Exosomes Released by Corneal Stromal Cells Show Molecular Alterations in Keratoconus Patients and Induce Different Cellular Behavior
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Isolation and Culture of Corneal Stromal Cells from Healthy Human Donor Corneas and Keratoconic Corneas
2.3. Corneal Epithelial Cell Lines
2.4. Exosome Purification
2.5. NanoSight Particle Size Analysis Using Dynamic Light Scattering
2.6. Transmission Electron Microscopy (TEM)
2.7. In-Gel Protein Digestion for Proteomic Analysis
2.8. Liquid Chromatography and Mass Spectrometer Analysis (LC-ESI-MS/MS)
2.9. Proteomics Data Analysis and Sequence Search
2.10. Differential Protein Expression and Bioinformatics Analysis
2.11. Small RNA Next Generation Sequencing (NGS)
2.12. Differential miRNA Expression and Bioinformatics Analysis
2.13. Cell Migration and Cell Proliferation Assays
2.14. Total RNA Isolation and cDNA Synthesis
2.15. qRT-PCR Reactions
3. Results
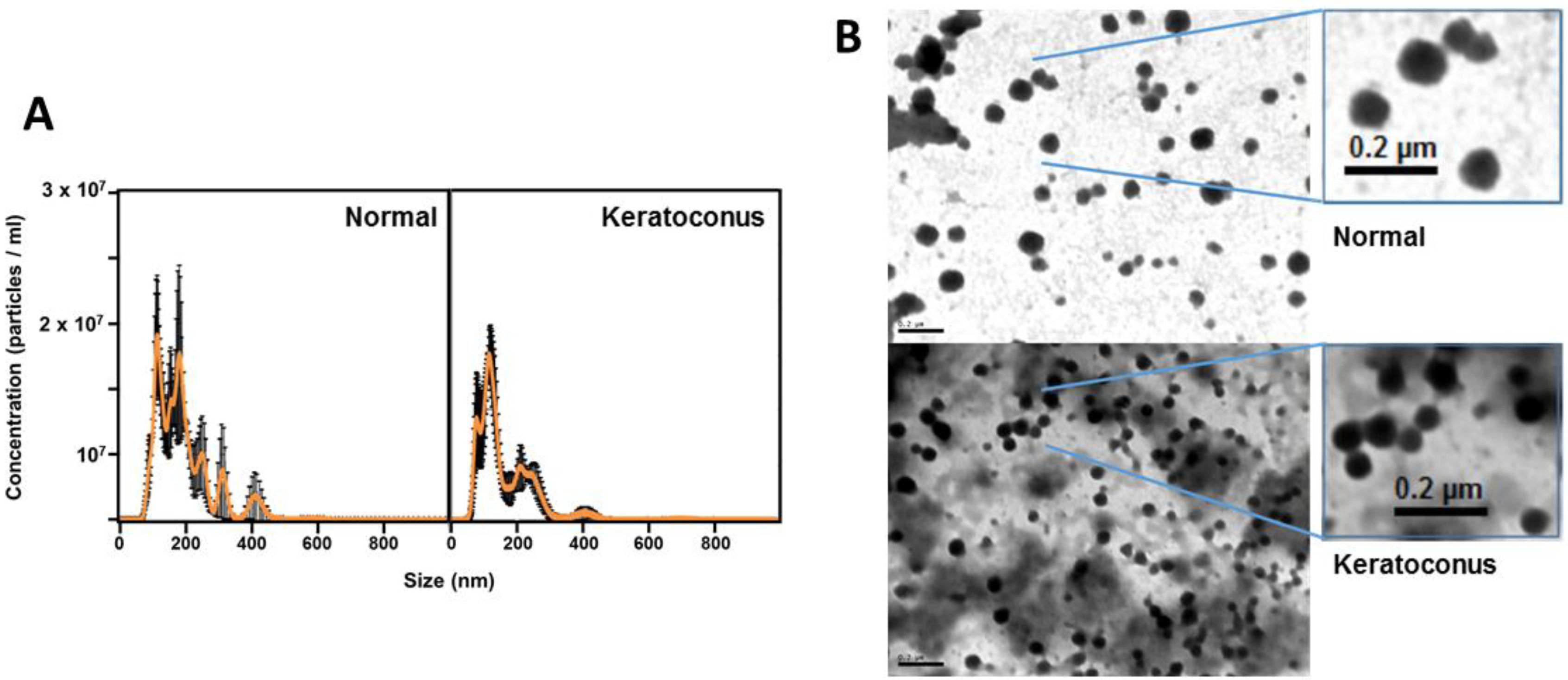
3.1. Characterization of Exosomes Derived from Corneal Stromal Cells
3.2. Proteomic Analysis of Normal Versus Keratoconus Stromal Cell Exosomes
3.3. miRNA Profile Analysis of Normal Versus Keratoconus Stromal Cell Exosomes
3.4. Effect of Exosomes on the Expression of Genes of Proteoglycans and Glycosaminoglycans in Corneal Cells
3.5. Effect of Exosomes on Proliferation and Migration of Corneal Epithelial and Stromal Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soiberman, U.; Foster, J.W.; Jun, A.S.; Chakravarti, S. Pathophysiology of Keratoconus: What Do We Know Today. Open Ophthalmol. J. 2017, 11, 252–261. [Google Scholar] [CrossRef]
- Ferrari, G.; Rama, P. The keratoconus enigma: A review with emphasis on pathogenesis. Ocul. Surf. 2020, 18, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Volatier, T.L.A.; Figueiredo, F.C.; Connon, C.J. Keratoconus at a Molecular Level: A Review. Anat. Rec. 2020, 303, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Khaled, M.L.; Helwa, I.; Drewry, M.; Seremwe, M.; Estes, A.; Liu, Y. Molecular and Histopathological Changes Associated with Keratoconus. BioMed Res. Int. 2017, 2017, 7803029. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Shaag, A.; Millodot, M.; Shneor, E.; Liu, Y. The genetic and environmental factors for keratoconus. BioMed Res. Int. 2015, 2015, 795738. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, L.; Hong, J.; Wu, D.; Xu, J. Association of Common Variants in LOX with Keratoconus: A Meta-Analysis. PLoS ONE 2015, 10, e0145815. [Google Scholar] [CrossRef]
- Ionescu, C.; Corbu, C.G.; Tanase, C.; Jonescu-Cuypers, C.; Nicula, C.; Dascalescu, D.; Cristea, M.; Voinea, L.M. Inflammatory Biomarkers Profile as Microenvironmental Expression in Keratoconus. Dis. Markers 2016, 2016, 1243819. [Google Scholar] [CrossRef]
- Mas Tur, V.; MacGregor, C.; Jayaswal, R.; O’Brart, D.; Maycock, N. A review of keratoconus: Diagnosis, pathophysiology, and genetics. Surv. Ophthalmol. 2017, 62, 770–783. [Google Scholar] [CrossRef]
- McMonnies, C.W. Mechanisms of rubbing-related corneal trauma in keratoconus. Cornea 2009, 28, 607–615. [Google Scholar] [CrossRef]
- Elder, M.J. Leber congenital amaurosis and its association with keratoconus and keratoglobus. J. Pediatr. Ophthalmol. Strabismus 1994, 31, 38–40. [Google Scholar] [CrossRef]
- Nemet, A.Y.; Vinker, S.; Bahar, I.; Kaiserman, I. The association of keratoconus with immune disorders. Cornea 2010, 29, 1261–1264. [Google Scholar] [CrossRef]
- Robertson, I. Keratoconus and the Ehlers-Danlos syndrome: A new aspect of keratoconus. Med. J. Aust. 1975, 1, 571–573. [Google Scholar] [CrossRef]
- Kalkan Akcay, E.; Akcay, M.; Uysal, B.S.; Kosekahya, P.; Aslan, A.N.; Caglayan, M.; Koseoglu, C.; Yulek, F.; Cagil, N. Impaired corneal biomechanical properties and the prevalence of keratoconus in mitral valve prolapse. J. Ophthalmol. 2014, 2014, 402193. [Google Scholar] [CrossRef]
- Woodward, E.G.; Morris, M.T. Joint hypermobility in keratoconus. Ophthalmic Physiol. Opt. 1990, 10, 360–362. [Google Scholar] [CrossRef]
- Ucakhan, O.O.; Kanpolat, A.; Ylmaz, N.; Ozkan, M. In vivo confocal microscopy findings in keratoconus. Eye Contact Lens 2006, 32, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Niederer, R.L.; Perumal, D.; Sherwin, T.; McGhee, C.N. Laser scanning in vivo confocal microscopy reveals reduced innervation and reduction in cell density in all layers of the keratoconic cornea. Investig. Ophthalmol. Vis. Sci. 2008, 49, 2964–2970. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, K.; Vorum, H.; Fagerholm, P.; Birkenkamp-Demtröder, K.; Honoré, B.; Ehlers, N.; Orntoft, T.F. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp. Eye Res. 2006, 82, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, O.P.; Chandrasekaran, D.; Pfister, R.R. Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas. Mol. Vis. 2006, 12, 1615–1625. [Google Scholar] [PubMed]
- Chaerkady, R.; Shao, H.; Scott, S.G.; Pandey, A.; Jun, A.S.; Chakravarti, S. The keratoconus corneal proteome: Loss of epithelial integrity and stromal degeneration. J. Proteom. 2013, 87, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.; Srivastava, O.P.; Pfister, R.R. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp. Eye Res. 2011, 92, 282–298. [Google Scholar] [CrossRef]
- García, B.; García-Suárez, O.; Merayo-Lloves, J.; Alcalde, I.; Alfonso, J.F.; Fernández-Vega Cueto, L.; Meana, Á.; Vázquez, F.; Quirós, L.M. Differential Expression of Proteoglycans by Corneal Stromal Cells in Keratoconus. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2618–2628. [Google Scholar] [CrossRef] [PubMed]
- García, B.; García-Suárez, O.; Merayo-Lloves, J.; Ferrara, G.; Alcalde, I.; González, J.; Lisa, C.; Alfonso, J.F.; Vazquez, F.; Quirós, L.M. Heparanase Overexpresses in Keratoconic Cornea and Tears Depending on the Pathologic Grade. Dis. Markers 2017, 2017, 3502386. [Google Scholar] [CrossRef]
- Whitelock, J.M.; Iozzo, R.V. Heparan sulfate: A complex polymer charged with biological activity. Chem. Rev. 2005, 105, 2745–2764. [Google Scholar] [CrossRef]
- Lema, I.; Durán, J.A. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology 2005, 112, 654–659. [Google Scholar] [CrossRef]
- Jun, A.S.; Cope, L.; Speck, C.; Feng, X.; Lee, S.; Meng, H.; Hamad, A.; Chakravarti, S. Subnormal cytokine profile in the tear fluid of keratoconus patients. PLoS ONE 2011, 6, e16437. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; Buermans, H.P.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.; ’t Hoen, P.A. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Klingeborn, M.; Dismuke, W.M.; Bowes Rickman, C.; Stamer, W.D. Roles of exosomes in the normal and diseased eye. Prog. Retin. Eye Res. 2017, 59, 158–177. [Google Scholar] [CrossRef]
- Li, N.; Zhao, L.; Wei, Y.; Ea, V.L.; Nian, H.; Wei, R. Recent advances of exosomes in immune-mediated eye diseases. Stem Cell Res. Ther. 2019, 10, 278. [Google Scholar] [CrossRef]
- De Toro, J.; Herschlik, L.; Waldner, C.; Mongini, C. Emerging roles of exosomes in normal and pathological conditions: New insights for diagnosis and therapeutic applications. Front. Immunol. 2015, 6, 203. [Google Scholar] [CrossRef]
- Anitua, E.; Sanchez, M.; Merayo-Lloves, J.; De la Fuente, M.; Muruzabal, F.; Orive, G. Plasma rich in growth factors (PRGF-Endoret) stimulates proliferation and migration of primary keratocytes and conjunctival fibroblasts and inhibits and reverts TGF-beta1-Induced myodifferentiation. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6066–6073. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007, 27, 91–105. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.C.; Park, A.Y.; Guan, J.L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vega, I.; García-Suárez, O.; García, B.; Crespo, A.; Astudillo, A.; Quirós, L.M. Heparan sulfate proteoglycans undergo differential expression alterations in right sided colorectal cancer, depending on their metastatic character. BMC Cancer 2015, 15, 742. [Google Scholar] [CrossRef] [PubMed]
- Dyrlund, T.F.; Poulsen, E.T.; Scavenius, C.; Nikolajsen, C.L.; Thøgersen, I.B.; Vorum, H.; Enghild, J.J. Human cornea proteome: Identification and quantitation of the proteins of the three main layers including epithelium, stroma, and endothelium. J. Proteome Res. 2012, 11, 4231–4239. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, H.; Sun, G.; Alwadani, S.; Semba, R.D.; Lutty, G.A.; Yiu, S.; Edward, D.P. Gene Expression Profile of Extracellular Matrix and Adhesion Molecules in the Human Normal Corneal Stroma. Curr. Eye Res. 2017, 42, 520–527. [Google Scholar] [CrossRef]
- Hughes, A.E.; Bradley, D.T.; Campbell, M.; Lechner, J.; Dash, D.P.; Simpson, D.A.; Willoughby, C.E. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet. 2011, 89, 628–633. [Google Scholar] [CrossRef]
- Jiang, P.; Zhang, S.; Cheng, C.; Gao, S.; Tang, M.; Lu, L.; Yang, G.; Chai, R. The Roles of Exosomes in Visual and Auditory Systems. Front. Bioeng. Biotechnol. 2020, 8, 525. [Google Scholar] [CrossRef]
- Samaeekia, R.; Rabiee, B.; Putra, I.; Shen, X.; Park, Y.J.; Hematti, P.; Eslani, M.; Djalilian, A.R. Effect of Human Corneal Mesenchymal Stromal Cell-derived Exosomes on Corneal Epithelial Wound Healing. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5194–5200. [Google Scholar] [CrossRef] [PubMed]
- Yeung, V.; Zhang, T.C.; Yuan, L.; Parekh, M.; Cortinas, J.A.; Delavogia, E.; Hutcheon, A.E.K.; Guo, X.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Myofibroblasts Promote Corneal Epithelial Cell Migration. Int. J. Mol. Sci. 2022, 23, 3136. [Google Scholar] [CrossRef]
- Kugeratski, F.G.; Hodge, K.; Lilla, S.; McAndrews, K.M.; Zhou, X.; Hwang, R.F.; Zanivan, S.; Kalluri, R. Quantitative proteomics identifies the core proteome of exosomes with syntenin-1 as the highest abundant protein and a putative universal biomarker. Nat. Cell Biol. 2021, 23, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Sharif, R.; Khaled, M.L.; McKay, T.B.; Liu, Y.; Karamichos, D. Transcriptional profiling of corneal stromal cells derived from patients with keratoconus. Sci. Rep. 2019, 9, 12567. [Google Scholar] [CrossRef]
- Nishtala, K.; Pahuja, N.; Shetty, R.; Nuijts, R.M.; Ghosh, A. Tear biomarkers for keratoconus. Eye Vis. 2016, 3, 19. [Google Scholar] [CrossRef]
- Lu, Y.; Vitart, V.; Burdon, K.P.; Khor, C.C.; Bykhovskaya, Y.; Mirshahi, A.; Hewitt, A.W.; Koehn, D.; Hysi, P.G.; Ramdas, W.D.; et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet. 2013, 45, 155–163. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef]
- Drewry, M.; Helwa, I.; Allingham, R.R.; Hauser, M.A.; Liu, Y. miRNA Profile in Three Different Normal Human Ocular Tissues by miRNA-Seq. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3731–3739. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Ng, T.K.; Choy, K.W.; Wong, H.K.; Chu, W.K.; Pang, C.P.; Jhanji, V. Histological and microRNA Signatures of Corneal Epithelium in Keratoconus. J Refract. Surg. 2018, 34, 201–211. [Google Scholar] [CrossRef]
- Li, W.; Chen, Y.-T.; Hayashida, Y.; Blanco, G.; Kheirkah, A.; He, H.; Chen, S.-Y.; Liu, C.-Y.; Tseng, S.C.G. Down-regulation of Pax6 is associated with abnormal differentiation of corneal epithelial cells in severe ocular surface diseases. J. Pathol. 2008, 214, 114–122. [Google Scholar] [CrossRef]
- Shalom-Feuerstein, R.; Serror, L.; De La Forest Divonne, S.; Petit, I.; Aberdam, E.; Camargo, L.; Damour, O.; Vigouroux, C.; Solomon, A.; Gaggioli, C.; et al. Pluripotent stem cell model reveals essential roles for miR-450b-5p and miR-184 in embryonic corneal lineage specification. Stem Cells 2012, 30, 898–909. [Google Scholar] [CrossRef] [PubMed]
- Hajrasouliha, A.R.; Jiang, G.; Lu, Q.; Lu, H.; Kaplan, H.J.; Zhang, H.G.; Shao, H. Exosomes from retinal astrocytes contain antiangiogenic components that inhibit laser-induced choroidal neovascularization. J. Biol. Chem. 2013, 288, 28058–28067. [Google Scholar] [CrossRef] [PubMed]
- Mansoor, H.; Ong, H.S.; Riau, A.K.; Stanzel, T.P.; Mehta, J.S.; Yam, G.H. Current Trends and Future Perspective of Mesenchymal Stem Cells and Exosomes in Corneal Diseases. Int. J. Mol. Sci. 2019, 20, 2853. [Google Scholar] [CrossRef] [PubMed]
- Tabak, S.; Schreiber-Avissar, S.; Beit-Yannai, E. Extracellular vesicles have variable dose-dependent effects on cultured draining cells in the eye. J. Cell. Mol. Med. 2018, 22, 1992–2000. [Google Scholar] [CrossRef]
- Leszczynska, A.; Kulkarni, M.; Ljubimov, A.V.; Saghizadeh, M. Exosomes from normal and diabetic human corneolimbal keratocytes differentially regulate migration, proliferation and marker expression of limbal epithelial cells. Sci. Rep. 2018, 8, 15173. [Google Scholar] [CrossRef] [PubMed]
- Zieske, J.D.; Hutcheon, A.E.K.; Guo, X. Extracellular Vesicles and Cell-Cell Communication in the Cornea. Anat. Rec. 2020, 303, 1727–1734. [Google Scholar] [CrossRef]







| Protein Symbol | UniPROT | Description/Name | Keratoconus 1 | Normal 1 |
|---|---|---|---|---|
| A2M | H0YFH1 | Alpha-2-macroglobulin | 0.143755 | 0.117508 |
| ACTB | G5E9R0 | Actin, cytoplasmic 1 | 3.044770 | 0.954318 |
| ACTBL2 | Q562R1 | Beta-actin-like protein 2 | 0.472427 | 0.416883 |
| AFP | J3KMX3 | Alpha-fetoprotein | 0.301108 | 0.197081 |
| AHSG | C9JV77 | Alpha-2-HS-glycoprotein | 0.652708 | 0.543988 |
| ANXA1 | Q5T3N0 | Annexin A1 | 0.952801 | 0.793662 |
| ANXA2 | H0YNB8 | Annexin A2 | 4.321061 | 4.004153 |
| ANXA5 | D6RBE9 | Annexin A5 | 1.281670 | 1.428309 |
| APOA1 | F8W696 | Apolipoprotein A-I | 0.243717 | 0.184766 |
| ART4 | H7C2G2 | NAD(P)(+)--arginine ADP-ribosyltransferase | 0.211927 | 0.191949 |
| AZGP1 | H7BZJ8 | Zinc-alpha-2-glycoprotein | 0.144309 | 0.000000 |
| C3 | M0QYC8 | Complement C3 | 0.271774 | 0.252169 |
| C4A | A0A140TA32 | Complement C4-A | 0.104644 | 0.090665 |
| CD44 | H0YE40 | CD44 antigen | 0.390023 | 0.203275 |
| CLEC3B | E9PHK0 | Tetranectin | 0.732630 | 0.423420 |
| COL1A2 | A0A087WTA8 | Collagen alpha-2(I) chain | 0.232500 | 0.127181 |
| COL5A1 | H7BY82 | Collagen alpha-1(V) chain | 0.028984 | 0.076454 |
| COL6A1 | P12109 | Collagen alpha-1(VI) chain | 0.444436 | 0.251104 |
| COMP | G3XAP6 | Cartilage oligomeric matrix protein | 0.088188 | 0.056545 |
| CTNNB1 | C9IZ65 | Catenin beta-1 | 0.048813 | 0.114816 |
| CTSD | A0A7I2V2N3 | Cathepsin D | 0.349379 | 0.472076 |
| DCD | P81605 | Dermcidin | 0.819703 | 0.595040 |
| DSC1 | Q08554 | Desmocollin-1 | 0.086888 | 0.065345 |
| DSG1 | Q02413 | Desmoglein-1 | 0.183173 | 0.328223 |
| DSP | P15924 | Desmoplakin | 0.116647 | 0.199771 |
| ENO1 | K7ERS8 | Alpha-enolase | 0.684927 | 0.537460 |
| FBLN1 | B1AHM9 | Fibulin-1 | 0.114234 | 0.150116 |
| FLG2 | Q5D862 | Filaggrin-2 | 0.022611 | 0.028105 |
| FN1 | H0Y7Z1 | Fibronectin | 0.553519 | 0.321627 |
| GAPDH | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 1.253172 | 1.073777 |
| GC | D6RF35 | Vitamin D-binding protein | 0.421269 | 0.310700 |
| GNA13 | Q14344 | Guanine nucleotide-binding protein subunit alpha-13 | 0.148349 | 0.108959 |
| GNAS | A0A590UJ47 | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas | 0.583043 | 1.155494 |
| GSN | Q5T0H8 | Gelsolin | 0.208645 | 0.369956 |
| HBA1 | P69905 | Hemoglobin subunit alpha | 1.045289 | 1.310054 |
| HGFAC | D6RAR4 | Hepatocyte growth factor activator | 0.103574 | 0.026398 |
| HRNR | Q86YZ3 | Hornerin | 0.013325 | 0.040701 |
| HSP90AA1 | G3V2J8 | Heat shock protein HSP 90-alpha | 0.091852 | 0.117342 |
| ITIH2 | Q5T987 | Inter-alpha-trypsin inhibitor heavy chain H2 | 0.190179 | 0.122103 |
| ITIH3 | E7ET33 | Inter-alpha-trypsin inhibitor heavy chain H3 | 0.112654 | 0.136175 |
| JUP | C9JPI2 | Junction plakoglobin | 0.342050 | 0.727787 |
| LTF | E7EQB2 | Lactotransferrin | 0.227806 | 0.382644 |
| LUM | P51884 | Lumican | 0.169542 | 0.164988 |
| MFGE8 | H0YKS8 | Lactadherin | 0.516690 | 0.682248 |
| MSN | P26038 | Moesin | 0.314202 | 0.178213 |
| ODF2 | S4R462 | Outer dense fiber protein 2 | 0.029924 | 0.028792 |
| PRDX2 | A6NIW5 | Peroxiredoxin-2 | 0.706698 | 0.840749 |
| PRDX4 | A6NG45 | Peroxiredoxin-4 | 0.614590 | 0.491306 |
| PTX3 | P26022 | Pentraxin-related protein PTX3 | 0.530573 | 0.309626 |
| PZP | P20742 | Pregnancy zone protein | 0.059333 | 0.059923 |
| RBP4 | Q5VY30 | Retinol binding protein 4, plasma, isoform CRA_b | 0.242353 | 0.167108 |
| RPL4 | P36578 | 60S ribosomal protein L4 | 0.054390 | 0.076779 |
| S100A7 | P31151 | Protein S100-A7 | 1.077940 | 1.636534 |
| S100A8 | P05109 | Protein S100-A8 | 1.135480 | 1.176553 |
| S100A9 | P06702 | Protein S100-A9 | 0.497486 | 1.321336 |
| SERPINA7 | P05543 | Thyroxine-binding globulin | 0.210750 | 0.244745 |
| SERPINB12 | Q96P63 | Serpin B12 | 0.177998 | 0.291514 |
| SERPINB3 | P29508 | Serpin B3 | 0.353509 | 1.231848 |
| SERPINC1 | P01008 | Antithrombin-III | 0.581404 | 0.580876 |
| SERPINE1 | P05121 | Plasminogen activator inhibitor 1 | 0.572236 | 0.286820 |
| SERPINF1 | A0A0J9YXF9 | Pigment epithelium-derived factor | 0.270115 | 0.268728 |
| SERPINF2 | A0A0J9YY65 | Alpha-2-antiplasmin | 0.137753 | 0.103781 |
| SLC1A5 | M0R144 | Neutral amino acid transporter B(0) | 0.266010 | 0.215608 |
| SLC3A2 | A0A7P0Z4P5 | 4F2 cell-surface antigen heavy chain | 0.235325 | 0.251178 |
| TF | P02787 | Serotransferrin | 0.288676 | 0.193706 |
| TGM3 | Q08188 | Protein-glutamine gamma-glutamyltransferase E | 0.303941 | 0.287802 |
| THBS1 | A8MZG1 | Thrombospondin-1 | 0.258995 | 0.222754 |
| THY1 | J3QRJ3 | Thy-1 membrane glycoprotein | 0.441508 | 0.244056 |
| TIMP1 | Q5H9B5 | Metalloproteinase inhibitor 1 | 1.168873 | 0.669724 |
| TRAPPC6A | O75865 | Trafficking protein particle complex subunit 6A | 0.000000 | 0.163156 |
| TUBA1C | Q9BQE3 | Tubulin alpha-1C chain | 0.470637 | 0.439214 |
| TUBB | P07437 | Tubulin beta chain | 0.215058 | 0.256739 |
| TXN | P10599 | Thioredoxin | 0.306628 | 0.249533 |
| VASH1 | Q7L8A9 | Vasohibin-1 | 0.074175 | 0.067182 |
| VIM | B0YJC4 | Vimentin | 0.262035 | 0.516628 |
| YWHAE | I3L3T1 | 14-3-3 protein epsilon | 0.250273 | 0.310520 |
| miRNA | Protein Symbol | UniPROT | Description/Name |
|---|---|---|---|
| hsa-miR-2355-3p | ACTN1 | P12814 | Alpha-actinin-1 |
| hsa-miR-183-5p | FNDC3B | Q53EP0 | Fibronectin type III domain-containing protein 3B |
| hsa-miR-455-3p | NFIB | O00712 | Nuclear factor 1 B-type |
| hsa-miR-3377 | SORL1 | Q92673 | Sortilin-related receptor |
| hsa-miR-3192-5p | TIMP3 | P35625 | Metalloproteinase inhibitor 3 |
| hsa-miR-34a-3p | TNF | P01375 | Tumor necrosis factor |
| hsa-miR-183-5p | ZEB1 | P37275 | Zinc finger E-box-binding homeobox 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozano, V.; Martín, C.; Blanco, N.; Alcalde, I.; Fernandez-Vega Cueto, L.; Merayo-Lloves, J.; Quirós, L.M. Exosomes Released by Corneal Stromal Cells Show Molecular Alterations in Keratoconus Patients and Induce Different Cellular Behavior. Biomedicines 2022, 10, 2348. https://doi.org/10.3390/biomedicines10102348
Lozano V, Martín C, Blanco N, Alcalde I, Fernandez-Vega Cueto L, Merayo-Lloves J, Quirós LM. Exosomes Released by Corneal Stromal Cells Show Molecular Alterations in Keratoconus Patients and Induce Different Cellular Behavior. Biomedicines. 2022; 10(10):2348. https://doi.org/10.3390/biomedicines10102348
Chicago/Turabian StyleLozano, Víctor, Carla Martín, Noelia Blanco, Ignacio Alcalde, Luis Fernandez-Vega Cueto, Jesús Merayo-Lloves, and Luis M. Quirós. 2022. "Exosomes Released by Corneal Stromal Cells Show Molecular Alterations in Keratoconus Patients and Induce Different Cellular Behavior" Biomedicines 10, no. 10: 2348. https://doi.org/10.3390/biomedicines10102348
APA StyleLozano, V., Martín, C., Blanco, N., Alcalde, I., Fernandez-Vega Cueto, L., Merayo-Lloves, J., & Quirós, L. M. (2022). Exosomes Released by Corneal Stromal Cells Show Molecular Alterations in Keratoconus Patients and Induce Different Cellular Behavior. Biomedicines, 10(10), 2348. https://doi.org/10.3390/biomedicines10102348






