A Systematic Review and Meta-Analysis of the Prevalence and Impact of Pulmonary Bacterial Colonisation in Stable State Chronic Obstructive Pulmonary Disease (COPD)
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy
2.2. Study Selection, Data Extraction and Quality Assessment
2.3. Statistical Analysis
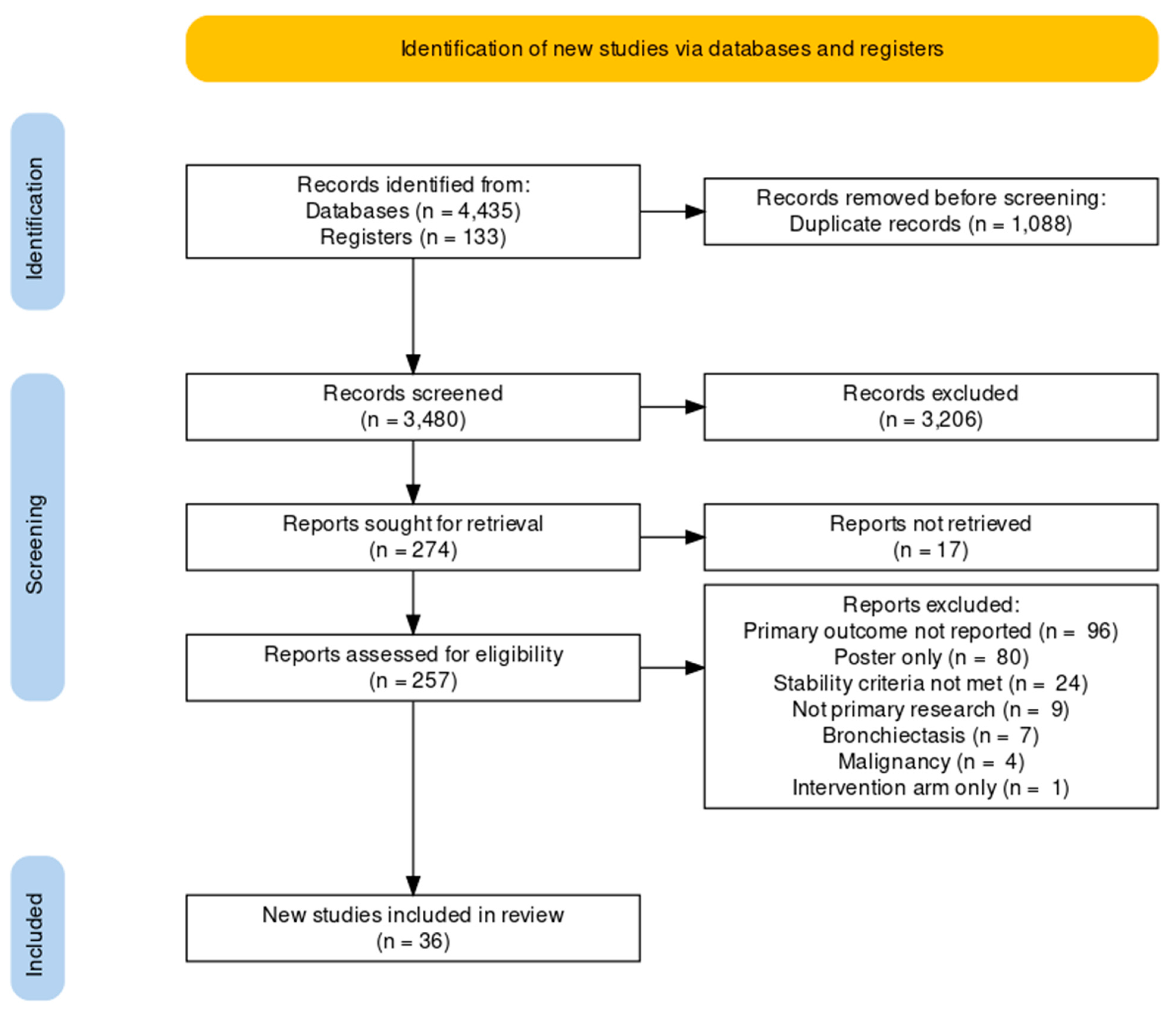
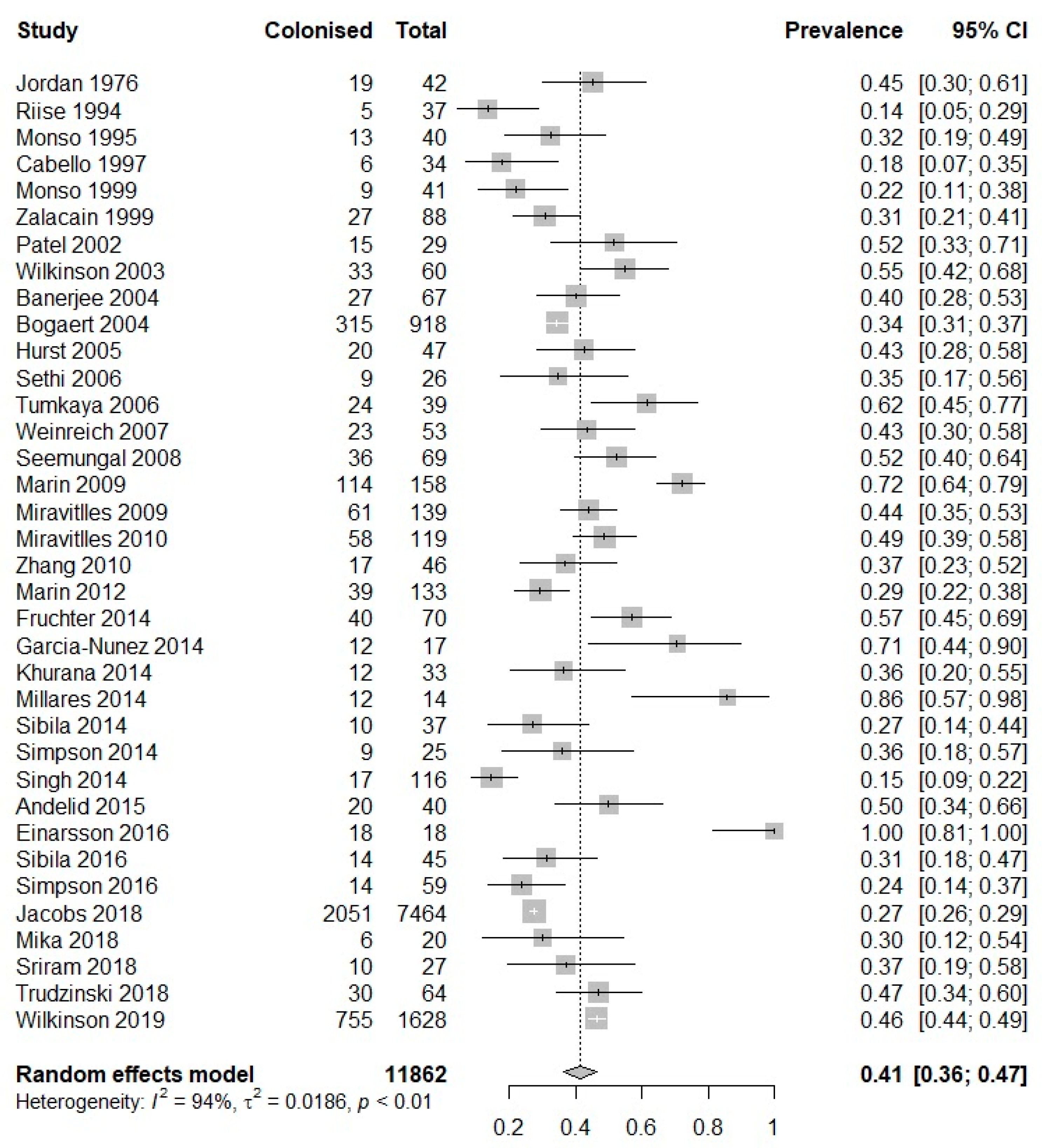
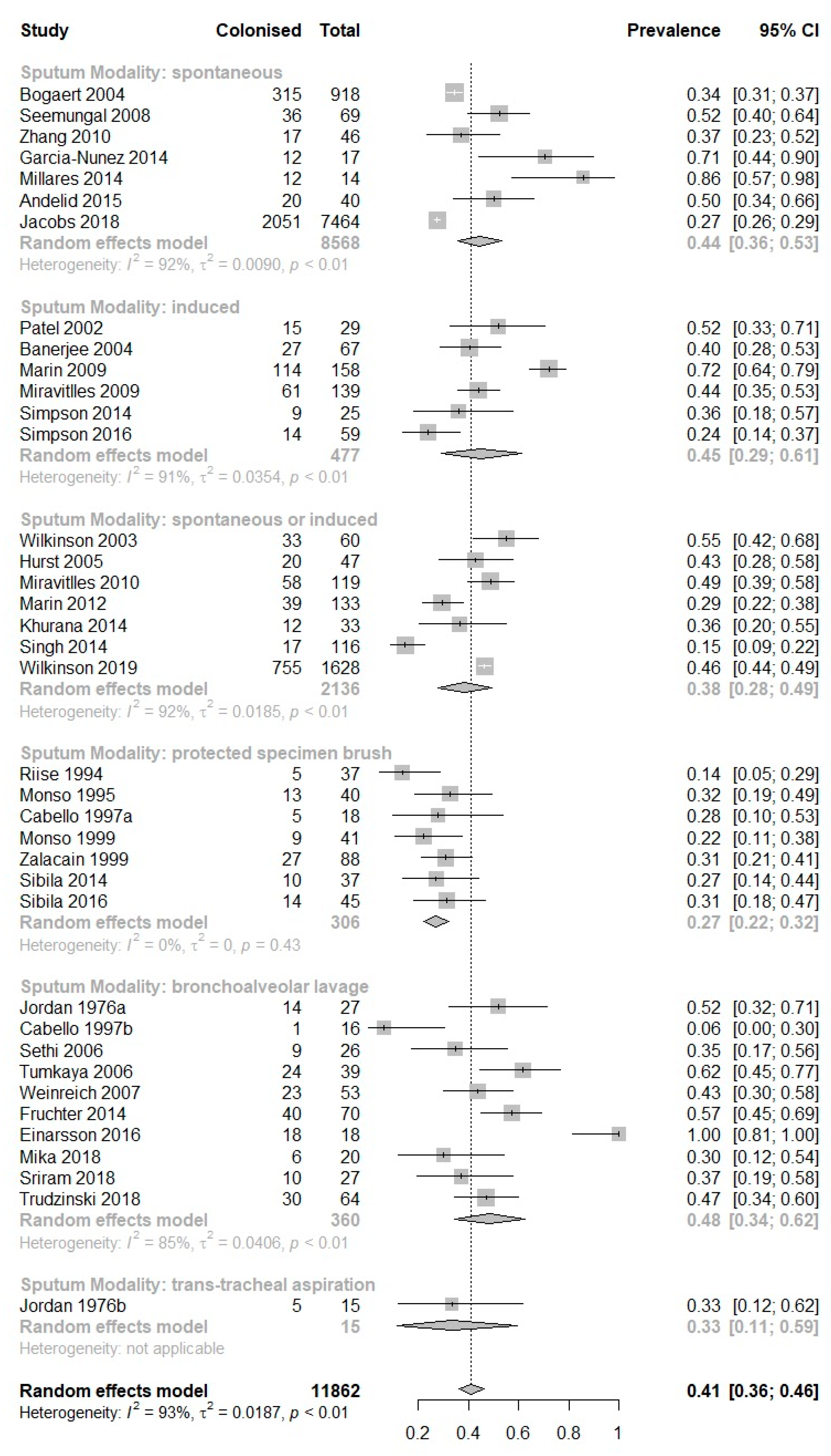
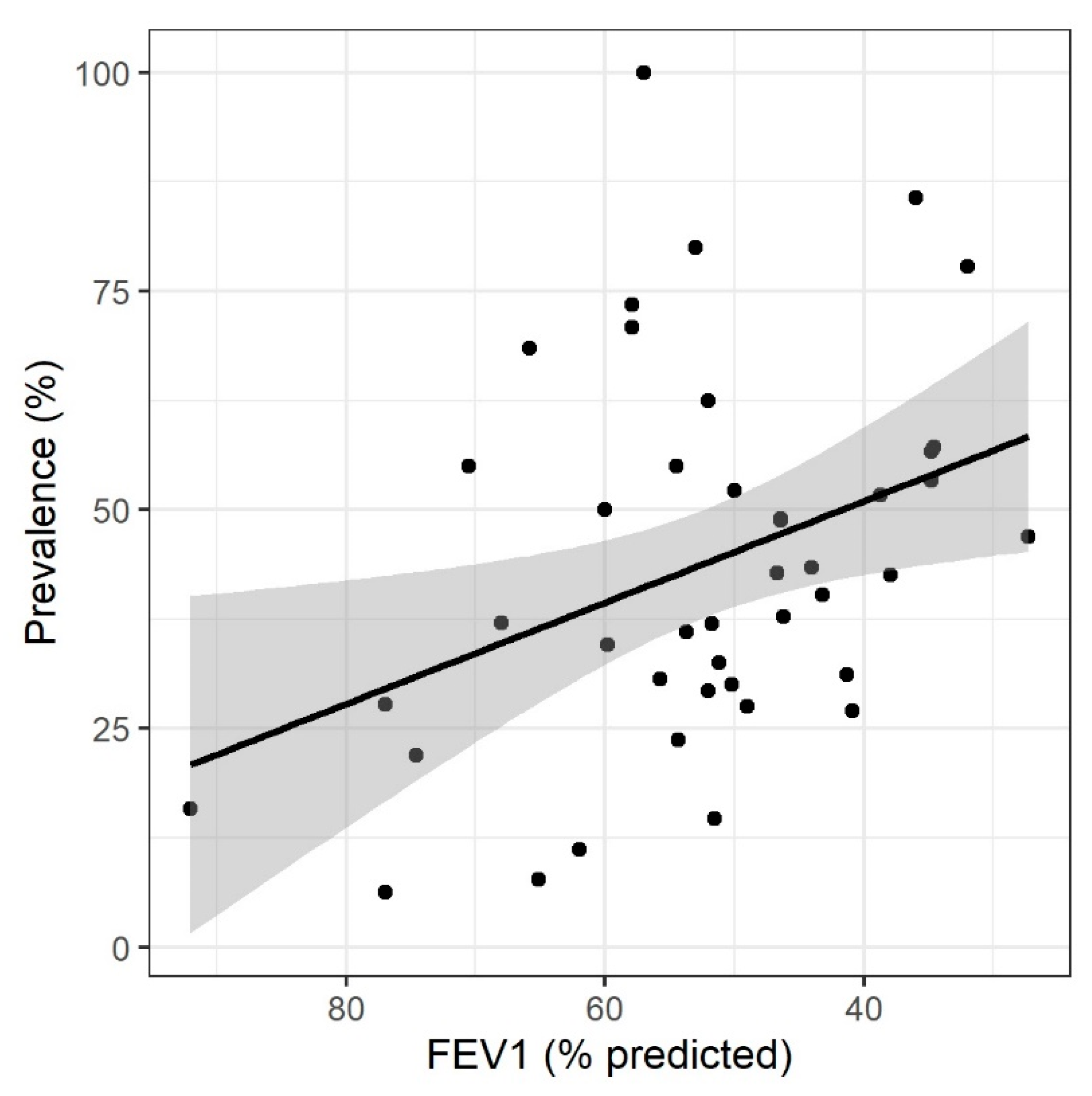
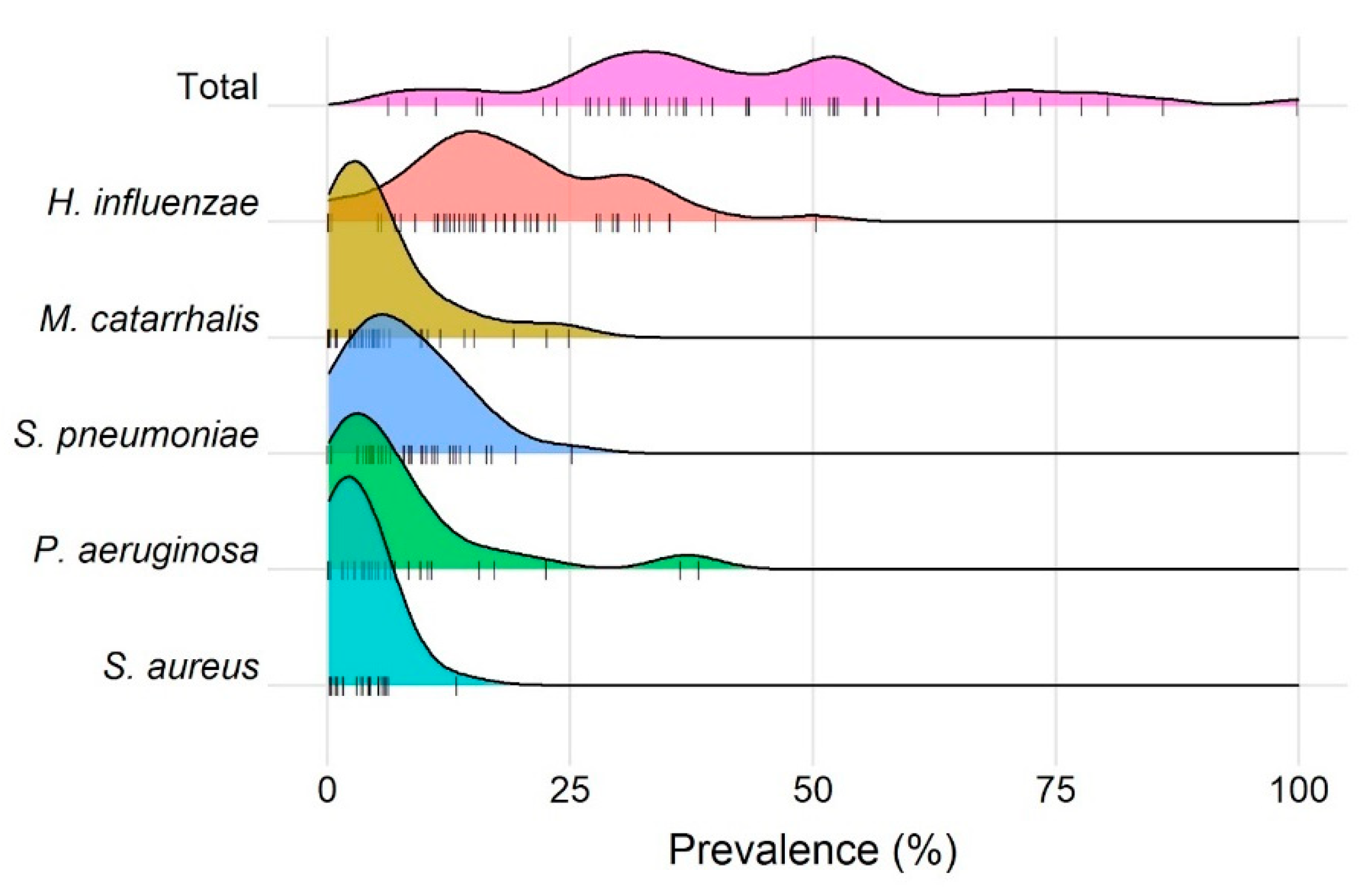
3. Results
| Study (Author, Year) | Country | Study Design | Population Description | Study Subgroup | No. Stable COPD Patients (% Female) | Age | Smoking, Pack-Years | FEV1, % Predicted | Stability Period Pre-Sampling |
|---|---|---|---|---|---|---|---|---|---|
| Andelid et al., 2015 [12] | Sweden | Prospective cohort | Smokers with obstructive disease and chronic bronchitis | - | 60 (24) | 62 {45–76} | 40 {14–156} | 60 {29–97} | 15 weeks |
| Banerjee et al., 2004 [13] | UK | Prospective cohort | Stable COPD outpatients | - | 67 (NA) | 66.7 (7.6) | 58.8 (25.1) | 43.2 (11.4) | 6 weeks |
| Bogaert et al., 2004 [14] | Netherlands | Prospective cohort | Stable COPD outpatients | - | 269 (NA) | {40–75} | NA | NA | Stable clinical condition |
| Cabello et al., 1997 [1] | Spain | Prospective cohort | Stable COPD outpatients with indication for bronchoscopy | PSB | 18 (17) | 60 (12) | NA | 77 (19) | 4 weeks |
| BAL | 18 (17) | 60 (12) | NA | 77 (19) | 4 weeks | ||||
| Einarsson et al., 2016 [15] | UK | Cross-sectional | COPD patients listed for bronchoscopy | - | 18 (22) | 60 {41–74} | NA | 57 {32–89} | 8 weeks |
| Fruchter et al., 2014 [16] | Israel | Prospective cohort | Severe COPD pre-BLVR | - | 70 (22) | 64 (8) | 28 (11) | 34.6 (7.3) | 90 days |
| Garcia-Nunez et al., 2014 [17] | Spain | Cross-sectional | Stable COPD outpatients | Moderate-to-severe disease | 17 (13) | 68 {62–69} | 75 {52–110} | 52.0 {41.5–69.0} | 4 weeks |
| Advanced disease | 17 (0) | 74 {68–77} | 55 {35–117} | 32.0 {29.5–35.0} | 4 weeks | ||||
| Hurst et al., 2005 [18] | UK | Prospective cohort | Stable COPD outpatients | Whole cohort | 47 (43) | 70.5 (7) | 46.1 (26.5) | 37.9 (13.6) | 12 weeks |
| Jacobs et al., 2018 [19] | USA | Prospective cohort | Stable COPD outpatients | - | 181 (NA) | 67 (9.2) | 79 (36) | 49 (18) | Stable clinical condition |
| Jordan et al., 1976 [20] | USA | Cross-sectional | Chronic bronchitis patients | BAL | 19 (NA) | NA | NA | NA | Stable clinical condition |
| Trans-tracheal aspiration | 19 (NA) | NA | NA | NA | Stable clinical condition | ||||
| Khurana et al., 2014 [21] | UK | Cross-sectional | Stable COPD outpatients | Non-persistent sputum | 52 (46) | 66.8 (6.5) | 35.3 {12.5–86} | 65.1 (16.3) | 6 weeks |
| Persistent sputum | 52 (54) | 65.7 (6.9) | 32.0 {18.5–122.2} | 54.5 (13.1) | 6 weeks | ||||
| Marin et al., 2009 [22] | Spain | Prospective cohort | Stable COPD outpatients | Baseline | 40 (3) | 66.5 (8.1) | NA | 57.9 (19.1) | 8 weeks |
| 9 month follow-up | 40 (3) | 66.5 (8.1) | NA | 57.9 (19.1) | 8 weeks | ||||
| Marin et al., 2012 [23] | Spain | Cross-sectional | COPD recruited on hospitalization for exacerbation | - | 133 (7) | 70 (9) | 67 {43–102} | 52 (16) | 12 weeks |
| Mika et al., 2018 [24] | Switzerland | Cross-sectional | COPD patients listed for bronchoscopy | - | 32 (31) | 65.7 (NA) | NA | 50.2 (24.9) | Stable clinical condition |
| Millares et al., 2014 [10] | Spain | Prospective cohort | COPD patients with >2 exacerbations per year | Whole cohort | 16 (0) | 71 (6) | 57 {57–110} | 36 {30–40} | >8 weeks |
| Miravitlles et al., 2009 [25] | Spain | Randomised control trial | COPD with sputum positive for PPM (p. aeruginosa excluded) | At randomisation | 119 (8) | 68 (9.1) | NA | 46.2 (14.1) | 16 weeks |
| Placebo 8 week follow-up | 119 (5) | 69 (10) | 43 (21) | 53 (16) | 16 weeks | ||||
| Miravitlles et al., 2010 [26] | Spain | Cross-sectional | Stable COPD outpatients | - | 119 (6) | 68 (9.1) | 40 (21.1) | 46.4 (14.1) | 12 weeks |
| Monso et al., 1995 [27] | Spain | Cross-sectional | COPD patients listed for bronchoscopy | - | 40 (0) | 61.1 (9.9) | NA | 51.2 (23) | 15 days |
| Monso et al., 1999 [28] | Spain | Cross-sectional | Stable chronic bronchitis | - | 41 (0) | 63.8 (9.1) | NA | 74.6 (23.7) | 15 days |
| Patel et al., 2002 [4] | UK | Prospective cohort | Stable COPD outpatients | - | 29 (28) | 66 {47–81} | 52.9 (42.2) | 38.7 (15.2) | 3 weeks |
| Riise et al., 1994 [29] | Sweden | Prospective cohort | Chronic bronchitis with and without COPD | Without COPD | 41 (NA) | 52 {36–68} | 36, 2 * | 92, 2 * | 4 weeks |
| With COPD | 41 (NA) | 57 {38–70} | 44, 4 * | 62, 2 * | 4 weeks | ||||
| Seemungal et al., 2008 [30] | UK | Randomised control trial | Stable COPD outpatients at baseline | - | 109 (37) | 67.2 (8.6) | 51.6 (33.9) | 50.0 (18.0) | 4 weeks |
| Sethi et al., 2006 [31] | USA | Prospective cohort | Ex-smokers with COPD | - | 26 (23) | 64.7 (1.7) | 66 (6.3) | 59.8 (4.1) | 4 weeks |
| Sibila et al., 2014 [32] | Spain | Cross-sectional | Stable COPD outpatients | - | 37 (24) | 67.9 (8.0) | 47.3 (12.7) | 40.9 (8.1) | 4 weeks |
| Sibila et al., 2016 [33] | Spain | Cross-sectional | Stable COPD outpatients | - | 45 (18) | 67.1 (8.5) | 54.3 (20.1) | 41.3 (10.2) | 4 weeks |
| Simpson et al., 2014 [34] | Australia | Randomised control trial | Stable COPD outpatients at randomisation | - | 30 (37) | 70.8 (7.6) | 46.1 (36.6) | 53.7 (13.7) | 4 weeks |
| Simpson et al., 2016 [35] | Australia | Cross-sectional | Stable COPD outpatients | - | 59 (51) | 69.7 (7.5) | 32.9 {17.0–53.8} | 54.3 (15.6) | Stable clinical condition |
| Singh et al., 2014 [36] | UK | Prospective cohort | Stable COPD outpatients | - | 99 (33) | 72.1 (8.9) | 48.4 {24.4–67.5} | 51.5 (21.6) | 4 weeks |
| Sriram et al., 2018 [37] | Australia | Cross-sectional | COPD patients listed for bronchoscopy | - | 27 (37) | 68 (9) | 43 (28) | 68 (25) | Excluded exacerbations |
| Trudzinski et al., 2018 [38] | Germany | Cross-sectional | COPD patients undergoing BLVR with EBV insertion | - | 64 (50) | 62.4 (8.7) | NA | 27.3 (9.5) | Excluded exacerbations |
| Tumkaya et al., 2006 [39] | Turkey | Prospective cohort | Stable COPD outpatients | Exacerbations (<3/year) | 39 (10) | 58.6 (7.7) | 46.2 (22.1) | 70.5 (12.0) | 4 weeks |
| Exacerbations (>3/year) | 39 (11) | 58.8 (7.7) | 50.26 (22.2) | 65.8 (12.8) | 4 weeks | ||||
| Weinreich et al., 2007 [40] | Denmark | Cross-sectional | COPD patients listed for bronchoscopy | - | 53 (49) | 67 {58–73} | 30 {21–45} | 44 {NA} | 4 weeks |
| Wilkinson et al., 2003 [41] | UK | Prospective cohort | Stable COPD outpatients | Baseline | 30 (27) | 66.4 (10.3) | 74.3 (66.5) | 34.8 (13.6) | 6 weeks |
| 12 month follow-up | 30 (27) | 66.4 (10.3) | 74.3 (66.5) | 34.8 (13.6) | 6 weeks | ||||
| Wilkinson et al., 2019 [11] | UK | Prospective cohort | Stable COPD outpatients | Year 1 | 127 (47) | 66.8 (8.6) | 47.0 {33.7–60.0} | 46.4 (15.2) | Stable clinical condition |
| Year 2 | 127 (44) | 66.7 (8.7) | 50.4 {34.0–60.0} | 46.7 (14.6) | Stable clinical condition | ||||
| Zalacain et al., 1999 [42] | Spain | Cross-sectional | Stable COPD outpatients | - | 88 (0) | 66.1 (7.2) | 53.6 (14.9) | 55.7 (12.9) | 4 weeks |
| Zhang et al., 2010 [43] | China | Prospective cohort | Stable COPD outpatients | - | 46 (17) | 70.9 (5.6) | NA | 51.8 (12.3) | 6 weeks |
| Study (Author, Year) | Study Subgroup | Sampling Modality | No. of Patients Producing Sputum | No. of Sputum Samples Produced | Prevalence of PPM Positive Sputum, Percent (95% CI) | Prevalence of H. influenzae in Sputum, Percent (95% CI) | Prevalence of M. catarrhalis in Sputum, Percent (95% CI) | Prevalence of S. pneumoniae in Sputum, Percent (95% CI) | Prevalence of P. aeruginosa in Sputum, Percent (95% CI) | Prevalence of S. aureus in Sputum, Percent (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| Andelid et al., 2015 [12] | - | Spontaneous | 40 | 40 | 50 (34–66) | 13 (4–27) | 3 (0–13) | 5 (1–17) | 3 (0–13) | 0 (0–9) |
| Banerjee et al., 2004 [13] | - | Induced | 67 | 67 | 40 (29–53) | 21 (12–33) | 15 (7–26) | 13 (6–24) | 2 (0–8) | 2 (0–8) |
| Bogaert et al., 2004 [14] | - | Spontaneous | 269 | 918 | 34 (31–38) | 19 (17–22) | 19 (17–22) | 13 (11–15) | NA | NA |
| Cabello et al., 1997 [1] | PSB | PSB | 18 | 18 | 28 (10–54) | 11 (1–35) | 0 (0–19) | 11 (1–35) | 0 (0–19) | 6 (0–27) |
| BAL | BAL | 16 | 16 | 6 (0–30) | 0 (0–20.6) | 0 (0–21) | 6 (0–30) | 0 (0–21) | 0 (0–21) | |
| Einarsson et al., 2016 [15] | - | BAL | 18 | 18 | 100 (82–100) | 28 (10–54) | 0 (0–19) | 17 (4–41) | 6 (0–27) | 6 (0–27) |
| Fruchter et al., 2014 [16] | - | BAL | 70 | 70 | 57 (45–69) | 7.1 (2–16) | 1 (0–8) | 4 (1–12) | 17 (9–28) | 13 (6–23) |
| Garcia-Nunez et al., 2014 [17] | Moderate-to-severe disease | Spontaneous | 8 | 8 | 63 (25–92) | 13 (0–53) | 25 (3–65) | 13 (0–53) | 38 (8–76) | NA |
| Advanced disease | Spontaneous | 9 | 9 | 78 (40–97) | 33 (8–70) | 0 (0–34) | 11 (0–48) | 22 (3–60) | NA | |
| Hurst et al., 2005 [18] | Whole cohort | Spontaneous or induced | 47 | 47 | 43 (28–58) | 19 (9–33) | 6 (1–18) | 6 (1–18) | 2 (0–11) | NA |
| Jacobs et al., 2018 [19] | - | Spontaneous | 181 | 7464 | 28 (27–29) | 14 (13–15) | 6 (5–6) | 6 (5–6) | 8 (7–8) | NA |
| Jordan et al., 1976 [20] | BAL | BAL | 19 | 27 | 52 (32–71) | 22 (9–42) | 0 (0–13) | NA | 11 (2–29) | 4 (0–19) |
| Trans-tracheal aspiration | Trans-tracheal aspiration | 11 | 15 | 33 (12–62) | 20 (4–48) | 0 (0–22) | NA | 0 (0–22) | 0 (0–22) | |
| Khurana et al., 2014 [21] | Non-persistent sputum | Spontaneous or induced | 13 | 13 | 8 (0–36) | 8 (0–36) | 0 (0–25) | 0 (0–25) | 0 (0–25) | 0 (0–25) |
| Persistent sputum | Spontaneous or induced | 20 | 20 | 55 (32–77) | 35 (15–59) | 5 (0–25) | 15 (3–38) | 0 (0–17) | 5 (0–25) | |
| Marin et al., 2009 [22] | Baseline | Induced | 40 | 79 | 73 (62–83) | 35 (25–47) | 5 (1–13) | 0 (0–5) | NA | NA |
| 9 month follow-up | Induced | 40 | 79 | 71 (60–81) | 32 (22–43) | 3 (0–9) | 0 (0–5) | NA | NA | |
| Marin et al., 2012 [23] | - | Spontaneous or induced | 133 | 133 | 2 (22–38) | 17 (11–24) | 5 (2–10) | 4 (1–9) | 6 (3–12) | NA |
| Mika et al., 2018 [24] | - | BAL | 20 | 20 | 30 (12–54) | 15 (3–38) | 10 (1–32) | 10 (1–32) | NA | NA |
| Millares et al., 2014 [10] | Whole cohort | Spontaneous | 14 | 14 | 86 (57–98) | 29 (8–58) | 14 (2–43) | 14 (2–43) | 36 (13–65) | 0 (0–23) |
| Miravitlles et al., 2009 [25] | At randomisation | Induced | 119 | 119 | 38 (29–47) | 16 (10–24) | 3 (1–8) | 3 (1–7) | 4 (1–10) | 0 (0–3) |
| Placebo 8 week follow-up | Induced | 20 | 20 | 80 (56–94) | 50 (27–73) | 5 (0–25) | 0 (0–17) | 0 (0–17) | 0 (0–17) | |
| Miravitlles et al., 2010 [26] | - | Spontaneous or induced | 119 | 119 | 49 (40–58) | 18 (11–26) | 3 (1–8) | 3 (1–8) | 4 (1–10) | 1 (0–5) |
| Monso et al., 1995 [27] | - | PSB | 40 | 40 | 33 (19–49) | 15 (6–30) | 3 (0–13) | 8 (2–20) | 3 (0–13) | 3 (0–13) |
| Monso et al., 1999 [28] | - | PSB | 41 | 41 | 22 (11–38) | 12 (4–26) | NA | 5 (1–17) | NA | NA |
| Patel et al., 2002 [4] | - | Induced | 29 | 29 | 52 (33–71) | 28 (13–47) | 10 (2–27) | NA | 10 (2–27) | NA |
| Riise et al., 1994 [29] | Without COPD | PSB | 19 | 19 | 16 (3–40) | 11 (1–33) | 0 (0–18) | 5 (0–26) | NA | 0 (0–18) |
| With COPD | PSB | 18 | 18 | 11 (1–35) | 0 (0–19) | 0 (0–19) | 11 (1–35) | NA | 0 (0–19) | |
| Seemungal et al., 2008 [30] | - | Spontaneous | 69 | 69 | 52 (40–64) | 32 (21–44) | 4 (1–12) | 9 (3–18) | NA | NA |
| Sethi et al., 2006 [31] | - | BAL | 26 | 26 | 35 (17–56) | 12 (3–30) | 0 (0–13) | 4 (0–20) | 4 (0–20) | 4 (0–20) |
| Sibila et al., 2014 [32] | - | PSB | 37 | 37 | 27 (14–44) | 14 (5–29) | 5 (1–18) | 5 (1–18) | 0 (0–10) | 0 (0–10) |
| Sibila et al., 2016 [33] | - | PSB | 45 | 45 | 31 (18–47) | 18 (8–32) | 4 (1–15) | 4 (1–15) | 0 (0–8) | NA |
| Simpson et al., 2014 [34] | - | Induced | 25 | 25 | 36 (18–58) | 40 (0–20) | 4 (0–20) | 8 (1–26) | 16 (5–36) | 4 (0–20) |
| Simpson et al., 2016 [35] | - | Induced | 59 | 59 | 24 (14–37) | 5 (1–14) | 12 (5–23) | NA | 7 (2–17) | 3 (0–12) |
| Singh et al., 2014 [36] | - | Spontaneous or induced | 99 | 116 | 11 (9–22) | 6 (3–12) | 1 (0–5) | 4 (1–10) | 2 (0–6) | 1 (0–5) |
| Sriram et al., 2018 [37] | - | BAL | 27 | 27 | 37 (19–58) | 22 (9–42) | NA | 4 (0–19) | 7 (1–24) | 4 (0–19) |
| Trudzinski et al., 2018 [38] | - | BAL | 64 | 64 | 47 (34–60) | 9 (4–19) | 2 (0–8) | 6 (2–15) | 5 (1–13) | 6 (2–15) |
| Tumkaya et al., 2006 [39] | Exacerbations (<3/year) | BAL | 20 | 20 | 55 (32–77) | 0 (0–17) | 0 (0–17) | 10 (1–32) | NA | 0 (0–17) |
| Exacerbations (>3/year) | BAL | 19 | 19 | 69 (44–78) | 11 (1–33) | 5 (0–26) | 0 (0–18) | NA | 5 (0–26) | |
| Weinreich et al., 2007 [40] | - | BAL | 53 | 53 | 43 (30–58) | 23 (12–36) | 4 (1–13) | 25 (14–38) | 4 (1–13) | 4 (1–13) |
| Wilkinson et al., 2003 [41] | Baseline | Spontaneous or induced | 30 | 30 | 53 (34–72) | 30 (15–49) | 10 (2–27) | 10 (2–27) | 10 (2–27) | NA |
| 12 month follow-up | Spontaneous or induced | 30 | 30 | 57 (37–75) | 23 (10–42) | 23 (10–42) | 0 (0–12) | 10 (2–27) | NA | |
| Wilkinson et al., 2019 [11] | Year 1 | Spontaneous or induced | 127 | 952 | 49 (46–52) | 30 (27–33) | 5 (4–7) | 19 (16–21) | 5 (4–7) | 4 (3–6) |
| Year 2 | Spontaneous or induced | 103 | 676 | 43 (39–47) | 23 (19–26) | 3 (2–5) | 16 (13–19) | 5 (3–7) | 6 (5–9) | |
| Zalacain et al., 1999 [42] | - | PSB | 88 | 88 | 31 (21–41) | 16 (9–25) | 5 (1–11) | 8 (3–16) | 0 (0–4) | 1 (0–6) |
| Zhang et al., 2010 [43] | - | Spontaneous | 46 | 46 | 37 (23–53) | 15 (6–29) | 2 (0–12) | 9 (2–21) | 4 (1–15) | 2 (0–12) |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cabello, H.; Torres, A.; Celis, R.; El-Ebiary, M.; De La Bellacasa, J.P.; Xaubet, A.; Gonzalez, J.; Agusti, C.; Soler, N. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: A bronchoscopic study. Eur. Respir. J. 1997, 10, 1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, H.; Eschberger, K.; Wrona, C.; Grove, L.; Agrawal, A.; Grant, B.; Yin, J.; Parameswaran, G.I.; Murphy, T.; Sethi, S. Bacterial Colonization Increases Daily Symptoms in Patients with Chronic Obstructive Pulmonary Disease. Ann. Am. Thorac. Soc. 2014, 11, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Bouquet, J.; Tabor, D.E.; Silver, J.S.; Nair, V.; Tovchigrechko, A.; Griffin, M.P.; Esser, M.T.; Sellman, B.R.; Jin, H. Microbial burden and viral exacerbations in a longitudinal multicenter COPD cohort. Respir. Res. 2020, 21, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, I.S.; Seemungal, T.A.R.; Wilks, M.; Lloyd-Owen, S.J.; Donaldson, G.C.; Wedzicha, J.A. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax 2002, 57, 759–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tangedal, S.; Aanerud, M.; Grønseth, R.; Drengenes, C.; Wiker, H.G.; Bakke, P.S.; Eagan, T.M. Comparing microbiota profiles in induced and spontaneous sputum samples in COPD patients. Respir. Res. 2017, 18, 164. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio 2017, 8, e02287-16. [Google Scholar] [CrossRef]
- Herath, S.C.; Normansell, R.; Maisey, S.; Poole, P. Prophylactic antibiotic therapy for chronic obstructive pulmonary disease (COPD). Cochrane Database Syst. Rev. 2018, 10, CD009764. [Google Scholar] [CrossRef]
- The University of Adelaide. Critical Appraisal Checklist for Prevalence Studies. 2020. Available online: https://jbi.global/critical-appraisal-tools (accessed on 18 August 2021).
- Barker, T.H.; Migliavaca, C.B.; Stein, C.; Colpani, V.; Falavigna, M.; Aromataris, E.; Munn, Z. Conducting proportional meta-analysis in different types of systematic reviews: A guide for synthesisers of evidence. BMC Med. Res. Methodol. 2021, 21, 189. [Google Scholar] [CrossRef]
- Millares, L.; Ferrari, R.; Gallego, M.; Garcia-Nuñez, M.; Pérez-Brocal, V.; Espasa, M.; Pomares, X.; Monton, C.; Moya, A.; Monsó, E. Bronchial microbiome of severe COPD patients colonised by Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1101–1111. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, T.M.; Aris, E.; Bourne, S.C.; Clarke, S.C.; Peeters, M.; Pascal, T.G.; Taddei, L.; Tuck, A.C.; Kim, V.L.; Ostridge, K.K.; et al. Drivers of year-to-year variation in exacerbation frequency of COPD: Analysis of the AERIS cohort. ERJ Open Res. 2019, 5, 00248-2018. [Google Scholar] [CrossRef]
- Andelid, K.; Tengvall, S.; Andersson, A.; Levänen, B.; Christenson, K.; Jirholt, P.; Åhrén, C.; Qvarfordt, I.; Ekberg-Jansson, A.; Lindén, A. Systemic cytokine signaling via IL-17 in smokers with obstructive pulmonary disease: A link to bacterial colonization? Int. J. Chronic Obstr. Pulm. Dis. 2015, 10, 689–702. [Google Scholar]
- Banerjee, D.; Khair, O.A.; Honeybourne, D. Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur. Respir. J. 2004, 23, 685. [Google Scholar] [CrossRef] [Green Version]
- Bogaert, D.; van der Valk, P.; Ramdin, R.; Sluijter, M.; Monninkhof, E.; Hendrix, R.; de Groot, R.; Hermans, P.W. Host-pathogen interaction during pneumococcal infection in patients with chronic obstructive pulmonary disease. Infect. Immun. 2004, 72, 818–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einarsson, G.G.; Comer, D.M.; McIlreavey, L.; Parkhill, J.; Ennis, M.; Tunney, M.M.; Elborn, J.S. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 2016, 71, 795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruchter, O.; Rosengarten, D.; Goldberg, E.; Ben-Zvi, H.; Tor, R.; Kramer, M.R. Airway bacterial colonization and serum C-reactive protein are associated with chronic obstructive pulmonary disease exacerbation following bronchoscopic lung volume reduction. Clin. Respir. J. 2014, 10, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Nuñez, M.; Millares, L.; Pomares, X.; Ferrari, R.; Pérez-Brocal, V.; Gallego, M.; Espasa, M.; Moya, A.; Monsó, E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 4217–4223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurst, J.R.; Wilkinson, T.M.A.; Perera, W.R.; Donaldson, G.C.; Wedzicha, J.A. Relationships Among Bacteria, Upper Airway, Lower Airway, and Systemic Inflammation in COPD. Chest 2005, 127, 1219–1226. [Google Scholar] [CrossRef]
- Jacobs, D.M.; Ochs-Balcom, H.M.; Zhao, J.; Murphy, T.F.; Sethi, S. Lower Airway Bacterial Colonization Patterns and Species-Specific Interactions in Chronic Obstructive Pulmonary Disease. J. Clin. Microbiol. 2018, 56, e00330-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, G.W.; Wong, G.A.; Hoeprich, P.D. Bacteriology of the Lower Respiratory Tract as Determined by Fiber-Optic Bronchoscopy and Transtracheal Aspiration. J. Infect. Dis. 1976, 134, 428–435. [Google Scholar] [CrossRef]
- Khurana, S.; Ravi, A.; Sutula, J.; Milone, R.; Williamson, R.; Plumb, J.; Vestbo, J.; Singh, D. Clinical characteristics and airway inflammation profile of COPD persistent sputum producers. Respir. Med. 2014, 108, 1761–1770. [Google Scholar] [CrossRef] [Green Version]
- Marin, A.; Monsó, E.; Garcia-Nunez, M.; Sauleda, J.; Noguera, A.; Pons, J.; Agustí, A.; Morera, J. Variability and effects of bronchial colonisation in patients with moderate COPD. Eur. Respir. J. 2010, 35, 295. [Google Scholar] [CrossRef]
- Marin, A.; Garcia-Aymerich, J.; Sauleda, J.; Belda, J.; Millares, L.; García-Núñez, M.; Serra, I.; Benet, M.; Agustí, A.; Antó, J.M.; et al. Effect of Bronchial Colonisation on Airway and Systemic Inflammation in Stable COPD. COPD J. Chronic Obstr. Pulm. Dis. 2012, 9, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Mika, M.; Nita, I.; Morf, L.; Qi, W.; Beyeler, S.; Bernasconi, E.; Marsland, B.J.; Ott, S.R.; von Garnier, C.; Hilty, M. Microbial and host immune factors as drivers of COPD. ERJ Open Res. 2018, 4, 00015-2018. [Google Scholar] [CrossRef] [Green Version]
- Miravitlles, M.; Marín, A.; Monsó, E.; Vilà, S.; de la Roza, C.; Hervás, R.; Esquinas, C.; García, M.; Millares, L.; Morera, J.; et al. Efficacy of moxifloxacin in the treatment of bronchial colonisation in COPD. Eur. Respir. J. 2009, 34, 1066. [Google Scholar] [CrossRef] [Green Version]
- Miravitlles, M.; Marín, A.; Monsó, E.; Vilà, S.; de la Roza, C.; Hervás, R.; Esquinas, C.; García, M.; Millares, L.; Morera, J.; et al. Colour of sputum is a marker for bacterial colonisation in chronic obstructive pulmonary disease. Respir. Res. 2010, 11, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monso, E.; Ruiz, J.; Rosell, A.; Manterola, J.; Fiz, J.; Morera, J.; Ausina, V. Bacterial infection in chronic obstructive pulmonary disease. A study of stable and exacerbated outpatients using the protected specimen brush. Am. J. Respir. Crit. Care Med. 1995, 152, 1316–1320. [Google Scholar] [CrossRef]
- Monso, E.; Rosell, A.; Bonet, G.; Manterola, J.; Cardona, P.J.; Ruiz, J.; Morera, J. Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur. Respir. J. 1999, 13, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riise, G.C.; Larsson, S.; Larsson, P.; Jeansson, S.; Andersson, B.A. The intrabronchial microbial flora in chronic bronchitis patients: A target for N-acetylcysteine therapy? Eur. Respir. J. 1994, 7, 94. [Google Scholar] [CrossRef]
- Seemungal, T.A.R.; Wilkinson, T.M.A.; Hurst, J.R.; Perera, W.R.; Sapsford, R.J.; Wedzicha, J.A. Long-term Erythromycin Therapy Is Associated with Decreased Chronic Obstructive Pulmonary Disease Exacerbations. Am. J. Respir. Crit. Care Med. 2008, 178, 1139–1147. [Google Scholar] [CrossRef]
- Sethi, S.; Maloney, J.; Grove, L.; Wrona, C.; Berenson, C.S. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006, 173, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Sibila, O.; Garcia-Bellmunt, L.; Giner, J.; Merino, J.L.; Suarez-Cuartin, G.; Torrego, A.; Solanes, I.; Castillo, D.; Valera, J.L.; Cosio, B.G.; et al. Identification of airway bacterial colonization by an electronic nose in Chronic Obstructive Pulmonary Disease. Respir. Med. 2014, 108, 1608–1614. [Google Scholar] [CrossRef] [Green Version]
- Sibila, O.; Garcia-Bellmunt, L.; Giner, J.; Rodrigo-Troyano, A.; Suarez-Cuartin, G.; Torrego, A.; Castillo, D.; Solanes, I.; Mateus, E.F.; Vidal, S.; et al. Airway Mucin 2 Is Decreased in Patients with Severe Chronic Obstructive Pulmonary Disease with Bacterial Colonization. Ann. Am. Thorac. Soc. 2016, 13, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Powell, H.; Baines, K.J.; Milne, D.; Coxson, H.O.; Hansbro, P.M.; Gibson, P.G. The effect of azithromycin in adults with stable neutrophilic COPD: A double blind randomised, placebo controlled trial. PLoS ONE 2014, 9, e105609. [Google Scholar] [CrossRef] [Green Version]
- Simpson, J.L.; Baines, K.J.; Horvat, J.C.; Essilfie, A.T.; Brown, A.C.; Tooze, M.; McDonald, V.M.; Gibson, P.G.; Hansbro, P.M. COPD is characterized by increased detection of Haemophilus influenzae, Streptococcus pneumoniae and a deficiency of Bacillus species. Respirology 2016, 21, 697–704. [Google Scholar] [CrossRef]
- Singh, R.; Mackay, A.J.; Patel, A.R.; Garcha, D.S.; Kowlessar, B.S.; Brill, S.E.; Donnelly, L.E.; Barnes, P.J.; Donaldson, G.C.; Wedzicha, J.A. Inflammatory thresholds and the species-specific effects of colonising bacteria in stable chronic obstructive pulmonary disease. Respir. Res. 2014, 15, 114. [Google Scholar] [CrossRef] [PubMed]
- Sriram, K.B.; Cox, A.J.; Sivakumaran, P.; Singh, M.; Watts, A.M.; West, N.P.; Cripps, A.W. Non-typeable Haemophilus Influenzae detection in the lower airways of patients with lung cancer and chronic obstructive pulmonary disease. Multidiscip. Respir. Med. 2018, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Trudzinski, F.C.; Seiler, F.; Wilkens, H.; Metz, C.; Kamp, A.; Bals, R.; Gärtner, B.; Lepper, P.M.; Becker, S.L. Microbiological airway colonization in COPD patients with severe emphysema undergoing endoscopic lung volume reduction. Int. J. Chronic Obstr. Pulm. Dis. 2017, 13, 29–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tumkaya, M.; Atis, S.; Ozge, C.; Delialioglu, N.; Polat, G.; Kanik, A. Relationship between airway colonization, inflammation and exacerbation frequency in COPD. Respir. Med. 2007, 101, 729–737. [Google Scholar] [CrossRef] [Green Version]
- Weinreich, U.M.; Korsgaard, J. Bacterial colonisation of lower airways in health and chronic lung disease. Clin. Respir. J. 2008, 2, 116–122. [Google Scholar] [CrossRef]
- Wilkinson, T.M.A.; Patel, I.S.; Wilks, M.; Donaldson, G.C.; Wedzicha, J.A. Airway Bacterial Load and FEV1 Decline in Patients with Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2003, 167, 1090–1095. [Google Scholar] [CrossRef]
- Zalacain, R.; Sobradillo, V.; Amilibia, J.; Barron, J.; Achotegui, V.; Pijoan, J.I.; Llorente, J.L. Predisposing factors to bacterial colonization in chronic obstructive pulmonary disease. Eur. Respir. J. 1999, 13, 343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Li, Q.; Zhang, X.Y.; Ding, X.; Zhu, D.; Zhou, X. Relevance of lower airway bacterial colonization, airway inflammation, and pulmonary function in the stable stage of chronic obstructive pulmonary disease. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1487–1493. [Google Scholar] [CrossRef]
- Choi, S.H.; Cha, S.I.; Choi, K.J.; Lim, J.K.; Seo, H.; Yoo, S.S.; Lee, J.; Lee, S.Y.; Kim, C.H.; Park, J.Y. Clinical Characteristics of Community-Acquired Viridans Streptococcal Pneumonia. Tuberc. Respir. Dis. 2015, 78, 196–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sethi, S.; Evans, N.; Grant, B.J.B.; Murphy, T.F. New Strains of Bacteria and Exacerbations of Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2002, 347, 465–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duim, B.; van Alphen, L.; Eijk, P.; Jansen, H.M.; Dankert, J. Antigenic drift of non-encapsulated Haemophilus influenzae major outer membrane protein P2 in patients with chronic bronchitis is caused by point mutations. Mol. Microbiol. 1994, 11, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Cole, P. The damaging role of bacteria in chronic lung infection. J. Antimicrob. Chemother. 1997, 40 (Suppl. S1), 5–10. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R. Bacteria, antibiotics and COPD. Eur. Respir. J. 2001, 17, 995. [Google Scholar] [CrossRef] [Green Version]
- Matkovic, Z.; Miravitlles, M. Chronic bronchial infection in COPD. Is there an infective phenotype? Respir. Med. 2013, 107, 10–22. [Google Scholar] [CrossRef] [Green Version]
- Stockley, R.A.; O’Brien, C.; Pye, A.; Hill, S.L. Relationship of Sputum Color to Nature and Outpatient Management of Acute Exacerbations of COPD. Chest 2000, 117, 1638–1645. [Google Scholar] [CrossRef]
| Outcomes | Data Collection Points | |
|---|---|---|
| Demographics | Age; sex; alpha-1 antitrypsin status; stability period; smoking status and pack years | |
| Primary Outcome | Determine prevalence of bacterial colonisation in stable-state COPD | Number of patients that produced a sample; number of samples collected; number of positive cultures; individual bacteriology (number positive for individual PPMs) |
| Secondary Outcomes | Assess the relationship between sampling modality and colonisation | Sampling modality (spontaneous, induced, PSB, bronchoscopy, trans-tracheal aspiration) |
| Assess relationship between bacterial colonisation and disease phenotype | FEV1; FEV1 category by GOLD criteria; quality of life (SGRQ/CAT/mMRC); exacerbation frequency; hospitalisation rate; mortality rate | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Armitage, M.N.; Spittle, D.A.; Turner, A.M. A Systematic Review and Meta-Analysis of the Prevalence and Impact of Pulmonary Bacterial Colonisation in Stable State Chronic Obstructive Pulmonary Disease (COPD). Biomedicines 2022, 10, 81. https://doi.org/10.3390/biomedicines10010081
Armitage MN, Spittle DA, Turner AM. A Systematic Review and Meta-Analysis of the Prevalence and Impact of Pulmonary Bacterial Colonisation in Stable State Chronic Obstructive Pulmonary Disease (COPD). Biomedicines. 2022; 10(1):81. https://doi.org/10.3390/biomedicines10010081
Chicago/Turabian StyleArmitage, Michael N., Daniella A. Spittle, and Alice M. Turner. 2022. "A Systematic Review and Meta-Analysis of the Prevalence and Impact of Pulmonary Bacterial Colonisation in Stable State Chronic Obstructive Pulmonary Disease (COPD)" Biomedicines 10, no. 1: 81. https://doi.org/10.3390/biomedicines10010081
APA StyleArmitage, M. N., Spittle, D. A., & Turner, A. M. (2022). A Systematic Review and Meta-Analysis of the Prevalence and Impact of Pulmonary Bacterial Colonisation in Stable State Chronic Obstructive Pulmonary Disease (COPD). Biomedicines, 10(1), 81. https://doi.org/10.3390/biomedicines10010081







