler, Tanja Gerlza, Kurt Zatloukal, and Andreas J. Kungl. 2022. "Enoxaparin and Pentosan Polysulfate Bind to the SARS-CoV-2 Spike Protein and Human ACE2 Receptor, Inhibiting Vero Cell Infection" Biomedicines 10, no. 1: 49. https://doi.org/10.3390/biomedicines10010049
APA StyleEnnemoser, M., Rieger, J., Muttenthaler, E., Gerlza, T., Zatloukal, K., & Kungl, A. J. (2022). Enoxaparin and Pentosan Polysulfate Bind to the SARS-CoV-2 Spike Protein and Human ACE2 Receptor, Inhibiting Vero Cell Infection. Biomedicines, 10(1), 49. https://doi.org/10.3390/biomedicines10010049







